Cortical signatures of precision grip force control in children, adolescents, and adults
Figures
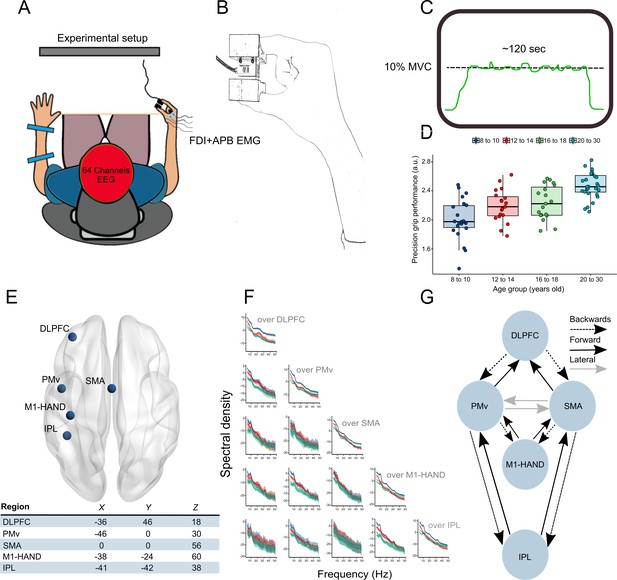
Experimental setup and framework of analysis.
(A) Participants were seated in front of a screen, and 64-channel EEG and EMG from the first dorsal interosseous (FDI) and abductor pollicis brevis (APB) was acquired. (B) Precision grip force transducer and hand configuration during the task. Part (C) displays the precision grip force-tracing task. A horizontal target line representing 10% of maximal voluntary contraction was centered in the middle of the screen, and participants were instructed to track this line as closely as possible by adjusting the force exerted between their index finger and thumb. (D) Boxplots representing performance in the precision grip task for the different age groups. (E) Regions shown to be involved in visually guided precision grip tasks and the corresponding MNI coordinates in mm used for the DCM analysis. (F) Auto and crossspectral densities for sensors located above the brain regions of interest. Solid lines represent group means, and shaded ribbons display 95% CIs. (G) Schematic representation of the extrinsic connections between brain regions specified in the DCMs.
-
Figure 1—source code 1
R-script used to analyze and plot motor performance data presented in Figure 1D.
- https://cdn.elifesciences.org/articles/61018/elife-61018-fig1-code1-v2.zip
-
Figure 1—source data 1
Motor performance data for behavioral analysis.
- https://cdn.elifesciences.org/articles/61018/elife-61018-fig1-data1-v2.xlsx
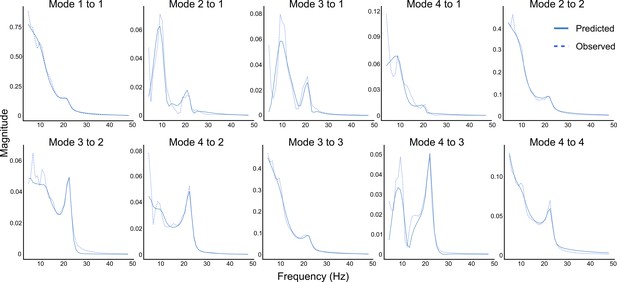
Observed and DCM-predicted auto- and cross-spectra.
Predicted and observed auto- and cross-spectra for the first four modes from an exemplary participant in the 8–10-year-old age group. See Figure 2—figure supplement 1 for exemplary data from individuals across all four age groups.
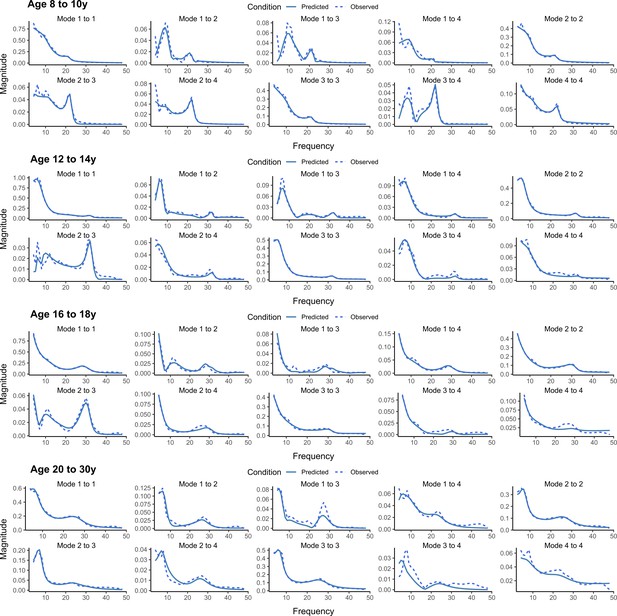
Examples of observed and predicted auto- and cross-spectra from all age groups.
As a supplement to the data presented in Figure 2, we present observed and DCM-predicted auto- and cross spectra from the first four principal modes of EEG data in representative subjects from each age group. The aim is to provide a visual presentation of principal data features from the Bayesian autoregressive model that were predicted by the DCMs for comparison between individuals from different age groups. Note that the four examples are characterized by prominent features in physiologically relevant frequency bands and that the DCMs have captured (predicted) these to a great extent.
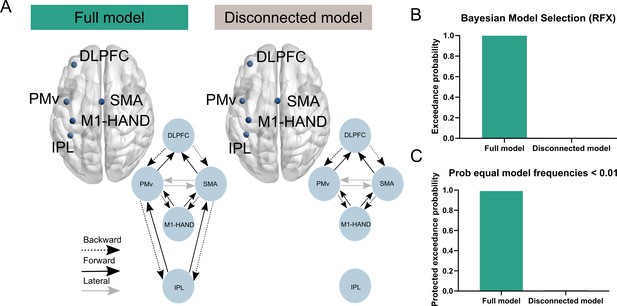
Model comparisons.
To ensure that the sources included in our a priori DCM were sufficiently spaced to minimize the influence of volume conduction, we performed a random effect (RFX) Bayesian model selection (BMS). (A) We compared two different DCMs. The full model was our a priori defined model of the contralateral grasping network, whereas the disconnected model included the same sources and thereby same data (via principal modes of the EEG data), but the IPL was modelled as a hidden source. This enabled a comparison of the two models using RFX BMS. The model comparison revealed greater evidence for the fully connected model as reflected in a greater exceedance probability and protected exceedance probability (B, C).
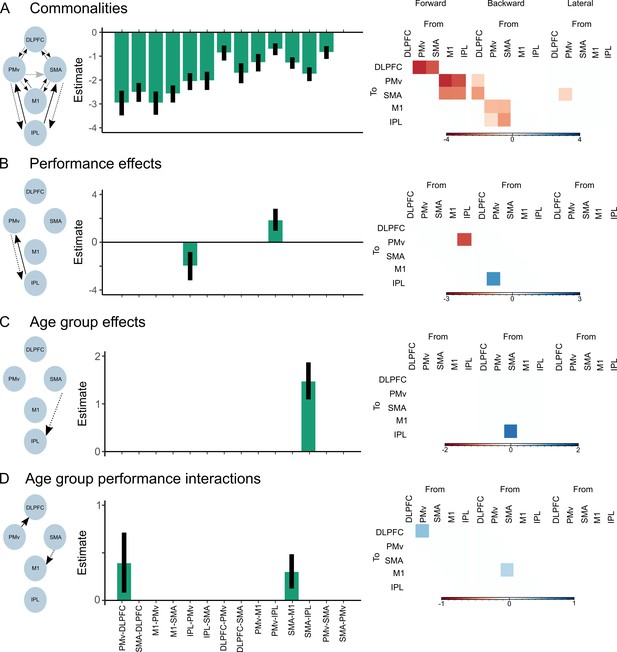
Parameters of the group-level parametric empirical Bayes following Bayesian model averaging (BMA).
Depicted are only those extrinsic connections surviving the 95% posterior probability threshold based on free energy. Left shows the schematic network presentation. Middle displays the posterior parameter estimates with the associated uncertainties. Right shows the same estimated parameters split into a matrix representing the forward, backward, and lateral extrinsic connections. (A) Commonalities (mean connectivity) across participants. (B) Differences in connectivity due to precision grip performance. (C) Differences in connectivity due to age group. (D) Differences in connectivity due to age group vs. precision grip performance interactions.
-
Figure 4—source code 1
R-script used to plot results from group-based parametric empirical Bayes analysis presented in Figure 4.
- https://cdn.elifesciences.org/articles/61018/elife-61018-fig4-code1-v2.zip
-
Figure 4—source data 1
Group-based parametric empirical Bayes results.
- https://cdn.elifesciences.org/articles/61018/elife-61018-fig4-data1-v2.xlsx
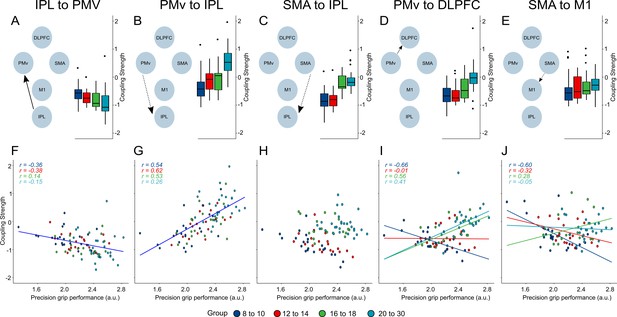
Main effects and interaction effects.
Schematic representing model for main effects and interaction effects (A–E). Boxplots representing the distribution of coupling strength faceted by age group for the respective connectivity parameter (A–E). Scatter plots (F–J) represent individual data points with one regression line for main effects and regression lines by age groups for interaction effects. Parts (A), (B), (F), and (G) display the association between precision grip performance and coupling strength. (C) and (H) depict the main effect of age group on coupling parameters. (D), (E), (I), and (J) display interactions between age group and performance. Coupling strength values are depicted as unit-less log-scaling entities.
-
Figure 5—source code 1
R-script used to plot associations between individual coupling parameters and motor performance presented in Figure 5.
- https://cdn.elifesciences.org/articles/61018/elife-61018-fig5-code1-v2.zip
-
Figure 5—source data 1
Individual coupling parameters and motor performance.
This data is also used for Figure 6.
- https://cdn.elifesciences.org/articles/61018/elife-61018-fig5-data1-v2.xlsx
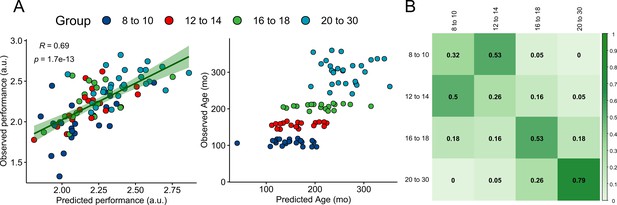
Actual and predicted precision grip performance and age.
(A) Associations between actual and predicted precision grip performance (left) and fractionized age (right) from leave-one-out cross-validations using multiple linear regression. (B) Confusion matrix normalized to group sample size representing the proportion of predicted labels based on a quadratic discriminant analysis in a LOO routine. The diagonal represents the proportion of correctly identified labels.
-
Figure 6—source code 1
R-script used to run and plot cross-validation and classification analyses presented in Figure 6.
- https://cdn.elifesciences.org/articles/61018/elife-61018-fig6-code1-v2.zip
Tables
Participant characteristics.
| 8–10 years | 12–14 years | 16–18 years | 20–30 years | |
|---|---|---|---|---|
| Number of participants | 22 | 19 | 19 | 28 |
| Sex (F/M) | 9/13 | 9/10 | 10/9 | 16/12 |
| Age (months) | 108 ± 7 | 156 ± 7 | 206 ± 7 | 301 ± 34 |
| Tanner developmental stage | 1.12 ± 0.27 | 2.56 ± 1.03 | 4.63 ± 0.55 | 5 ± 0 |
-
Data presented as counts or as means ± standard deviations (SDs).
Additional files
-
Source code 1
MATLAB script used to preprocess the EEG data used for the dynamic causal modeling analysis.
- https://cdn.elifesciences.org/articles/61018/elife-61018-code1-v2.zip
-
Supplementary file 1
Table representing the pipeline used in EEGLAB (v.14.1.1) interfaced in MATLAB R2017b.
- https://cdn.elifesciences.org/articles/61018/elife-61018-supp1-v2.docx
-
Supplementary file 2
Table representing steps applied during conversion of files to SPM data files in SPM12 (v7487) interfaced in MATLAB R2017b.
- https://cdn.elifesciences.org/articles/61018/elife-61018-supp2-v2.docx
-
Supplementary file 3
Table representing the pipeline used for estimating single-subject dynamic causal modelings in SPM12 (v7487).
- https://cdn.elifesciences.org/articles/61018/elife-61018-supp3-v2.docx
-
Supplementary file 4
Represents the pipeline used for the group-level (second level) analyses of dynamic causal modelings in SPM12 (v7487).
*The parametric empirical Bayes analysis was run three times, with the design matrix reordered so that the main regressor of interest was listed just after the commonalities (column 2) as per https://en.wikibooks.org/wiki/SPM/Parametric_Empirical_Bayes_(PEB)#Search_over_nested_PEB_models.
- https://cdn.elifesciences.org/articles/61018/elife-61018-supp4-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61018/elife-61018-transrepform-v2.docx





