SUMOylation contributes to proteostasis of the chloroplast protein import receptor TOC159 during early development
Figures
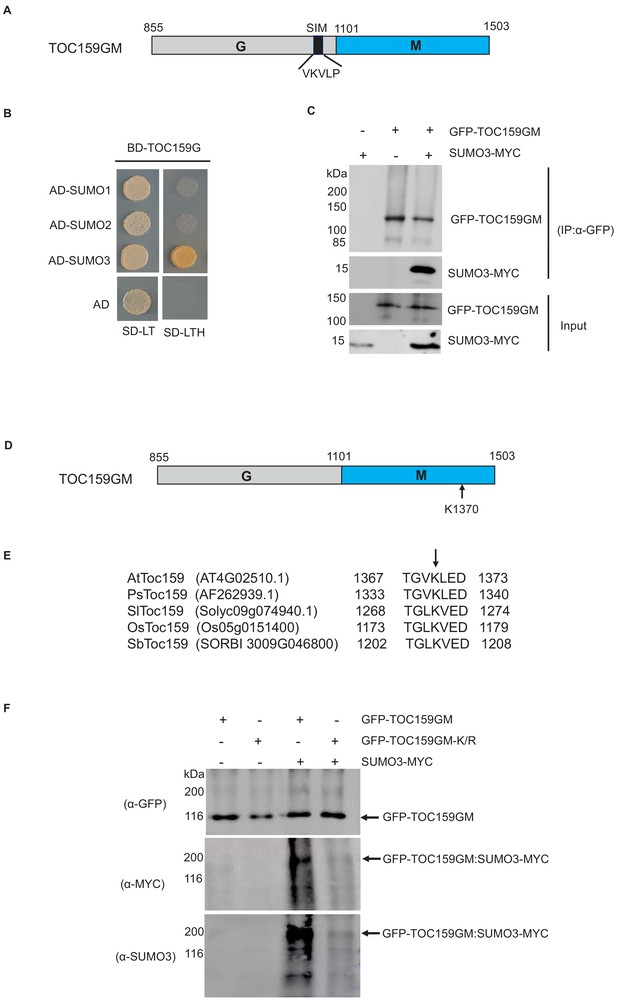
Small ubiquitin-related modifier (SUMO) interaction and SUMOylation of TOC159GM.
(A) Schematic representation of TOC159GM indicating the predicted SUMO-interacting motif (SIM) in the G-domain. (B) Yeast two-hybrid interaction assay of TOC159 G with SUMO proteins on –Leu, –Trp and –Leu, –Trp, –His medium. AD, activation domain; BD, binding domain. (C) Transient expression of SUMO3-MYC, GFP-TOC159GM, and the combination of both in Nicotiana benthamiana leaves. Total protein extracts were subjected to immunoprecipitation with anti-GFP beads. The immunoprecipitated proteins from the expression of SUMO3-MYC (lane 1) and GFP-TOC159GM (lane 2) alone and the co-expression both (lane 3) were analyzed by western blotting using anti-GFP and anti-MYC antibodies. (D) Schematic representation of TOC159GM with indication of the predicted SUMOylation site K1370 (Lysine) at the M-domain. (E) Alignment of the conserved predicted K1370 SUMOylation sites in the M-domain of multiple species: Arabidopsis thaliana (At), Pisum sativum (Ps), Solanum lycopersicum (Sl), Oryza sativa (Os), and Sorghum bicolor (Sb) by using CLUSTAL Omega (1.2.4) multiple sequence alignment tool. (F) Transient expression of GFP-TOC159GM and GFP-TOC159GM-K/R (SUMO mutant, K1370 replaced with R) with and without SUMO3-MYC in N. benthamiana leaves. Total protein extracts were subjected to immunoprecipitation with anti-GFP beads. The immunoprecipitated proteins from the expression of GFP-TOC159GM (lane 1) and GFP-TOC159GM-K/R (lane 2) alone as well as the co-expression with SUMO3 (lanes 3 and 4) were analyzed by western blotting using anti-GFP, anti-MYC and anti-SUMO3 antibodies.
-
Figure 1—source data 1
Source data for Figure 1E.
- https://cdn.elifesciences.org/articles/60968/elife-60968-fig1-data1-v2.docx

Yeast two-hybrid interaction assay of TOC159 M-domain with SUMO proteins on –Leu, –Trp and –Leu, –Trp, and –His medium.
AD, activation domain; BD, binding domain; empty vector was used as a control.
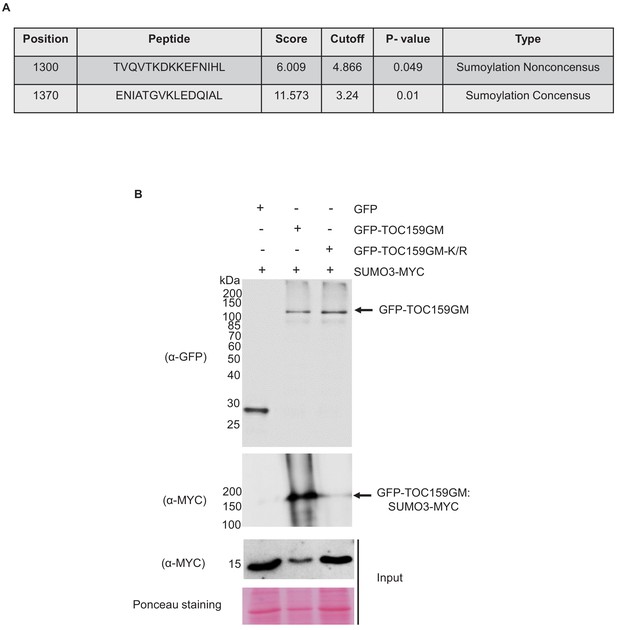
Predicted SUMOylation sites at TOC159GM and in planta SUMOylation assay.
(A) Predicted SUMOylation sites at TOC159GM domain using the GPS-SUMO prediction algorithm with a high threshold (http://sumosp.biocuckoo.org/online.php). (B) Transient expression of GFP, GFP-TOC159GM, and GFP-TOC159GM-K/R (SUMO mutant, K1370 replaced with R) with SUMO3-MYC in Nicotiana benthamiana leaves. Total protein extracts were subjected to immunoprecipitation with anti-GFP beads. The immunoprecipitated proteins from the expression of GFP (lane 1), GFP-TOC159GM (lane 2), and GFP-TOC159GM-K/R (lane 3) co-expression with SUMO3 were analyzed by western blotting using anti-GFP and anti-MYC antibodies.
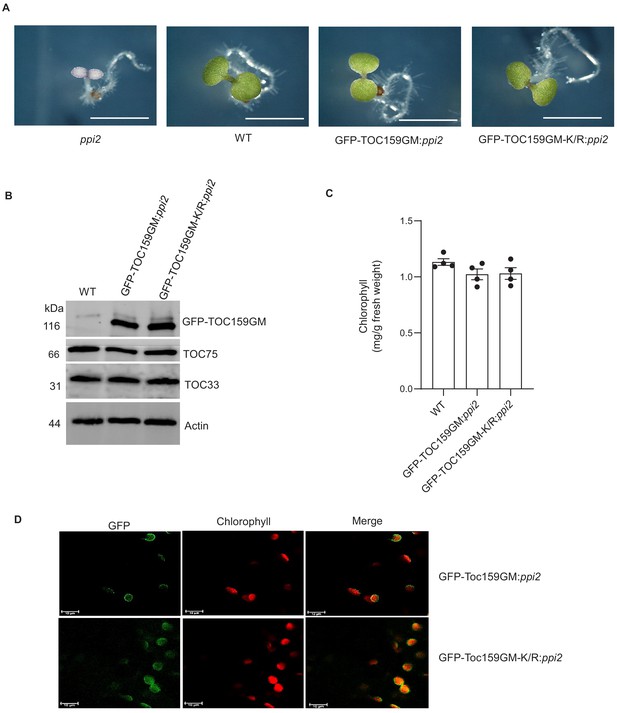
Complementation of ppi2 (toc159 mutant) by non-SUMOylatable TOC159GM-K/R.
(A) Phenotypes of 7 days old seedling of ppi2, WT (Ws), GFP-TOC159GM:ppi2, and GFP-TOC159GM-K/R:ppi2. (B) Western blot analysis of total protein extracts from 7 days old seedlings of WT (Ws), GFP-TOC159GM:ppi2 and GFP-TOC159GM-K/R:ppi2. The blot was probed with anti-GFP, -TOC75, and TOC33 antibodies. Actin was used as a loading control. (C) Chlorophyll levels of wild type, GFP-TOC159GM:ppi2, and GFP-TOC159GM-K/R:ppi2 from 7 days old seedlings. Error bars indicate ± SEM (n = 4). (D) Confocal microscopy analysis of 7 days old GFP-TOC159GM:ppi2 and GFP-TOC159GM-K/R:ppi2 seedlings. Green fluorescence (GFP, left-hand panel), red fluorescence (chlorophyll, middle pane), and the overlay of the two (right-hand panel) are shown.
-
Figure 2—source data 1
Source data for Figure 2C.
- https://cdn.elifesciences.org/articles/60968/elife-60968-fig2-data1-v2.xlsx
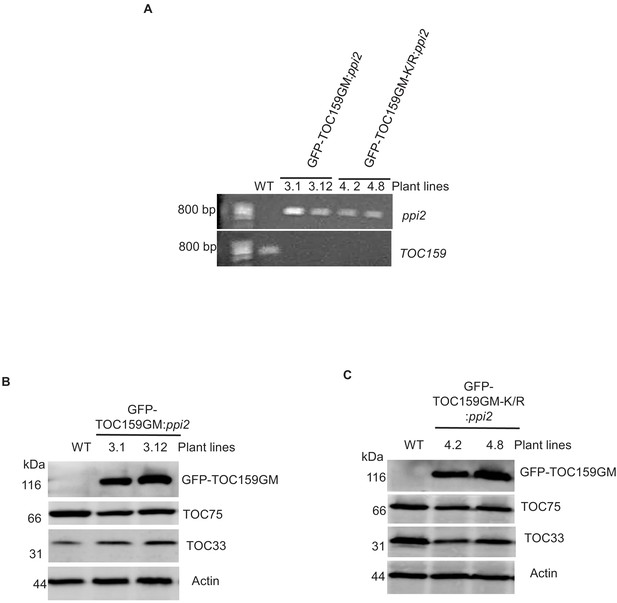
Screening of ppi2 plants for complementation with GFP-TOC159GM and GFP-TOC159GM-K/R.
(A) Genotyping of two independent plant lines (3.1 and 3.12) expressing GFP-TOC159GM in the ppi2 background (GFP-TOC159GM:ppi2) and two independent plant lines (4.2 and 4.8) expressing GFP-TOC159GM-K/R in the ppi2 background (GFP-TOC159GM-K/R:ppi2). WT was used as the control for primers. The ppi2 and TOC159 primer sets were used. (B) Immunoblotting of total protein extracts of seedlings from two independent plant lines (3.1 and 3.12) expressing GFP-TOC159GM in the ppi2 background (GFP-TOC159GM:ppi2). WT was used as the control for antibody specificity. The following antibodies were used: GFP, TOC75, and TOC33 proteins. Actin was used as a loading control. (C) Immunoblotting of total protein extracts of seedlings from two independent plant lines (4.2 and 4.8) expressing GFP-TOC159GM-K/R in ppi2 background (GFP-TOC159GM-K/R:ppi2). WT was used as the control for antibody specificity. The following antibodies were used: GFP, TOC75, and TOC33 proteins. Actin was used as a loading control.
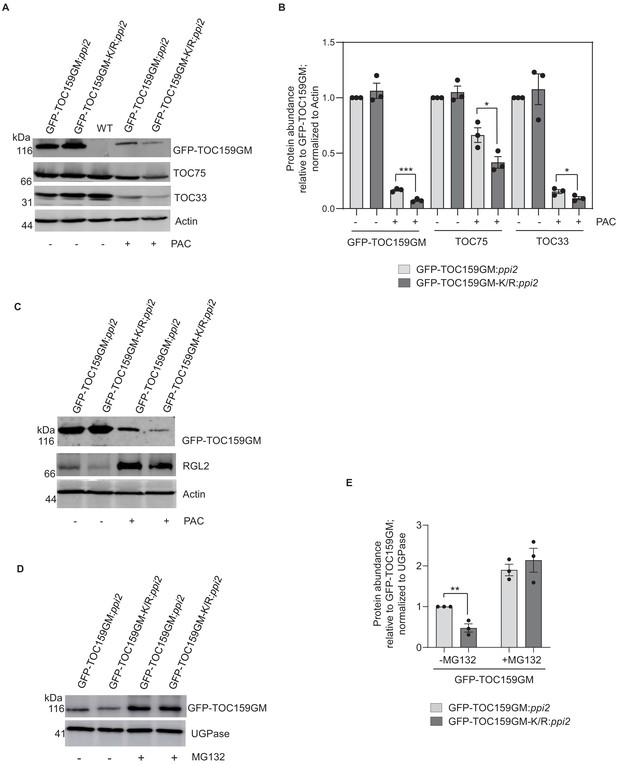
SUMOylation partially protects TOC159 from UPS-mediated degradation.
(A) Immunoblotting of total protein extracts from 3 days old seedling of GFP-TOC159GM:ppi2 and GFP-TOC159GM-K/R:ppi2 grown in the presence or absence of paclobutrazol (PAC) (5 µM). WT (Ws) was used as the control for antibody specificity. The blot was probed with anti-GFP, -TOC75, and -TOC33 antibodies. Anti-actin was used for a loading control. (B) Specific bands corresponding to GFP, TOC75, TOC33, and actin were quantified from three independent experiments (A). The quantified bands were normalized to GFP-TOC159GM in GFP-TOC159GM:ppi2 plants grown in the absence of PAC. Error bars indicate ± SEM (n = 3). two-tailed t test; ∗p<0.05; ∗∗∗p<0.005. (C) Immunoblotting of total protein extracts from 3 days old GFP-TOC159GM:ppi2 and GFP-TOC159GM-K/R:ppi2 seedlings grown in the presence or absence of PAC (5 µM). The blot was probed with anti-GFP and -RGL2 antibodies. Anti-actin was used for a loading control. (D) Total protein extracts of 3 days old GFP-TOC159GM:ppi2 or GFP-TOC159GM-K/R:ppi2 grown seedling grown on PAC and subsequently treated with or without MG132 were analyzed by immunoblotting using anti-GFP antibodies and anti-UGPase for a loading control. (E) The specific bands corresponding to GFP and UGPase were quantified from three independent experiments (C). The quantified bands were normalized to GFP-TOC159GM in GFP-TOC159GM:ppi2 without MG132. Error bars indicate ± SEM (n = 3). two-tailed t test; ∗∗p<0.01.
-
Figure 3—source data 1
Source data for Figure 3B, E, Figure 3—figure supplements 3 and 5.
- https://cdn.elifesciences.org/articles/60968/elife-60968-fig3-data1-v2.xlsx
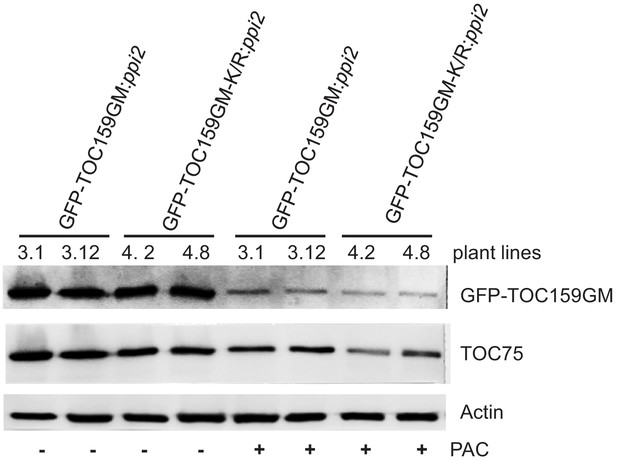
Immunoblotting of total protein extracts from two independent GFP-TOC159GM:ppi2 lines (3.1 and 3.12) and two independent GFP-TOC159GM-K/R:ppi2 lines (4.2 and 4.8).
Total protein was extracted from 3 days old seedlings grown in the presence or absence of PAC (5 µM). WT was used as the control for antibody specificity. The following antibodies were used GFP, TOC75, and TOC33 proteins. Actin was used as a loading control.
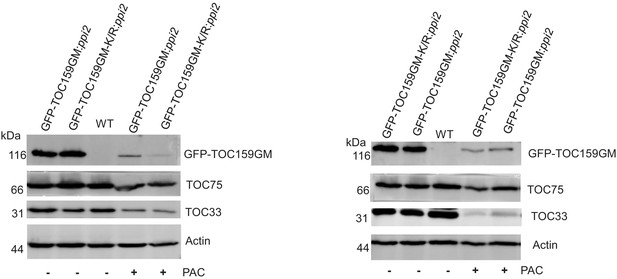
Immunoblotting of total protein extracts from GFP-TOC159GM:ppi2 and GFP-TOC159GM-K/R:ppi2 grown in the presence or absence of PAC (5 µM).
WT was used as the control for antibody specificity. The following antibodies were used: GFP, TOC75, and TOC33 proteins. Actin was used as a loading control. The specific bands corresponding to GFP, TOC75, TOC33, and actin were quantified. The quantified bands were normalized to GFP-TOC159GM in GFP-TOC159GM:ppi2 grown in the absence of PAC. These data were used in Figure 3B.
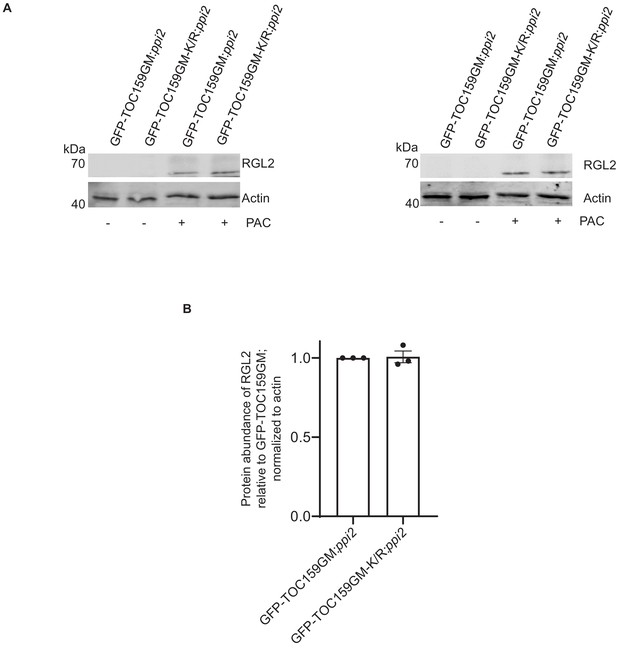
RGL2 protein accumulation in GFP-TOC159GM:ppi2and GFP-TOC159GM-K/R:ppi2under low GA conditions.
(A) Immunoblotting of total protein extracts from two independent biological replicates of 3 days old GFP-TOC159GM:ppi2 and GFP-TOC159GM-K/R:ppi2 seedlings grown in the presence or absence of PAC (5 µM). The blot was probed with anti-GFP and -RGL2 antibodies. Anti-actin was used as a loading control. (B) Specific bands corresponding to GFP, RGL2, and actin were quantified from three independent experiments (A) and Figure 3C.
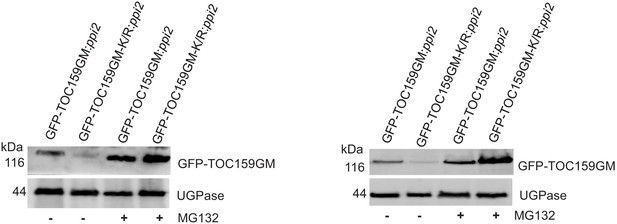
Total protein extracts of 3 days old GFP-TOC159GM:ppi2 or GFP-TOC159GM-K/R:ppi2 grown in the presence of PAC and further treated with or without MG132, analyzed by immunoblotting using antibodies against GFP.
UGPase was used as a loading control. The specific bands corresponding to GFP and UGPase were quantified. The quantified bands were normalized to GFP-TOC159GM in GFP-TOC159GM:ppi2 without MG132. These data were used for Figure 3E.
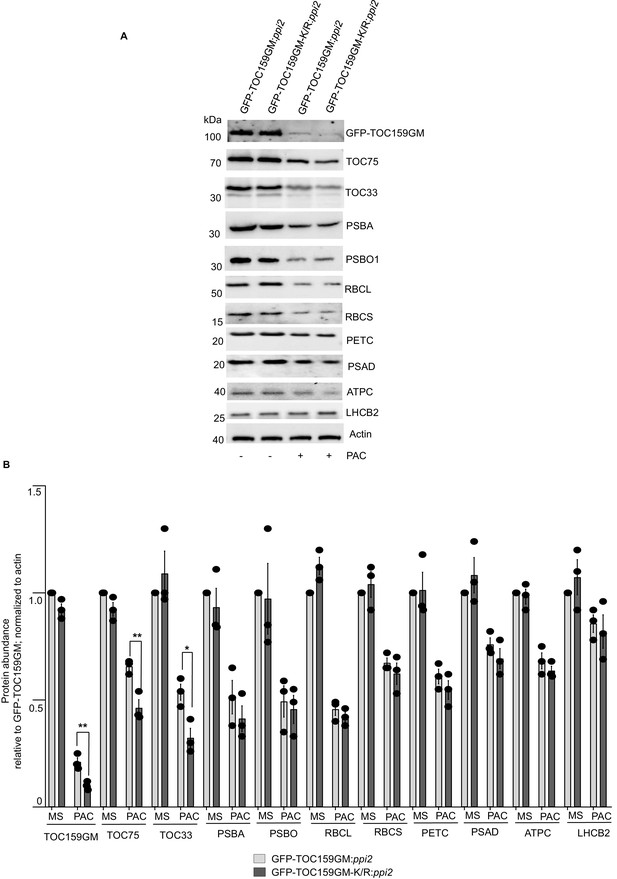
The TOC159 SUMOylation-deficient line TOC159GM-K/R:ppi2 accumulates photosynthesis-associated proteins trended lower compared to TOC159GM:ppi2 under low GA conditions.
(A) Immunoblotting of total protein extracts from 3 days old seedling of GFP-TOC159GM:ppi2 and GFP-TOC159GM-K/R:ppi2 grown in the presence or absence of PAC (2 µM). The blot was probed with anti-GFP, -TOC75, -TOC33, -PSBA, and -PSBO1 antibodies. Anti-actin was used as a loading control. (B) The specific bands corresponding to GFP, TOC75, TOC33, PSBA, PSBO1, RBCL, RBCS, PETC, PSAD, ATPC, LHCB2, and actin were quantified from three independent experiments (A). The quantified bands were normalized to GFP-TOC159GM in GFP-TOC159GM:ppi2 grown in the absence of PAC. Error bars indicate ± SEM (n = 3). two-tailed t test; ∗p<0.05; ∗∗p<0.01.
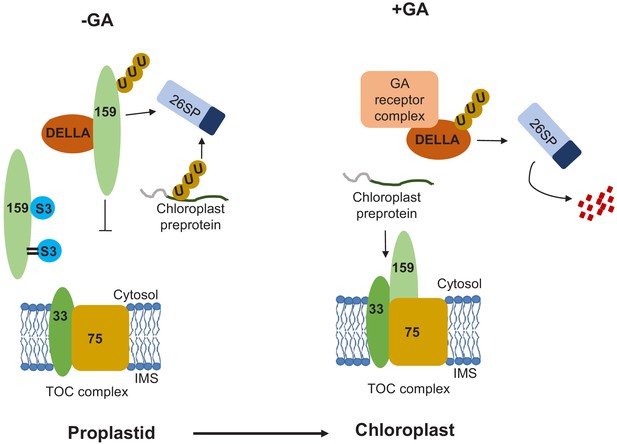
Hypothetical model for the SUMOylation-dependent fine-tuning of chloroplast biogenesis at the level of the TOC159 import receptor in early plant development.
Environmental conditions influence the concentrations of gibberellic acid (GA) that accumulate upon seed imbibition. When active GA levels are reduced by stress (-GA; left-hand panel), DELLA (RGL2) accumulates and sequesters TOC159, which is then degraded via the UPS. In addition, TOC159 interacts and is also covalently SUMOylated by SUMO3. Covalent SUMOylation protects TOC159 against UPS-mediated degradation and supports the accumulation of photosynthesis-associated proteins in the chloroplast. Any unimported preproteins are degraded in the cytosol via the UPS. When GA concentrations increase (+GA, right-hand panel), the GA receptor complex binds to DELLA, which is degraded by the UPS. TOC159 is then free to assemble into the TOC complex. Protein import is thus enabled, allowing proplastids to differentiate into chloroplasts.
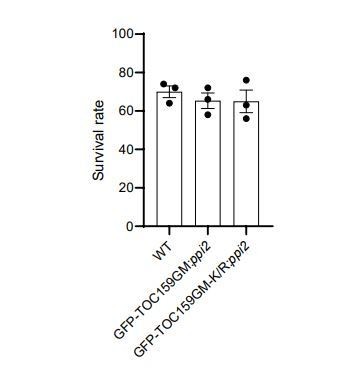
A de-etiolation assay was carried out with WT, GFP-TOC159GM:ppi2 and GFP-TOC159GM-K/R:ppi2 plant lines (three independent biological replicates with at least 50 seedlings).
The seeds were stratified and exposed for 3h to 120 µmol x m-2 x s-1 of white light, then grown in the dark for 6 days. The etiolated seedlings were transferred to continuous light for two more days. The survival rate of de-etiolated seedlings based on cotyledon greening. Error bars indicate ± SEM (n = 3).
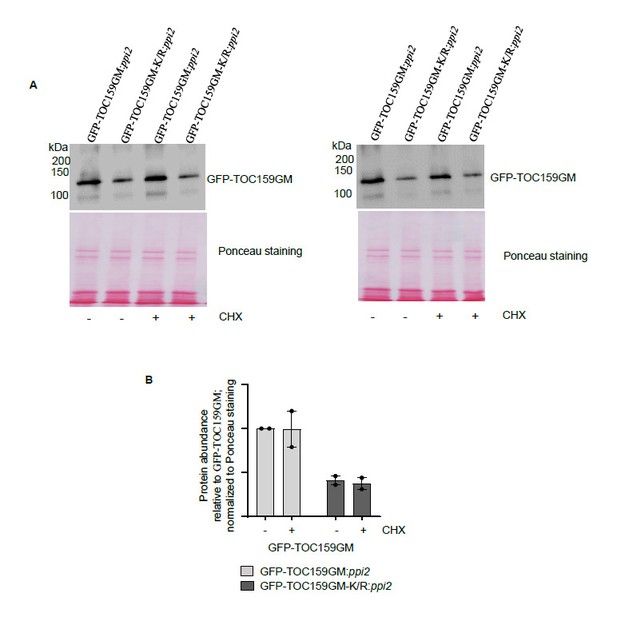
The TOC159 protein accumulation from GFP-TOC159GM:ppi2and GFP-TOC159GM-K/R:ppi2under low GA conditions and further treated with cycloheximide.
(A) Total protein extracts of three days old GFP-TOC159GM:ppi2 or GFP-TOC159GM-K/R:ppi2 grown seedlings grown on PAC and subsequently treated with or without cycloheximide were analyzed by immunoblotting using anti-GFP antibodies and Ponceau staining for a loading control.(B) The specific bands corresponding to GFP were quantified from two independent experiments (A). The quantified bands were normalized to GFP-TOC159GM in GFP-TOC159GM:ppi2 without cycloheximide. Error bars indicate ± SEM (n = 2).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Arabidopsis thaliana) | pTOC159-GFP-TOC159GM:ppi2 | Shanmugabalaji et al., 2018 | ||
| Genetic reagent (Arabidopsis thaliana) | pTOC159-GFP-TOC159GM-K/R | This paper | ||
| Recombinant DNA reagent | pGBKT7-TOC159G | Shanmugabalaji et al., 2018 | Yeast two-hybrid assay | |
| Recombinant DNA reagent | pGBKT7-TOC159M | Shanmugabalaji et al., 2018 | Yeast two-hybrid assay | |
| Recombinant DNA reagent | pGADT7-SUMO1 | This paper | Yeast two-hybrid assay | |
| Recombinant DNA reagent | pGADT7-SUMO2 | This paper | Yeast two-hybrid assay | |
| Recombinant DNA reagent | pGADT7-SUMO3 | This paper | Yeast two-hybrid assay | |
| Recombinant DNA reagent | 35S-GFP-TOC159GM | Shanmugabalaji et al., 2018 | In planta CoIP | |
| Recombinant DNA reagent | 35S-GFP | This paper | In planta CoIP | |
| Recombinant DNA reagent | 35S-SUMO3-MYC | This paper | In planta CoIP | |
| Recombinant DNA reagent | 35S-GFP-TOC159GM-K/R | This paper | In planta CoIP | |
| Antibody | Anti-TOC75, rabbit polyclonal | Hiltbrunner et al., 2001 | 1:1000 | |
| Antibody | Anti-TOC33, rabbit polyclonal | Rahim et al., 2009 | 1:1000 | |
| Antibody | Anti-RGL2, rabbit polyclonal | Piskurewicz et al., 2008 | 1:1000 | |
| Antibody | Anti-SUMO3, rabbit polyclonal | Agrisera | Cat# AS08349 | 1:1000 |
| Antibody | Anti-UGPase, rabbit polyclonal | Agrisera | Cat# AS05086 RRID:AB_1031827 | 1:5000 |
| Antibody | Anti-PSBA, rabbit polyclonal | Agrisera | Cat# AS05084 RRID:AB_2172617 | 1:10,000 |
| Antibody | Anti-PSBO1, rabbit polyclonal | Agrisera | Cat# AS142824 RRID:AB_1031788 | 1:5000 |
| Antibody | Anti-RBCL, rabbit polyclonal | Agrisera | Cat# AS03037 RRID:AB_2175406 | 1:15,000 |
| Antibody | Anti-RBCS, rabbit polyclonal | Agrisera | Cat# AS07259 RRID:AB_1031806 | 1:5000 |
| Antibody | Anti-PETC, rabbit polyclonal | Agrisera | Cat# AS08330 RRID:AB_2162102 | 1:5000 |
| Antibody | Anti-PSAD, rabbit polyclonal | Agrisera | Cat# AS09461 RRID:AB_1832088 | 1:5000 |
| Antibody | Anti-ATPC, rabbit polyclonal | Agrisera | Cat# AS08312 RRID:AB_2290280 | 1:5000 |
| Antibody | Anti-LHCB2, rabbit polyclonal | Agrisera | Cat# AS01003 RRID:AB_1832080 | 1:10,000 |
| Antibody | Anti-actin, mouse monoclonal antibody | Sigma | Cat# A0480 RRID:AB_476670 | 1:2000 |
| Antibody | Anti-GFP, mouse monoclonal antibody | Takara | Cat#632380 RRID:AB_10013427 | 1:1000 |
| Antibody | Anti-MYC, mouse monoclonal antibody | Cell Signaling | Cat#2276 RRID:AB_331783 | 1:1000 |
| Antibody | Anti-rabbit IgG conjugated with horseradish peroxidase | Millipore | Cat# AP132P RRID:AB_90264 | 1:10,000 |
| Antibody | Anti-mouse IgG conjugated with horseradish peroxidase | Sigma | Cat# A5278 RRID:AB_258232 | 1:10,000 |
| Strain, strain background (Escherichia coli) | DH5α | Invitrogen | Cat# 18265017 | |
| Strain, strain background (Agrobacterium tumefaciens) | C58 | Community resource | ||
| Chemical compound, drug | MG132 | AbMole | Cat# M1902 | |
| Chemical compound, drug | Paclobutrazol | Sigma | Cat# 43900 | |
| Commercial assay or kit | µMACS GFP-tagged beads | Miltenyi Biotech | Cat# 130091125 | |
| Software, algorithm | ImageQuant TL | GE Healthcare | RRID:SCR_014246 | |
| Software, algorithm | GraphPad Prism | Graphpad | RRID:SCR_015807 |
Additional files
-
Supplementary file 1
List of primers used in this study.
- https://cdn.elifesciences.org/articles/60968/elife-60968-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60968/elife-60968-transrepform-v2.docx





