Conformation of the nuclear pore in living cells is modulated by transport state
Figures
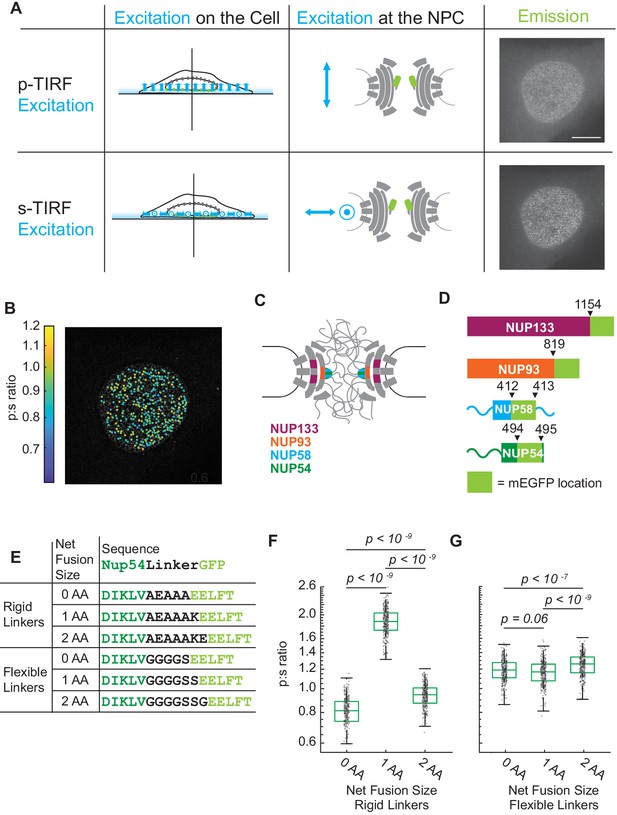
The orientation of Nup-mEGFP fusion proteins can be measured in individual NPCs with pol-TIRFM.
(A) Using pol-TIRFM, the bottom of the nucleus is illuminated and Nup-mEGFP fusion proteins are excited with -polarized or -polarized light. -excitation is parallel and -excitation is perpendicular to the nucleocytoplasmic axis of the NPC. The emission from Nup54-mEGFP fusion proteins in HeLa cells in response to each excitation is shown in the right column. (Scale bar = 10 µm). (B) p:s ratios, a measurement of orientation of mEGFP, are calculated for each NPC and represented with a color scale. (C) Schematic of the NPC indicating the Nups studied. (D) Schematic of the mEGFP placement within the Nups. (E) Nup54-mEGFP constructs with flexible or rigid linkers in between the Nup and the mEGFP. With each additional amino acid in a rigid alpha helix, the mEGFP rotates 103° relative to the Nup, which does not happen with a flexible linker. (F) The p:s ratios of Nup54-mEGFP494 fusion proteins with a rigid linker shift with the addition of each amino acid. (G) The p:s ratios of Nup54-mEGFP494 fusion proteins with a flexible linker do not shift upon amino acid additions. (n = 300 NPCs, 10 cells, boxes indicate quartiles, center bars indicate medians, one-way ANOVA with post-hoc Tukey test).
-
Figure 1—source data 1
Source data for Figure 1.
- https://cdn.elifesciences.org/articles/60654/elife-60654-fig1-data1-v1.xlsx
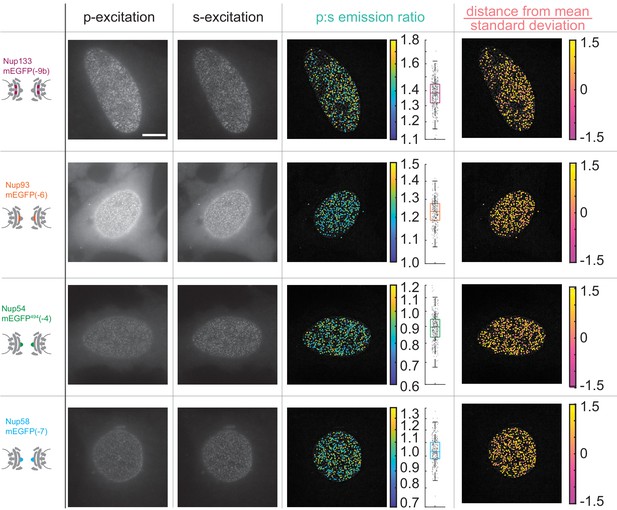
The orientation of Nup-mEGFP fusion proteins can be monitored with pol-TIRFM.
Four different Nup-mEGFP (Nup133, Nup93, Nup54, and Nup58) were transiently expressed for 48 hr in HeLa cells. (Columns 1–2) The constructs were alternately excited with -polarized (perpendicular to the coverslip) and \hat{s}-polarized (parallel to the coverslip) light (Scale bar = 10 µm). (Column 3) The p:s ratios, a measurement of orientation of mEGFP relative to the coverslip are calculated for each NPC. The color of the dot represents the p:s ratio of the NPC upon which it is superimposed. The box plot represents the data directly taken from the cell next to it. Each dot represents the p:s measurement of a single NPC. (Column 4) The difference from the mean divided by the standard deviation is shown in the final column, illustrating how the p:s ratio does not vary with respect to the basal surface of the nucleus and that the cell holds its nuclear envelope parallel to the coverslip.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/60654/elife-60654-fig1-figsupp1-data1-v1.xlsx
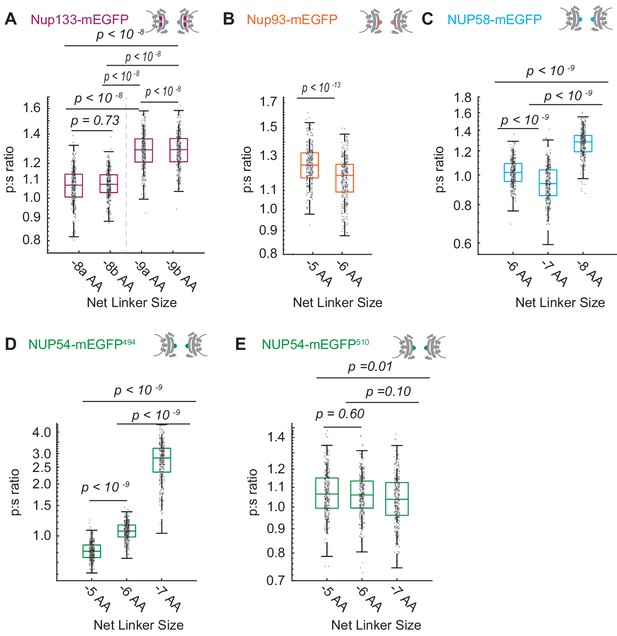
Varying the length of the linker between the Nup and the mEGFP by single amino acids to test validity of the orientational sensors.
(A) Nup133-mEGFP, with linkers of different lengths at its carboxyl terminus, conjugated to mEGFP. A change in the linker length by a single amino acid changes the p:s ratio. Pairs of different rigid alpha-helical linkers of the same length generate indistinguishable p:s ratios. (B) Nup93-mEGFP with different linker lengths to mEGFP at the carboxyl terminus. One deleted amino acid shifts the p:s ratio. (C) Nup58 with the mEGFP at the carboxyl terminus of the coiled-coiled domain. Each subsequent deletion of a single amino acid alters the p:s ratio. (D) Nup54-mEGFP494 with the mEGFP at the carboxyl terminus of the coiled-coiled domain (Amino Acid: 494). Each subsequent deletion of a single amino acid changes the p:s ratio. (E) Nup54-mEGFP510 with the mEGFP at the carboxyl-terminus of the protein (Amino Acid: 510). Each subsequent deletion of a single amino acid does not alter the p:s ratio. Detailed linker descriptions are available in Supplementary file 1. (n = 300 NPCs, 10 cells, boxes indicate quartiles, center bars indicate medians, one-way ANOVA with post-hoc Tukey test for A, C-E, Student's t-test for B).
-
Figure 2—source data 1
Source data for Figure 1.
- https://cdn.elifesciences.org/articles/60654/elife-60654-fig2-data1-v1.xlsx
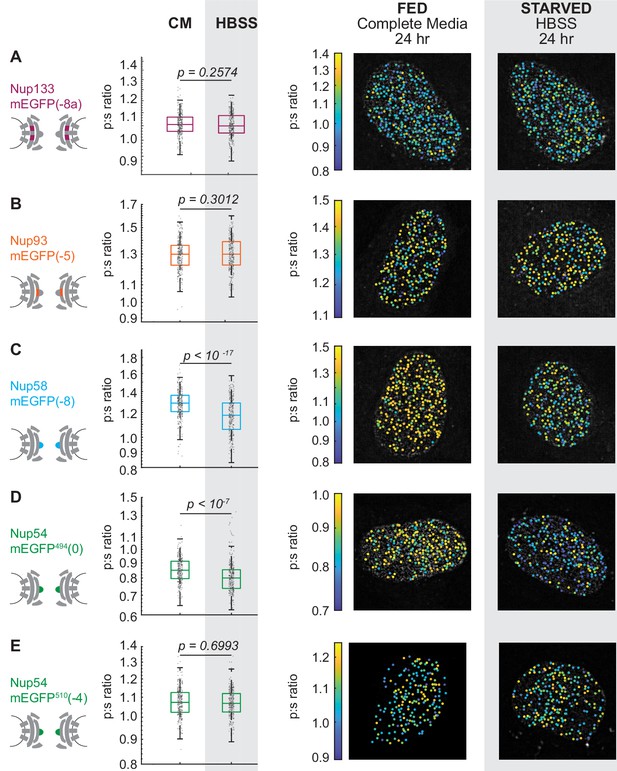
The Inner Ring Nups, Nup54 and Nup58, are reorganized with respect to the NPC after starvation.
Cells were maintained in complete media (CM) or starved for 24 hr in HBSS prior to imaging. The p:s ratios and representative images are presented for: (A) Nup133-mEGFP(−8a), (B) Nup93-mEGFP(–5), (C) Nup58-mEGFP(−8), (D) Nup54-mEGFP494(0), and (E) Nup54-mEGFP510(−4). The orientation changed for Nup58-mEGFP and Nup54-mEGFP494. For additional linker lengths, see Figure 3—figure supplement 2. HeLa cells were imaged 48 hr post transfection. (n = 300 NPCs, 10 cells, boxes indicate quartiles, center bars indicate medians, Student's t-test).
-
Figure 3—source data 1
Source data for Figure 3.
- https://cdn.elifesciences.org/articles/60654/elife-60654-fig3-data1-v1.xlsx
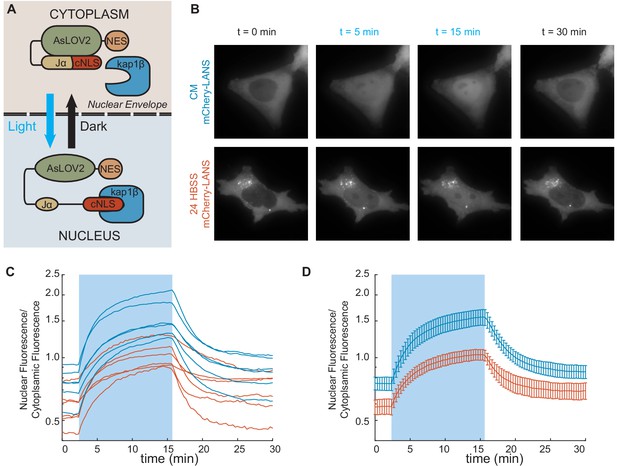
Nuclear transport is attenuated post-starvation.
(A) The NLS domain of the LANS construct normally is masked by the AsLOV2 domain and the protein remains predominantly in the cytosol. Upon stimulation with blue light, the NLS is unmasked and the protein translocates into the nucleus. Figure adapted from Yumerefendi et al., 2015. (B) Representative images for light activation and reversion for HeLa cells in complete media (CM) and HBSS for 24 hr. (Scale bar = 10 µm). Images are all scaled to the same color axis. (C) The change in the ratio of nuclear/cytoplasmic fluorescence in HeLa cells in CM (blue) and HBSS (red). The shaded region represents the time of blue light LANS activation. (D) The average change in the ratio of nuclear/cytoplasmic fluorescence in HeLa cells in CM (blue) and HBSS (red) (n = 6, mean reported ± SEM with error bars). The shaded region represents the time of blue light LANS activation. Note: In starved cells, there were some bright cytoplasmic regions, which might be autophagosomes, that were excluded from analysis.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/60654/elife-60654-fig3-figsupp1-data1-v1.xlsx
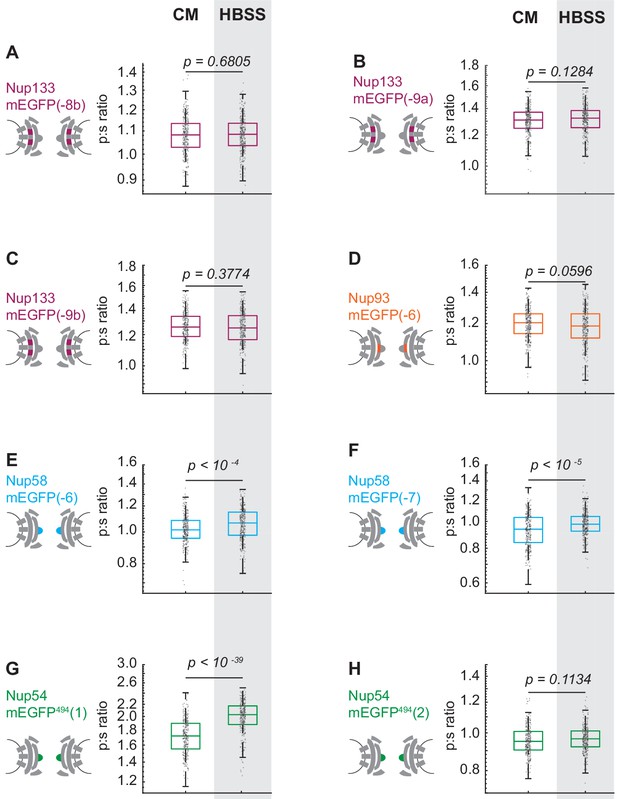
The Inner Ring Nups, Nup54 and Nup58, are reorganized with respect to the NPC after starvation.
Cells were maintained in complete media (CM) or starved for 24 hr in HBSS prior to imaging. The p:s ratios are presented for: (A) Nup133-mEGFP(−8b), (B) Nup133-mEGFP(−9a), (C) Nup133-mEGFP(−9b), (D) Nup93-mEGFP(−6), (E) Nup58-mEGFP(−6), (F) Nup58-mEGFP(−7), (G) Nup54-mEGFP494(1), and (H) Nup54-mEGFP494(2). The orientation changed for Nup58-mEGFP and Nup54-mEGFP494. There was no change of orientation for Nup133-mEGFP(−8a) after starvation (Figure 3). Here, the results are expanded to include Nup133-mEGFP(−8b), Nup133-mEGFP(−9a) and Nup133-mEGFP(−9b), none of which show a change of orientation after starvation. No change in orientation of Nup93-mEGFP(−5) was seen after starvation (Figure 3); here, Nup93-mEGF(−6) also shows no change in orientation post-starvation. Starvation shifted the orientation of Nup58-mEGFP(−8) (Figure 3), and here it shifts the orientation of Nup58-mEGFP(−6) and Nup58-mEGFP(−7). Starvation shifts the orientation of Nup54-mEGFP494(0) (Figure 3), and here starvation shifts the orientation of Nup54-mEGFP494(1). A significant change in orientation is not seen on Nup54-mEGFP494(2). It is expected that the magnitude of the shift of the orientation will differ between different probe locations. For example, if a starvation induces a rotation of the excitation dipole of the mEGFP from −52° to +52° relative to the axis normal to the coverslip, then p:s ratio will not change. This result underscores the importance of testing different linker lengths. HeLa cells were imaged 48 hr post transfection. (n = 300 NPCs, 10 cells, boxes indicate quartiles, center bars indicate medians, Student's t-test).
-
Figure 3—figure supplement 2—source data 1
Source data for Figure 3—figure supplement 2.
- https://cdn.elifesciences.org/articles/60654/elife-60654-fig3-figsupp2-data1-v1.xlsx
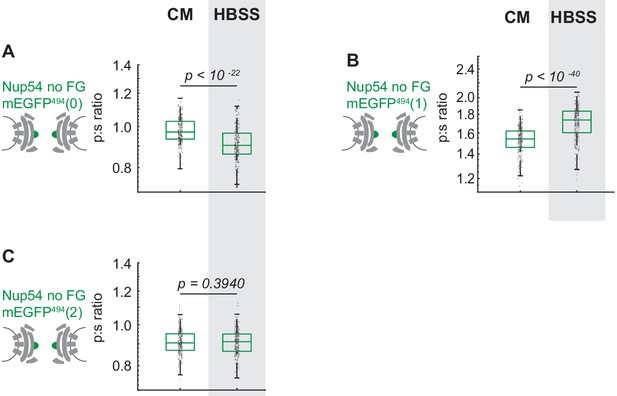
Cells were maintained in complete media (CM) or starved for 24 hr in HBSS prior to imaging.
The p:s ratios are presented for: (A) Nup54 no FG-mEGFP494(0), (B) Nup54 no FG-mEGFP494(1), (C) Nup54 no FG-mEGFP494(2) (n = 300 NPCs, 10 cells, boxes indicate quartiles, center bars indicate medians, Student's t-test).
-
Figure 3—figure supplement 3—source data 1
Source data for Figure 3—figure supplement 3.
- https://cdn.elifesciences.org/articles/60654/elife-60654-fig3-figsupp3-data1-v1.xlsx
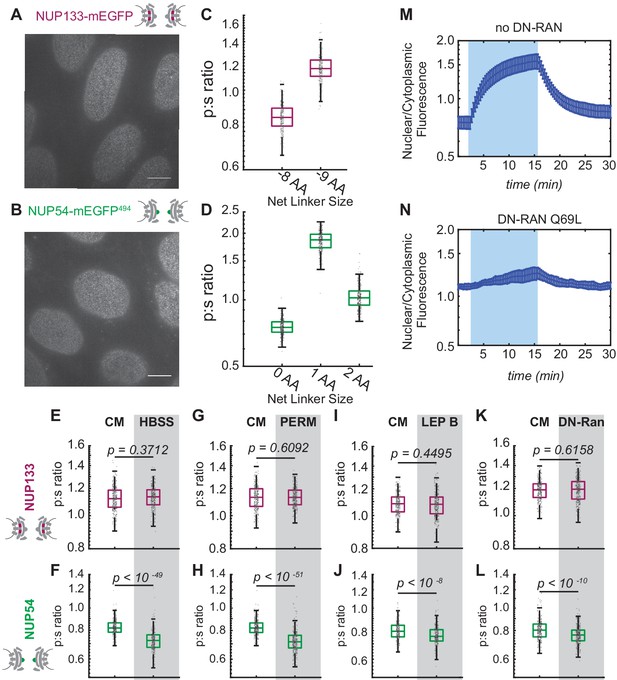
Conformational changes of the Inner Ring of the NPC revealed by perturbations of cargo state in CRISPR cell lines.
(A-B) No morphological distortions are detected in cell lines endogenously expressing orientational sensors. (C) Nup133-mEGFP cell lines with the mEGFP at the carboxyl-terminus of the protein with different linker lengths. One deleted amino acid shifts the p:s ratio. (D) Nup54-mEGFP494 cell lines with the mEGFP at the carboxyl-end of the coiled-coiled domain of the protein with constructs of different linker lengths. One deleted amino acid shifts the p:s ratio. Detailed linker descriptions are available in Supplementary file 2. (E-F) Orientational sensor cell lines were maintained in CM or starved for 24 hr in HBSS. (G-H) CRISPR cell lines mock permeabilized or digitonin-permeabilized prior to imaging. (I-J) CRISPR cell lines mock treated or treated with leptomycin B prior to imaging. (K-L) CRISPR cell lines with or without transient expression of dominant-negative Ran. (n = 300 NPCs, 10 cells, boxes indicate quartiles, center bars indicate medians, Student's t-test). (M-N) The average change in the ratio of nuclear/cytoplasmic fluorescence in HeLa cells with and without dominant-negative Ran transiently expressed (n = 6, mean reported ± SEM with error bars). The shaded region represents the time of blue light LANS activation.
-
Figure 4—source data 1
Source data for Figure 4C–L.
- https://cdn.elifesciences.org/articles/60654/elife-60654-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Source data for Figure 4M–N.
- https://cdn.elifesciences.org/articles/60654/elife-60654-fig4-data2-v1.xlsx
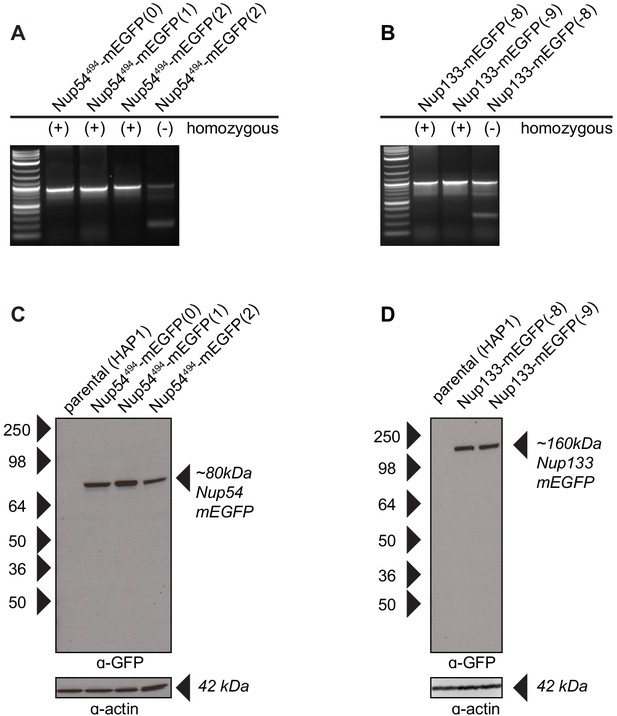
Validation of CRISPR cell lines.
(A–B) PCR amplification of the Nup54 and Nup133 region reveal one band when the Nup-mEGFP region is homozygous for mEGFP incorporation and two bands when the region is heterozygous. The bands were sequenced to verify proper incorporation of the mEGFP. The validated homozygous clones were used for our experiments. (C–D) Western blots probed for anti-GFP detects Nup54 and Nup133 fusion proteins as expected. The blots were stripped of the anti-GFP and probed with anti-actin to confirm protein loading.
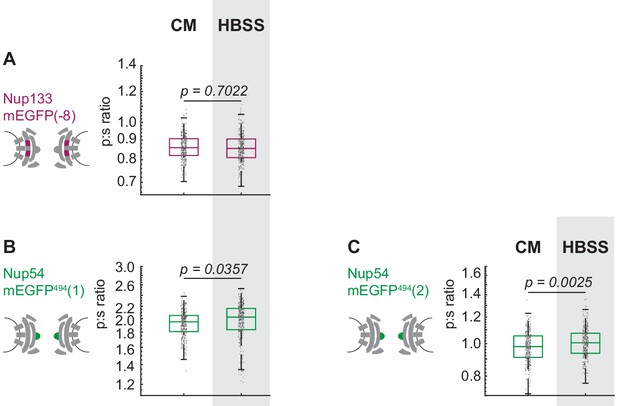
The Inner Ring Nup Nup54 is reorganized with respect to the NPC after starvation.
Hap1 cell lines that had been engineered with CRISPR/Cas9 were maintained in complete media (CM) or starved for 24 hr in HBSS prior to imaging. The p:s ratios are presented for: (A) Nup133-mEGFP(−8), (B) Nup54-mEGFP494(1), (C) Nup54-mEGFP494(2). These results are similar to what was observed for the transiently transfected cells in Figure 3 and Figure 3S and the CRISPR line in Figure 4E. The notable exception is that the effect of starvation is more significant in the transiently transfected Nup54-mEGFP494(1) than in the CRISPR cell line of Nup54-mEGFP494(1). This difference in severity of the effect of treatment could be due to a difference in signal-noise between the transfection and the CRISPR cell line or due to a difference in ‘steady-state’ conformation in the two different cell types. The cell line used in Figure 4E was Nup133-mEGFP(−9), and in this figure the cell line used was Nup133-mEGFP(−8). The CRISPR cell line used in Figure 4F was Nup54-mEGFP494(0); and in this figure, the cell lines used are Nup54-mEGFP494(1) and Nup54-mEGFP494(2). (n = 300 NPCs, 10 cells, boxes indicate quartiles, center bars indicate medians, Student's t-test).
-
Figure 4—figure supplement 2—source data 1
Source data for Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/60654/elife-60654-fig4-figsupp2-data1-v1.xlsx
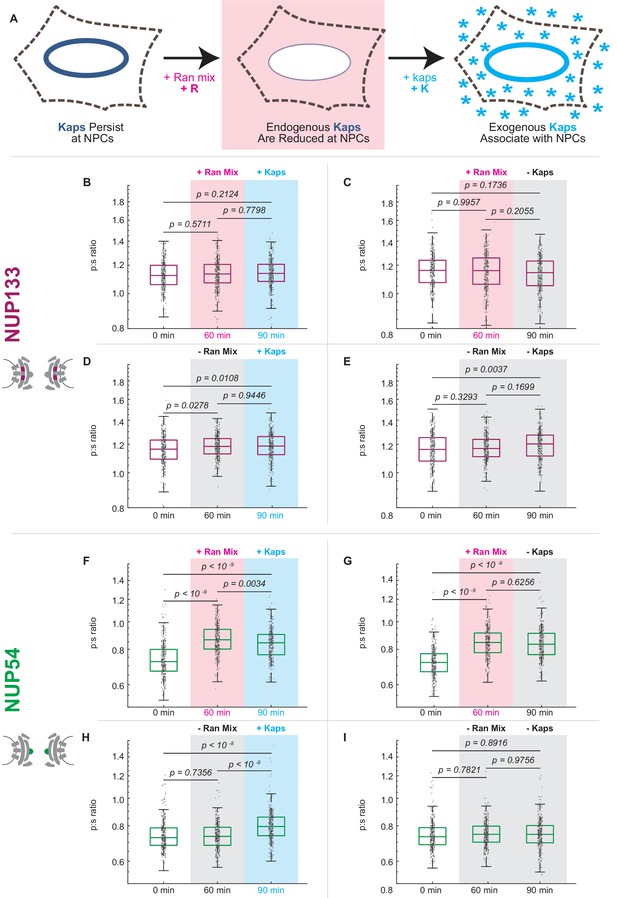
Karyopherin content at the nuclear periphery induces conformational changes in Nup54-mEGFP494 but not Nup133-mEGFP.
(A) Digitonin permeabilization allows the introduction of transport factors to the nuclear periphery. (B-E) Nup133-mEGFP does not experience a shift in orientation after removal of endogenous kaps or addition of exogenous kaps. (F-I) The orientation of Nup54-mEGFP494 changes after removal of endogenous kaps or introduction of exogenous kaps. Pink boxes indicate the addition of Ran mix, blue boxes indicate the addition of kaps, and gray boxes indicate a buffer change with no additional transport factors. (n = 300 NPCs, 30 cells, boxes indicate quartiles, center bars indicate medians, one-way ANOVA with post-hoc Tukey test).
-
Figure 5—source data 1
Source data for Figure 5.
- https://cdn.elifesciences.org/articles/60654/elife-60654-fig5-data1-v1.xlsx
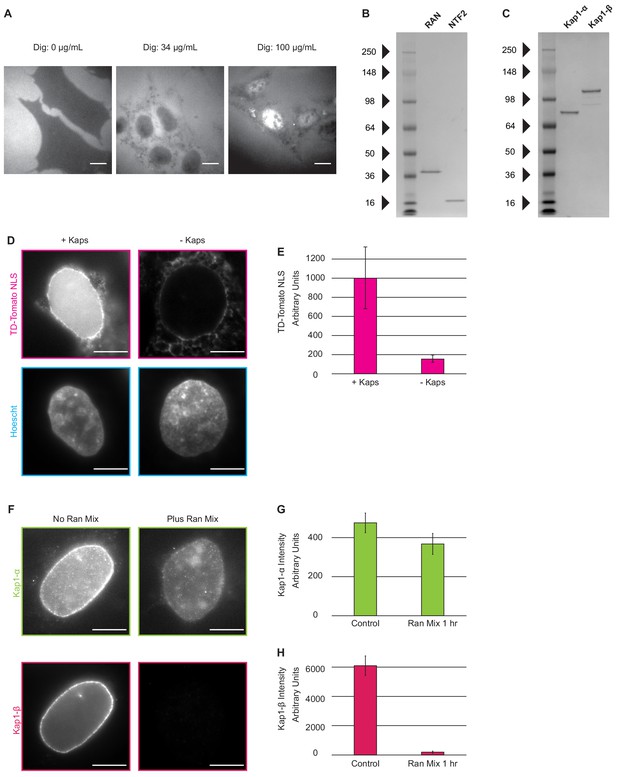
Introduction of nuclear transport factors to the nuclear periphery using digitonin permeabilization of the plasma membrane.
(A) Cells were incubated with R-phycoerythrin after the digitonin permeabilization protocol was performed with (left) 0 µg/mL digitonin, for which no R-phycoerythrin is detected in the cell, (middle) 34 µg/mL, for which R-phycoerythrin has entered the cytosol but is excluded from the nucleus, and (right) 100 µg/mL digitonin for which R-phycoerythrin is seen throughout the cytosol and nucleus. (Scale Bar = 10 µm). (B–C) The Coomassie-stained gels show purified Ran, NTF2, Kap1-α, Kap1-β proteins with minimal contaminants. (D) Cells were incubated with an ATP-regenerating system, Ran-GTP, NTF2, and an NLS-TD-Tomato cargo in the presence and absence of Kap1- α and Kap1-β. (E) Amount of NLS-TD-Tomato accumulated in the nucleus was measured by quantifying the average fluorescence signal in the nucleus, as defined by Hoechst staining. (n = 5 cells, error bars represent standard deviation). (F–G) Removal of kaps was confirmed with quantitative immunofluorescence. (Scale Bar = 10 µm; error bars represent standard deviation; n = 5 cells).
-
Figure 5—figure supplement 1—source data 1
Source data for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/60654/elife-60654-fig5-figsupp1-data1-v1.xlsx
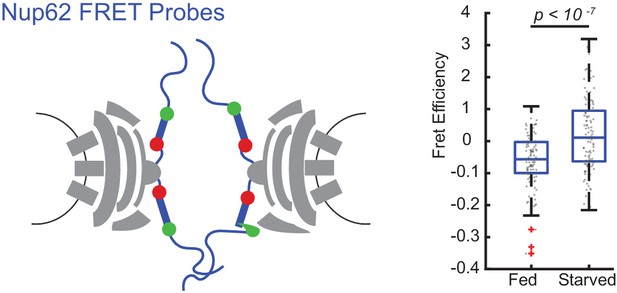
FRET between Nup62 ‘finger’ domains increases after starvation.
Under starvation conditions, FRET increased between Nup62 ‘finger’ domains. (A) Schematic of Nup62 FRET probe labeling scheme. (B) FRET efficiency for HeLa cells were imaged 48 hr post transfection. Cells were kept in CM or starved for 24 hr in HBSS (n = 100 NPCs, 10 cells, boxes indicate quartiles, bars indicate medians, Student's t-test).
-
Figure 6—source data 1
Source data for Figure 6.
- https://cdn.elifesciences.org/articles/60654/elife-60654-fig6-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | α- karyopherin α1/6 (2D9) (rat monoclonal) | Santa Cruz | sc-101540 RRID:AB_2133549 | IF 1:500 |
| Antibody | α-kap1ß/impß −1 (3E9) (mouse monoclonal) | Abcam | ab2811 RRID:AB_2133989 | IF 1:1000 |
| Antibody | anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (goat polyclonal) | Invitrogen | CAT # A-11008 RRID:AB_2534074 | IF 1:2000 |
| Antibody | anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 (goat polyclonal) | Invitrogen | CAT # A-11005 RRID:AB_141372 | IF 1:2000 |
| Antibody | anti-GFP: Living Colors A.v. Monoclonal Antibody (JL-8) | Clontech | CAT # 632381 RRID:AB_2313808 | WB 1:1000 |
| Antibody | anti-Mouse IgG (Fab specific)–Peroxidase antibody (goat polyclonal) | Sigma-Aldrich | CAT # A9917 RRID:AB_258476 | WB 1:50,000 |
| Antibody | Anti-β-Actin antibody, Ac-74 (mouse monoclonal) | Sigma-Aldrich | CAT # A5316 RRID:AB_476743 | WB 1:1000 |
| Strain, strain background (Escherichia coli) | BL21-CodonPlus (DE3)-RIL | Agilent | CAT # 230245 | Chemically competent cells |
| Chemical compound, drug | Dulbecco’s Modified Eagle’s Medium | Gibco | CAT # 11995–065 | |
| Chemical compound, drug | Fetal Bovine Serum | Sigma-Aldrich | CAT #F4135 | |
| Chemical compound, drug | Hanks Balanced Salt Solution with Calcium and Magnesium | Gibco | CAT # 14025076 | |
| Chemical compound, drug | Leptomycin B | Sigma-Aldrich | CAT # L2913 | 25 nM |
| Chemical compound, drug | Fibronectin | Gibco | CAT # 33010018 | |
| Chemical compound, drug | Paraformaldehyde | Electron Microscopy Sciences | CAT #15711 | 4% w/v |
| Chemical compound, drug | PBS, pH 7.4 | Gibco | CAT # 10010023 | |
| Chemical compound, drug | Normal Donkey Serum | Sigma-Aldrich | CAT # 566460 | 2.5% v/v |
| Chemical compound, drug | Normal Goat Serum | Sigma-Aldrich | CAT # NS02L | 2.5% v/v |
| Chemical compound, drug | Bovine Serum Albumin | Sigma-Aldrich | CAT #A2153 | 1% v/v |
| Chemical compound, drug | SlowFade Diamond Antifade Mountant | Invitrogen | CAT # S36972 | |
| Chemical compound, drug | FuGENE 6 Transfection Reagent | Promega | CAT #E2691 | |
| Chemical compound, drug | Opti-MEM I Reduced Serum Medium, no phenol red | Gibco | CAT # 11058021 | |
| Chemical compound, drug | Digitonin, High Purity – Calbiochem | Millipore | CAT # 300410; CAS 11024-24-1 | 34 µg/mL |
| Chemical compound, drug | 360kD polyvinylpyrrolidone (PVP) | Sigma-Aldrich | CAT #PVP360; CAS 9003-39-8 | 1.5% w/v |
| Chemical compound, drug | R-phycoerythrin | ThermoFisher | CAT # P801 | 500 ng/mL |
| Chemical compound, drug | isopropyl β-D-1-thiogalactopyranoside (IPTG) | Sigma-Aldrich | CAT # I5502 | 0.5 mM |
| Chemical compound, drug | Benzonase Nuclease | EMD Millipore | CAT # 70746 | 25 U/mL |
| Chemical compound, drug | rLysozyme Solution | EMD Millipore | CAT # 71110 | 12 U/mL |
| Chemical compound, drug | cOmplete, EDTA-free Protease Inhibitor Cocktail | Roche | CAT # 11873580001 | |
| Chemical compound, drug | Imidazole | Alfa Aesar | CAT #47274; CAS 288-32-4 | |
| Chemical compound, drug | Ni-NTA Agarose | Qiagen | CAT # 30250 | |
| Chemical compound, drug | Guanosine-5'-Triphosphate Disodium Salt | Fisher Scientific | CAT # AAJ16800MC; CAS 56001-37-7 | 0.1 mM |
| Chemical compound, drug | Adenosine 5'-triphosphate disodium salt (ATP disodium salt) hydrate | VWR | CAT # TCA0157; CAS 34369-07-8 | 1 mM |
| Chemical compound, drug | Creatine phosphate | Sigma-Aldrich | CAT # CRPHO-RO; CAS 71519-72-7 | 1 mg/mL |
| Chemical compound, drug | Creatine Phosphokinase, Porcine Heart | Sigma-Aldrich | CAT # 238395; CAS 9001-15-4 | 15 U/mL |
| Chemical compound, drug | NucBlue Live ReadyProbes Reagent (Hoechst 33342) | ThermoFisher | CAT # R37605 | |
| Chemical compound, drug | FuGENE HD Transfection Reagent | Promega | CAT # E2311 | |
| Chemical compound, drug | Puromycin | Invivogen | CAT # ant-pr-5 | |
| Cell line (Homo-sapiens) | HeLa Cells | ATCC | CCL-2 RRID:CVCL_0030 | |
| Cell line (Homo-sapiens) | Hap1 Cells | Horizon | N/A | |
| Cell line (Homo-sapiens) | Nup133_mEGFP(−9) | This Paper | N/A | CRISPR-edited Hap1 cell line expressing Nup133_mEGFP(−9) |
| Cell line (Homo-sapiens) | Nup133_mEGFP(−8) | This Paper | N/A | CRISPR-edited Hap1 cell line expressing Nup133_mEGFP(−8) |
| Cell line (Homo-sapiens) | Nup54-mEGFP494(0) | This Paper | N/A | CRISPR-edited Hap1 cell line expressing Nup54-mEGFP494(0) |
| Cell line (Homo-sapiens) | Nup54-mEGFP494(1) | This Paper | N/A | CRISPR-edited Hap1 cell line expressing Nup54-mEGFP494(1) |
| Cell line (Homo-sapiens) | Nup54-mEGFP494(2) | This Paper | N/A | CRISPR-edited Hap1 cell line expressing Nup54-mEGFP494(2) |
| Cell line (Homo-sapiens) | Nup62_mCherry290_mEGFP321 | This Paper | N/A | CRISPR-edited Hap1 cell line expressing Nup62_mCherry290_mEGFP321 |
| Sequence-based reagent | Cloning Primer Nup54 CRISPR Forward | This Paper | PCR primers | CACCCGATCTAGAAGATATAAAGC (guide bolded) |
| Sequence-based reagent | Cloning Primer Nup54 CRISPR Reverse | This Paper | PCR primers | AAACGCTTTATATCTTCTAGATCG |
| Sequence-based reagent | Cloning Primer Nup133 CRISPR Forward | This Paper | PCR primers | CACCGCTCAGTGAGTACTTACCGG (guide bolded) |
| Sequence-based reagent | Cloning Primer Nup133 CRISPR Reverse | This Paper | PCR primers | AAACCCGGTAAGTACTCACTGAGC |
| Sequence-based reagent | PCR Primer Nup54 Forward | This Paper | PCR primers | CCTGTGACTAGCTTGCAGTT |
| Sequence-based reagent | PCR Primer Nup54 Reverse | This Paper | PCR primers | ACCTCTGATGTGGATGGTTTC |
| Sequence-based reagent | PCR Primer Nup133 Forward | This Paper | PCR primers | AGTCCAATCCTTACTTCGAGTTT |
| Sequence-based reagent | PCR Primer Nup133 Reverse | This Paper | PCR primers | AGGAACAACAACTGACACATTTC |
| Recombinant DNA reagent | Nup133_mEGFP(−8a) (plasmid) | Kampmann et al., 2011 | Addgene # 163417 | Mammalian expression of Nup133 fused at carboxy-terminus to mEGFP with total net fusion of (−8 amino acids) |
| Recombinant DNA reagent | Nup133_mEGFP(−8b) (plasmid) | Kampmann et al., 2011 | Addgene #163418 | Mammalian expression of Nup133 fused at carboxy-terminus to mEGFP with total net fusion of (−8 amino acids) |
| Recombinant DNA reagent | Nup133_mEGFP(−9a) (plasmid) | Kampmann et al., 2011 | Addgene # 163419 | Mammalian expression of Nup133 fused at carboxy-terminus to mEGFP with total net fusion of (−9 amino acids) |
| Recombinant DNA reagent | Nup133_mEGFP(−9b) (plasmid) | Kampmann et al., 2011 | Addgene # 163420 | Mammalian expression of Nup133 fused at carboxy-terminus to mEGFP with total net fusion of (−9 amino acids) |
| Recombinant DNA reagent | Nup93_mEGFP(−5) (plasmid) | This paper | Addgene # 163421 | Mammalian expression of Nup93 fused at carboxy-terminus to mEGFP with total net fusion of (−5 amino acids) |
| Recombinant DNA reagent | Nup93_mEGFP(−6) (plasmid) | This paper | Addgene # 163422 | Mammalian expression of Nup93 fused at carboxy-terminus to mEGFP with total net fusion of (−6 amino acids) |
| Recombinant DNA reagent | Nup58_mEGFP(−6) (plasmid) | This paper | Addgene # 163423 | Mammalian expression of Nup58 with mEGFP (missing first six amino acids) at position 412 |
| Recombinant DNA reagent | Nup58_mEGFP(−7) (plasmid) | This paper | Addgene # 163424 | Mammalian expression of Nup58 with mEGFP (missing first seven amino acids) at position 412 |
| Recombinant DNA reagent | Nup58_mEGFP(−8) (plasmid) | This paper | Addgene # 163425 | Mammalian expression of Nup58 with mEGFP (missing first eight amino acids) at position 412 |
| Recombinant DNA reagent | Nup54-mEGFP494(0) (plasmid) | This paper | Addgene # 163426 | Mammalian expression of Nup54 with mEGFP (missing first five amino acids) at amino acid 494 with five amino acid rigid alpha helical linker |
| Recombinant DNA reagent | Nup54-mEGFP494(1) (plasmid) | This paper | Addgene # 163427 | Mammalian expression of Nup54 with mEGFP (missing first five amino acids) at amino acid 494 with six amino acid rigid alpha-helical linker |
| Recombinant DNA reagent | Nup54-mEGFP494(2) (plasmid) | This paper | Addgene # 163428 | Mammalian expression of Nup54 with mEGFP (missing first five amino acids) at amino acid 494 with seven amino acid rigid alpha helical linker |
| Recombinant DNA reagent | Nup54-mEGFP494(flex0)(plasmid) | This paper | Addgene # 163429 | Mammalian expression of Nup54 with mEGFP (missing first five amino acids) at amino acid 494 with five amino acid flexible alpha-helical linker |
| Recombinant DNA reagent | Nup54-mEGFP494(flex1)(plasmid) | This paper | Addgene # 163430 | Mammalian expression of Nup54 with mEGFP (missing first five amino acids) at amino acid 494 with six amino acid flexible alpha-helical linker |
| Recombinant DNA reagent | Nup54-mEGFP494(flex2)(plasmid) | This paper | Addgene # 163431 | Mammalian expression of Nup54 with mEGFP (missing first five amino acids) at amino acid 494 with seven amino acid flexible alpha helical linker |
| Recombinant DNA reagent | Nup54_mEGFP494(−4) (plasmid) | This paper | Addgene # 163432 | Mammalian expression of Nup54 with mEGFP (missing first four amino acids) at amino acid 494 |
| Recombinant DNA reagent | Nup54_mEGFP494(−5) (plasmid) | This paper | Addgene # 163433 | Mammalian expression of Nup54 with mEGFP (missing first five amino acids) at amino acid 494 |
| Recombinant DNA reagent | Nup54_mEGFP494(−6) (plasmid) | This paper | Addgene # 163434 | Mammalian expression of Nup54 with mEGFP (missing first six amino acids) at amino acid 494 |
| Recombinant DNA reagent | Nup54_mEGFP510(−4) (plasmid) | This paper | Addgene # 163435 | Mammalian expression of Nup54 with mEGFP (missing first five amino acids) at the carboxy-terminus with total net fusion of (−4) amino acids |
| Recombinant DNA reagent | Nup54_mEGFP510(−5) (plasmid) | This paper | Addgene # 163436 | Mammalian expression of Nup54 with mEGFP (missing first five amino acids) at the carboxy-terminus with total net fusion of (−5) amino acids |
| Recombinant DNA reagent | Nup54_mEGFP510(−6) (plasmid) | This paper | Addgene # 163437 | Mammalian expression of Nup54 with mEGFP (missing first five amino acids) at the carboxy-terminus with total net fusion of (−6) amino acids |
| Recombinant DNA reagent | pSpCas9(BB)−2A-Puro (PX459) V2.0 (plasmid) | Ran et al., 2013 | Addgene # 62988 | |
| Recombinant DNA reagent | pTriEx-mCherry::LANS4 (plasmid) | Yumerefendi et al., 2015 | Addgene #60785 | |
| Recombinant DNA reagent | BFP-RanQ69L (plasmid) | This paper | Addgene # 163438 | Mammalian expression of RanQ69L with tag-BFP |
| Recombinant DNA reagent | pET28-RAN (plasmid) | Günter Blobel | Addgene # 163439 | Ran in pET28 protein expression backbone |
| Recombinant DNA reagent | pET28_KPNA1 (plasmid) | This paper | Addgene #163440 | KPNA1 in pET28 protein expression backbone |
| Recombinant DNA reagent | pET28-KPNB1 (plasmid) | This paper | Addgene # 163441 | KPNB1 in pET28 protein expression backbone |
| Recombinant DNA reagent | pET28-NTF2 (plasmid) | This paper | Addgene # 163442 | NTF2 in pET28 protein expression backbone |
| Recombinant DNA reagent | Nup54 no FG-mEGFP494(0) | This paper | Addgene # 164269 | Mammalian expression of Nup54 without the FG-Nup domain, with mEGFP (missing the first 5 amino acids) at amino acid position 494 with a rigid alpha helix of 5 amino acids at the carboxyl end of mEGFP |
| Recombinant DNA reagent | Nup54 no FG-mEGFP494(1) | This paper | Addgene # 164270 | Mammalian expression of Nup54 without the FG-Nup domain, with mEGFP (missing the first 5 amino acids) at amino acid position 494 with a rigid alpha helix of 6 amino acids at the carboxyl end of mEGFP |
| Recombinant DNA reagent | Nup54 no FG-mEGFP494(2) | This paper | Addgene # 164271 | Mammalian expression of Nup54 without the FG-Nup domain, with mEGFP (missing the first 5 amino acids) at amino acid position 494 with a rigid alpha helix of 7 amino acids at the carboxyl end of mEGFP |
| Recombinant DNA reagent | NLS-tdTomato | This paper | Addgene # 163443 | Bacterial expression of His-tagged, SV40 NLS-tagged tdTomato in the modified pRSETB protein expression backbone |
| Software, algorithm | Metamorph Ver 7.7.8 | Molecular Devices | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy#gref | |
| Software, algorithm | MatLab 2019A | Mathworks | https://www.mathworks.com/ | |
| Software, algorithm | Fiji | Schindelin et al., 2012 | https://imagej.net/Fiji/Downloads | |
| Software, algorithm | CRISPR Guide RNA Design | Benchling | https://www.benchling.com/crispr/ | |
| Software, algorithm | Adobe Illustrator | Adobe | https://www.adobe.com/products/illustrator.html |
Additional files
-
Source code 1
Analysis code for quantification of orientation.
- https://cdn.elifesciences.org/articles/60654/elife-60654-code1-v1.zip
-
Supplementary file 1
Table of Nup-mEGFP transfected fusion protein linkage identities.
The size of mEGFP deletion describes the number of amino acids deleted from the amino terminus of mEGFP and the net linker size describes the number of amino acids in the linker minus the deletions from the Nup and mEGFP.
- https://cdn.elifesciences.org/articles/60654/elife-60654-supp1-v1.docx
-
Supplementary file 2
Table of Nup-mEGFP cell line linkage identities.
The size of mEGFP deletion describes the number of amino acids deleted from the amino terminus of mEGFP and the net linker size describes the number of amino acids in the linker minus the deletions from the Nup and mEGFP.
- https://cdn.elifesciences.org/articles/60654/elife-60654-supp2-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60654/elife-60654-transrepform-v1.pdf





