Antibody escape by polyomavirus capsid mutation facilitates neurovirulence
Figures
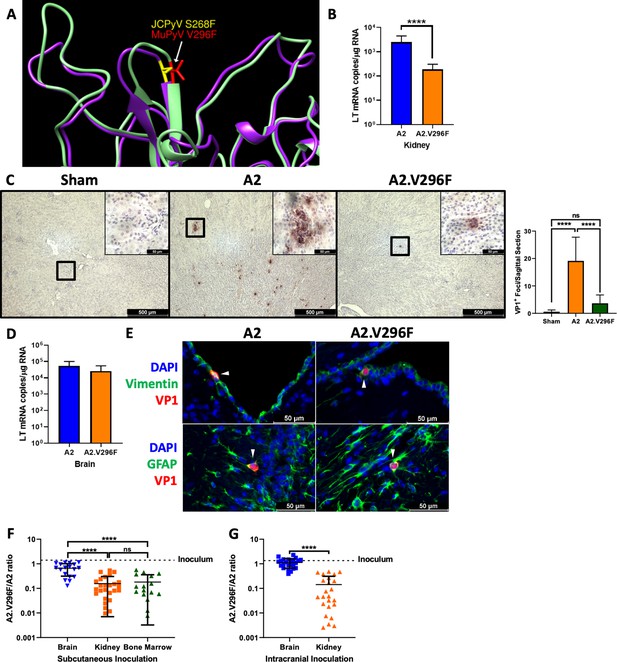
The V296F VP1 mutation in MuPyV impairs kidney, but not brain, infection.
(A) Structural comparison of JCPyV.S268 (PDB 3NXG) and MuPyV.V296 (PDB 5CPU) VP1 residues (Buch et al., 2015; Neu et al., 2010). (B) A2 and A2.V296F LT mRNA levels 4 dpi in the kidneys of mice infected s.c. Data are from three independent experiments, n = 17 mice (p<0.0001). (C) Left: 40x images of kidneys from CD8 T cell-depleted, Stat1–/– mice 7 dpi stained for VP1. Inset is a 400x image of the region outlined in black. Right: Quantification of VP1+ foci per sagittal kidney section. Data are the average of two sagittal kidney sections per mouse from three independent experiments, n = 6–11. For Sham vs. A2 p<0.0001, Sham vs. A2.V296F p=0.5801, A2 vs. A2.V296F p<0.0001. (D) A2 and A2.V296F LT mRNA levels four dpi in the brains of mice infected i.c. Data are from three independent experiments, n = 12–13 mice (p=0.1366). (E) 400x images of brains 4 dpi with A2 or A2.V296F i.c. VP1+ cells are indicated with white arrows. Representative of three independent experiments. (F and G) Ratio of A2.V296F to A2 in various organs of mice 14 dpi with a 1:1 PFU inoculum of A2:A2.V296F s.c. (F) or i.c. (G). The dotted line indicates the ratio of A2:A2.V296F DNA in the inoculum. Data are from 2 to 3 independent experiments, n = 16–26 mice. For (F) Brain vs. Kidney p<0.0001, Brain vs. Bone Marrow p<0.0001, Kidney vs. Bone Marrow p=0.9443; for (G) p<0.0001. Data were analyzed by Mann–Whitney U test (B, D, G) or one-way ANOVA (F). ****p<0.0001.
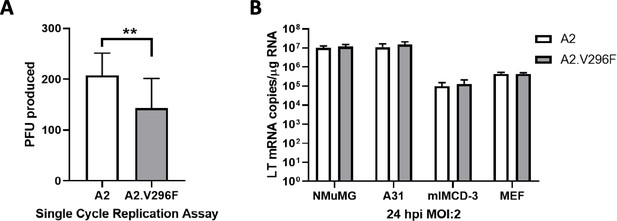
The A2.V296F VP1 mutant virus retains infectivity in vitro.
(A) Single-cycle replication assay in A31 fibroblasts. Cells were infected with an multiplicity of infection (MOI) of 0.1 and virus was collected 60 hpi and measured by plaque assay. Data are from two independent experiments, n = 15 (p=0.0013).(B) LT mRNA levels in cell lines and primary MEFs 24 hpi with an MOI of 2. Data are from two independent experiments, n = 6. For NMuMG p=0.8088, A31 p=0.5975, mIMCD-3 p=0.8088, MEF p=0.9081. Data analyzed by Mann–Whitney U test (A) or multiple t tests (B). **p<0.01.
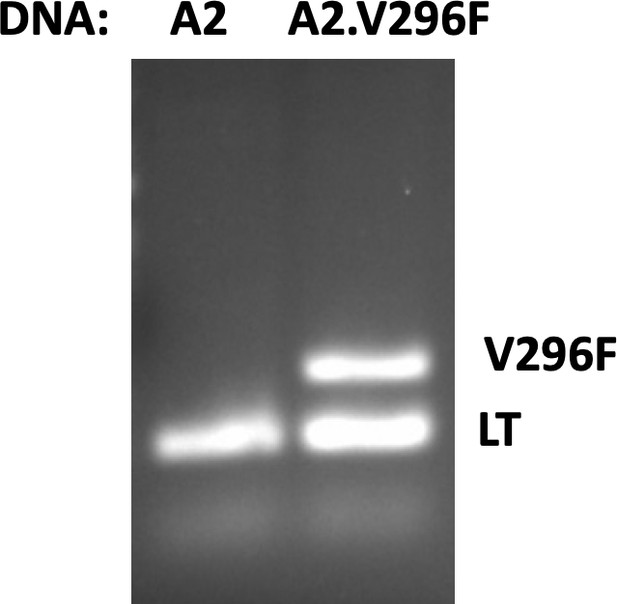
Discrimination of A2 and A2.V296F DNA by PCR.
PCR products after amplification of A2 or A2.V296F DNA with a combination of PCR primers for LT or the V296F-VP1 mutation.
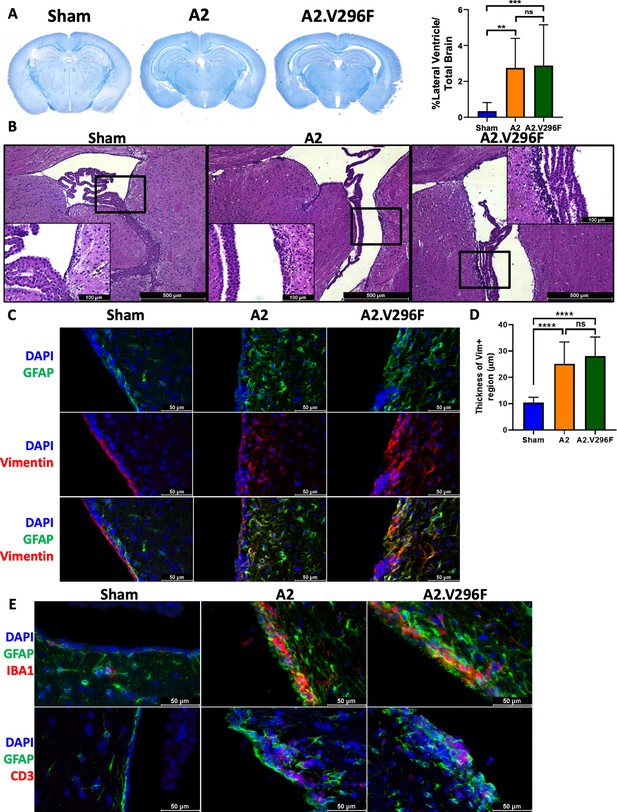
Persistent infection with either A2 or A2.V296F results in CNS pathology.
(A) Left: LFB-PAS-stained brain sections 30 dpi with A2 or A2.V296F. Right: Hydrocephalus was quantified as the size of the lateral ventricle compared to total brain size. Data are from three independent experiments, n = 10–14 mice. For Sham vs A2 p=0.0022, Sham vs A2.V296F p=0.0010, and A2 vs. A2.V296F p=0.9806. (B) Representative 40x H & E images of the ependyma and choroid plexus of the lateral ventricle in mice 30 dpi. Inset image is 200x. (C) 400x fluorescence images of GFAP and vimentin expression in the lining of the lateral ventricles in mice 30 dpi. (D) Quantification of the thickness of the vimentin+ region shown in (C). Data are from three independent experiments, n = 13–15 mice. For Sham vs. A2 p<0.0001, Sham vs. A2.V296F p<0.0001, and A2 vs. A2.V296F p=0.4626. (E) 400x fluorescence images of Iba1+ and CD3+ cells in the lateral ventricles 30 dpi. Data were analyzed by one-way ANOVA (A, D). **p<0.01, ***p<0.001, ****p<0.0001.
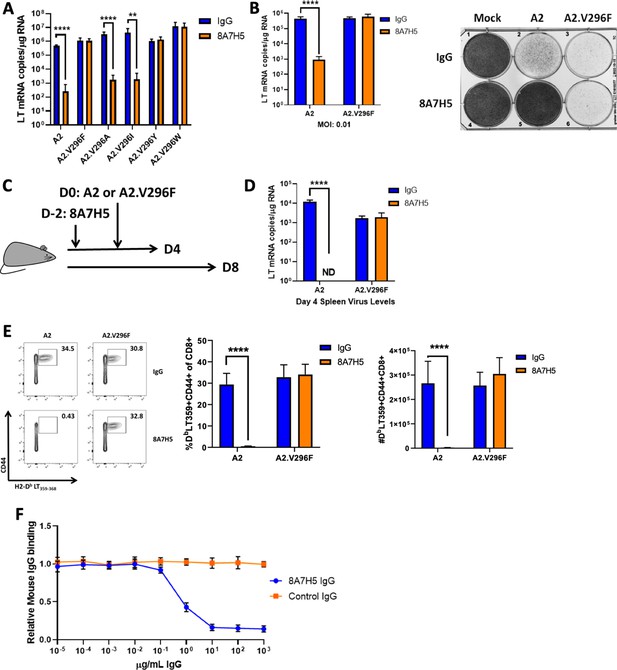
The V296F VP1 mutation confers resistance to a neutralizing mAb.
(A) LT mRNA levels in NMuMG cells 24 hr pi with A2 or V296 mutant viruses preincubated with 8A7H5 or control IgG. Data are from two independent experiments, n = 12. For A2 p<0.0001, A2.V296F p=0.9626, A2.V296A p<0.0001, A2.V296I p=0.0076, A2.296Y p=0.5580, and A2.V296W p=0.9626. (B) Left: LT mRNA levels in A31 fibroblasts 96 hpi with A2 or A2.V296F at 0.01 MOI, 8A7H5 or control IgG was added to the media 24 hpi. Data are from two independent experiments, n = 6. For A2 p<0.0001 and A2.V296F p=0.3947. Right: Protection from virus-induced cell death. A31 fibroblasts were treated as in left panel, fixed with formaldehyde and stained with crystal violet 7 dpi. (C) Experimental design for in vivo neutralization experiments. (D) LT mRNA levels in the spleens of mice injected with 8A7H5 or control IgG followed by infection with A2 or A2.V296F. Data are from two independent experiments, n = 6 mice. For A2 p<0.0001 and A2.V296F p=0.7812. (E) Splenic T cell responses 8 dpi in mice treated and infected as in (D). Data are from two independent experiments, n = 6 mice. Middle: A2 p<0.0001, A2.V296F p=0.6758; Right: A2 p<0.0001, A2.V296F p=0.1679. (F) Competition for binding to VP1 pentamers between immune sera and 8A7H5 IgG. Sera from mice 30 dpi with A2 were diluted to 2 μg/mL of VP1-specific IgG and combined with increasing concentrations of 8A7H5 or control IgG. The serum/8A7H5 was then incubated with VP1 pentamers, and a mouse IgG-specific secondary antibody was used to measure the amount of serum bound to VP1 pentamers by ELISA. Each sample was normalized to binding in the absence of exogenous IgG. Data are from three independent experiments, n = 12 mice. Data were analyzed by multiple t tests (A, B, D, E). **p<0.01, ****p<0.0001.
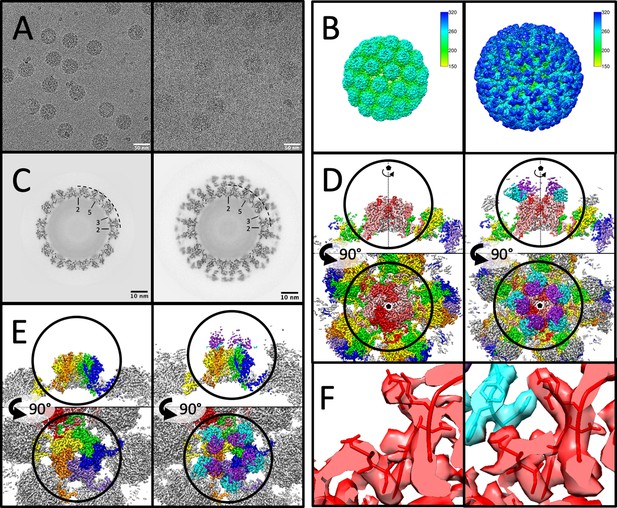
Cryo-EM image sub-particle refinement reconstructions showing architecture of MuPyV-Fab complexes.
(A) Micrographs of virus and virus–Fab complex (shown left and right, throughout figure) illustrate particle diameter difference due to bound Fab. (B) Surface rendered icosahedrally averaged maps. (C) Central sections demonstrate the quality of the maps and show the Fab and capsid densities are of comparable magnitude. (D and E) Sub-particle refinement of pentavalent and hexavalent capsomers with sections through the maps (upper) and top-down views (lower) show the overall architecture. Pentavalent capsomers (D, VP1 density in shades of red) have fivefold symmetry (pentagon), whereas hexavalent capsomers have pseudo-symmetry (E, VP1 in OYGBV) most apparent in the contribution of VP1 C-terminal extensions to neighboring capsomers. Epitopes for the Fab molecules (light chain: purple; heavy chain: cyan) bridge adjacent VP1 molecules. (F) Local refinement of capsomer sub-particles resulted in interpretable sidechain density at the MuPyV-Fab interface (colors as in D and E).
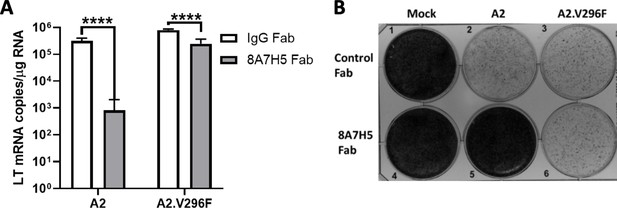
8A7H5 Fab neutralizes A2 but not A2.V296F.
(A) LT mRNA levels in NMuMG cells 24 hpi with A2 or A2.V296F preincubated with 8A7H5 Fab or control Fab. Data are from two independent experiments, n = 6. For A2 p<0.0001, A2.V296F p<0.0001. (B) A31 fibroblasts were infected with A2 or A2.V296F at 0.01 MOI, with 8A7H5 or control Fab added 24 hpi. Cells were fixed with formaldehyde and stained with crystal violet 7 dpi. Data analyzed by multiple t tests (A). ****p<0.0001.
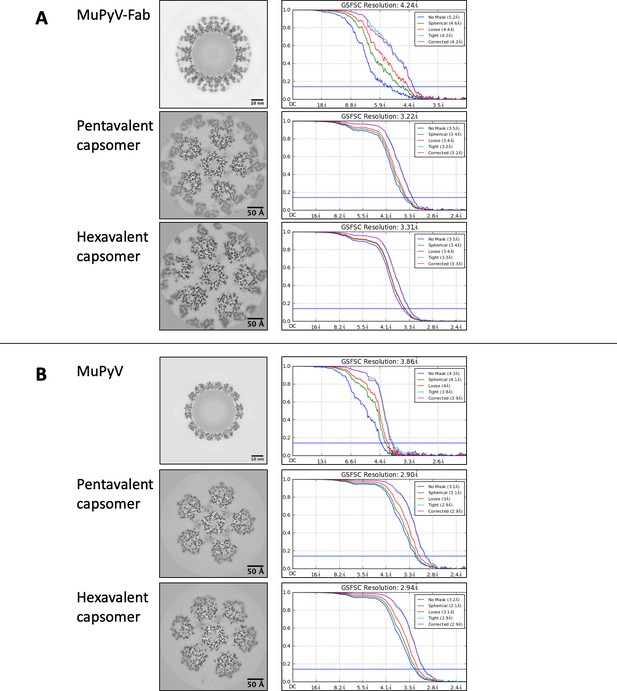
Fourier Shell Correlation (FSC).
Gold standard FSC curves were generated in cryoSPARC from half-maps, displayed as central sections with scale bars for the MuPyV-Fab complex (A, top), pentavalent capsomer complex (A, middle), and hexavalent capsomer complex (A, bottom). FSC curves were also generated for the MuPyV particle (B, top), pentavalent capsomer (B, middle) and hexavalent capsomer (B, bottom). For the sub-particle maps, FSCs were calculated after applying a soft tight mask to isolate the central capsomer-Fab complex or capsomer.

Asymmetric Unit.
(A) The surface rendered MuPyV capsid map overlaid with an asymmetric unit (ROYGBV) consisting of one VP1 chain from the pentavalent capsomer (red), together with five VP1 chains from a neighboring hexavalent capsomer (gray). (B) The MuPyV-Fab complex map with Fab and VP1 colored as in (A). Each asymmetric unit contains six nearly identical copies of the Fab epitope.

Refinement workflow.
(A) Icosahedral refinement produces a moderate resolution map of the full capsid and allows for designation of x,y,z coordinates of pentavalent (red) and hexavalent capsomers (blue). (B) Coordinates and orientations for all pentavalent and hexavalent capsomers are calculated, which enables sub-particle extraction and local refinement of capsomer centers and orientations, significantly improving map resolution compared to the initial icosahedral reconstruction.
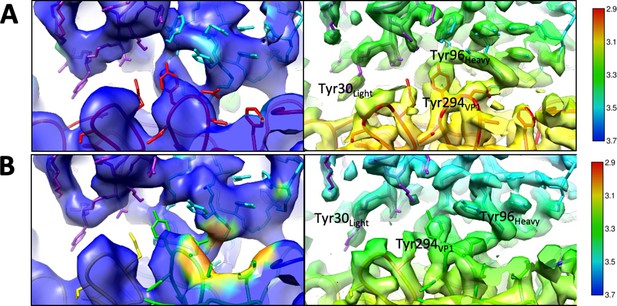
Local resolution.
(A and B) Equivalent zoomed views with density colored according to local resolution at the interface for the pentavalent (A) and hexavalent (B) capsomers show resolution improved after subvolume refinement (right) compared to the initial icosahedral refinement (left).
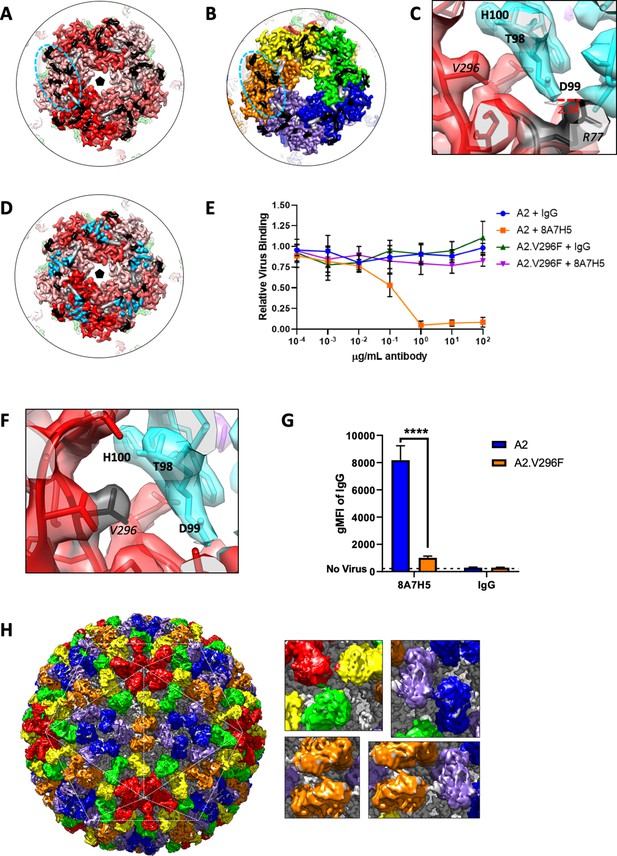
Cryo-EM reconstruction of MuPyV identifies mechanism of VP1 antibody escape by the V296F mutation.
(A and B) The Fab epitope bridges adjacent copies of VP1 on the pentavalent capsomer (A, shades of red) and hexavalent capsomer (B, OYGBV). Neighboring epitopes abut directly against each other. Contact residues from the main VP1 chain are noted in black, with minor contributions from the adjacent VP1 in gray. (C) A salt-bridge is formed between R77 and Fab heavy chain residue D99. This interaction is near key residue V296, despite the large distance in linear sequence. (D) The Fab epitope and receptor-binding residues (sky blue) overlap (PDB ID 5CPY) (Buch et al., 2015). (E) Increasing concentrations of 8A7H5 prevent the attachment of A2, but not A2.V296F. H2B-GFP labeled virus was incubated with antibody prior to incubation with NMuMG cells. GFP fluorescence was measured by flow cytometry. Data are from two independent experiments, n = 6. (F) The V296F mutation would place a bulky residue at the MuPyV-Fab interface, disrupting the Fab heavy chain residue T98, D99, and H100 (cyan) interactions. (G) V296F prevents the binding of 8A7H5 to VP1. NMuMG cells were incubated with A2 or A2.V296F followed by incubation with 8A7H5. Bound 8A7H5 was detected with an anti-IgG secondary. Data are from two independent experiments, n = 6 For 8A7H5 p<0.0001, IgG p=0.9693. (H) Six quasi-equivalent Fab molecules (red, orange, yellow, blue, green, purple) are contained within the asymmetric unit without clashes, despite the close proximity of Fab constant domains (inset). Data were analyzed by multiple t tests (G). ****p<0.0001.
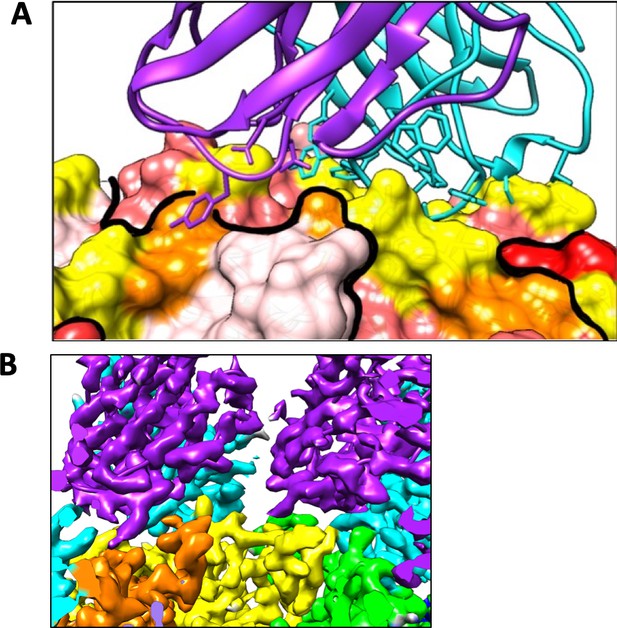
Heavy and light chain interactions with the capsid surface.
(A) The heavy chain (cyan) interacts with one copy of VP1 (yellow residues) with the light chain (purple) making additional contacts with the neighboring VP1 (orange residues). Symmetry-related VP1 molecules are shown in shades of red, divided by a thick black line. (B) Variable domains of adjacent bound Fab do not clash (heavy: cyan; light: purple). VP1 chains are colored as in Figure 4D,E.
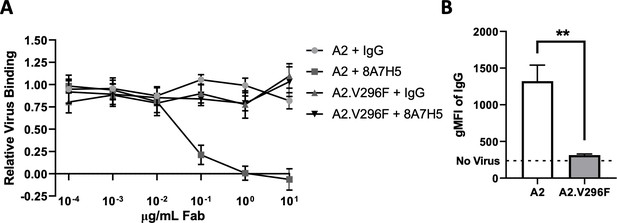
8A7H5 Fab blocks A2 attachment and fails to bind A2.V296F.
(A) Increasing concentrations of 8A7H5 Fab prevent the attachment of A2, but not A2.V296F, to NMuMG cells. Data from two independent experiments, n = 5–6. (B) Binding of 8A7H5 Fab to VP1 is prevented by V296F. NMuMG cells were incubated with A2 or A2.V296F followed by incubation with 8A7H5 Fab. Bound 8A7H5 was detected with an anti-IgG secondary. Data are from two independent experiments, n = 6 (p=0.0022). Data analyzed by Mann–Whitney U test (B). **p<0.01.
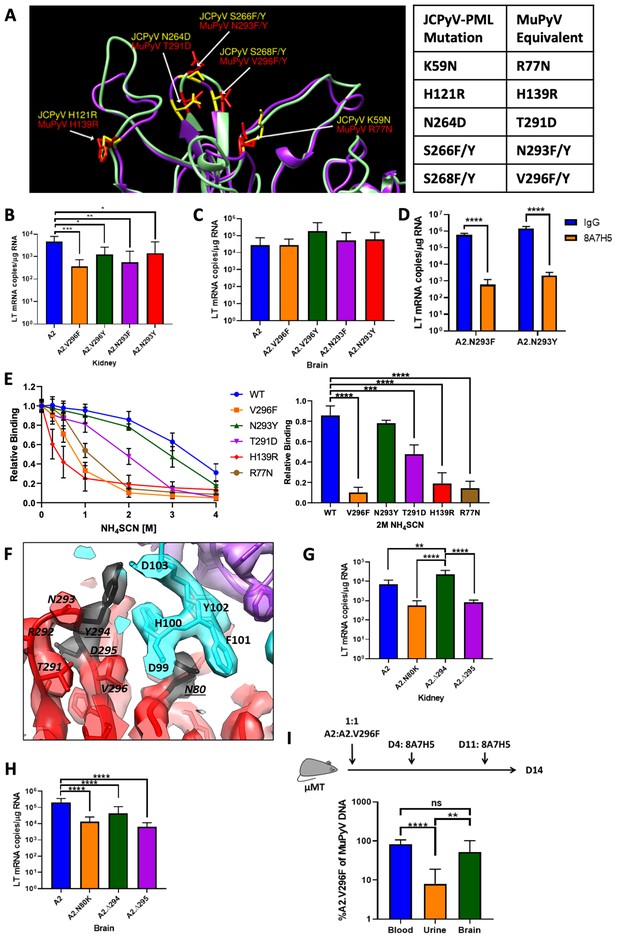
Additional JCPyV-PML mutations in MuPyV impair kidney, but not brain infection, and have varying effects on VP1 mAb neutralization.
(A) Structural comparison of PML mutation sites in JCPyV VP1 (PDB 3NXG) with MuPyV VP1 (PDB 5CPU) residues (Buch et al., 2015; Neu et al., 2010). (B) Kidney LT mRNA levels in mice 4 dpi with A2 or mutant viruses s.c. Data are from two independent experiments, n = 9–10 mice. For A2 vs. A2.V296F p=0.0008, A2 vs. A2.V296Y p=0.0100, A2 vs. A2.N293F p=0.0014, and A2 vs .A2.N293Y p=0.0168. (C) Brain LT mRNA levels in mice 4 dpi with A2 or mutant viruses i.c. Data are from four independent experiments, n = 13–14 mice. For A2 vs. A2.V296F p>0.9999, A2 vs. A2.V296Y p=0.2576, A2 vs. A2.N293F p=0.9974, and A2 vs. A2.N293Y p=0.9928. (D) LT mRNA levels in NMuMG cells 24 hpi with A2.N293F or A2.N293Y preincubated with 8A7H5 or control IgG. Data are from two independent experiments, n = 12. For A2.N293F p<0.0001 and A2.N293Y p<0.0001. (E) Analysis of 8A7H5 avidity for mutant VP1’s using NH4SCN. Left: Relative binding of 8A7H5 to recombinant VP1 with increasing concentrations of NH4SCN. Right: Relative binding of 8A7H5 to recombinant VP1 at 2M NH4SCN. Each point is the average of two technical replicates in an independent repeat, n = 3. For WT vs. V296F p<0.0001, WT vs. N293Y p=0.6440, WT vs. T291D p=0.0003, WT vs. H139R p<0.0001, and WT vs. R77N p<0.0001. (F) Location of novel changes in the VP1 escape mutations indicated by underlines. (G) LT mRNA levels 4 dpi in the kidneys of mice infected s.c. Data are from two independent experiments, n = 8 mice. For A2 vs. A2.N80K p=0.3374, A2 vs. A2.∆294 p=0.0010, A2 vs. A2.∆295 p=0.3724, A2.N80K vs. A2.∆294 p<0.0001, A2.N80K vs. A2.∆295 p=0.9999, and A2.∆294 vs. A2.∆295 p<0.0001. (H) LT mRNA levels 4 dpi in the brains of mice infected i.c. Data are from 2 to 4 independent experiments, n = 8–16 mice. For A2 vs. A2.N80K p<0.0001, A2 vs. A2.∆294 p<0.0001, A2 vs. A2.∆295 p<0.0001, A2.N80K vs. A2.∆294 p=0.8811, A2.N80K vs. A2.∆295 p=0.9979, and A2.∆294 vs. A2.∆295 p=0.6869. (I) Viral shedding in the urine is impaired by V296F, despite antibody escape in the blood. Top: Experimental design for infection of μMT mice and 8A7H5 treatment. Bottom: Frequency of A2.V296F DNA in blood, urine, and brain tissue of mice. Data are from three independent experiments, n = 13–14 mice. For Blood vs. Urine p<0.0001, Blood vs. Brain p=0.0500, and Urine vs. Brain p=0.0028. Data were analyzed by one-way ANOVA (B, C, E Right, G, H, I) or multiple t tests (D). In E, comparisons were only between WT and each mutant. **p<0.01, ***p<0.001, ****p<0.0001.
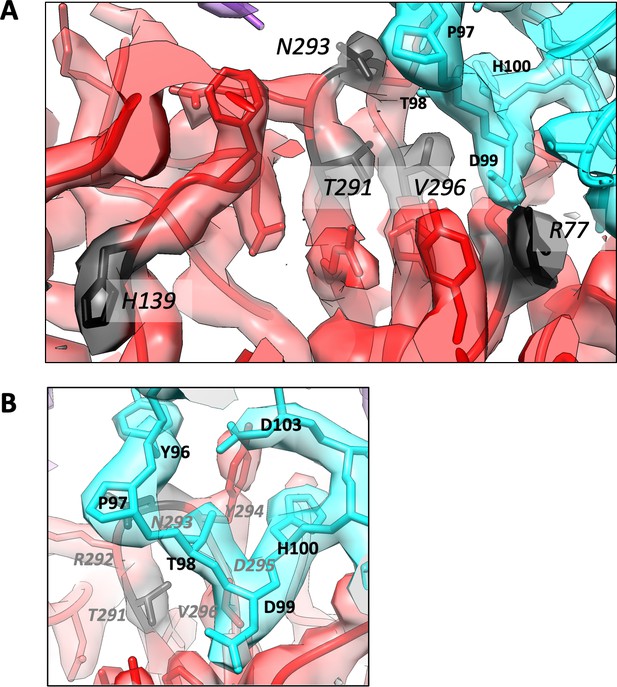
Relation of other VP1 mutations to virus–Fab interface.
(A) Cryo-EM density at the interface between the pentavalent capsomer (red) and Fab (cyan, purple). Location of the residues mutated to mimic those found in JCPyV-PML (H139, T291, N293, V296, R77) are noted in black. Residues T291, N293, V296 and R77 are located at the interface whereas H139 is a buried residue. (B) An alternate view of the interface shows the interaction between the HI loop and the Fab heavy chain CDR loop H3.
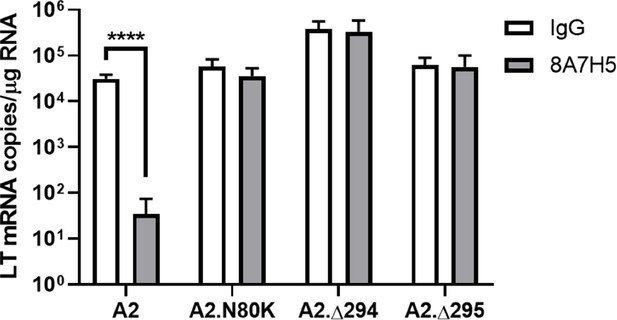
Resistance of escape mutations to 8A7H5 neutralization.
LT mRNA levels in NMuMG cells 24 hpi with A2, A2.N80K, A2.∆294, A2.∆295 preincubated with 8A7H5 or control IgG. Data are from two independent experiments, n = 6 (A2 p<0.0001, A2.N80K p=0.2374, A2.∆294 p=0.9403, A2.∆295 p=0.9403). Data were analyzed by multiple t tests. ****p<0.0001.
Tables
VP1 contact residues within −0.4 Å van der Waal’s overlap.
The conformational epitope spans three loops over two copies of VP1. Contributions from the adjacent VP1 are denoted with `.
| Loop | Residue | |
|---|---|---|
| BC | THR | 67 |
| GLU | 68 | |
| ARG | 77 | |
| GLY | 78 | |
| ASN | 80 | |
| THR | 83 | |
| GLU | 91 | |
| DE | PHE | 141’ |
| LYS | 151’ | |
| HI | ARG | 292’ |
| ASN | 293 | |
| TYR | 294 | |
| VAL | 296 | |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-VP1 (Rat Clone 8A7H5) | Swimm et al., 2010 | Clone 8A7H5 | See Materials and methods for concentrations |
| Antibody | ChromPure Rat IgG | Jackson ImmunoResearch | Cat#012-000-003 | See Materials and methods for concentrations |
| Antibody | Anti-CD8β (Rat monoclonal) | Pierres et al., 1982 | Clone H35-17.2 | 250 μg per injection |
| Antibody | Anti-VP1 (Rabbit polyclonal) | Provided by Robert Garcea (University of Colorado Boulder) | IF(1:1000) | |
| Antibody | Anti-Vimentin (Rat monoclonal) | R & D Systems | Cat#MAB2105 | IF(1:100) |
| Antibody | Anti-GFAP (Goat polyclonal) | Abcam | Cat#ab53554 | IF(1:1000) |
| Antibody | Anti-Iba1 (Rabbit polyclonal) | FUJIFILM Wako | Cat#019–19741 | IF(1:500) |
| Antibody | Anti-CD3 (Rabbit monoclonal) | Abcam | Cat#ab16669 | IF(1:100) |
| Antibody | Anti-Goat IgG AF488 (Bovine polyclonal) | Jackson ImmunoResearch | Cat#805-545-180 | IF(1:500) |
| Antibody | Anti-Rat IgG AF568 (Donkey polyclonal) | Abcam | Cat#ab175475 | IF(1:500) |
| Antibody | Anti-Rabbit IgG AF647 (Donkey polyclonal) | Jackson ImmunoResearch | Cat#711-605-152 | IF(1:500) |
| Antibody | Anti-CD8α-AF700 (Rat monoclonal) | Biolegend | Cat#100730 | FC(1:200) |
| Antibody | Anti-CD44-FITC (Rat monoclonal) | Biolegend | Cat#103006 | FC(1:200) |
| Antibody | Anti-Rat IgG-APC (Goat polyclonal) | BD | Cat#551019 | FC(1:200) |
| Antibody | Anti-Mouse IgG-HRP (Goat polyclonal) | Biolegend | Cat#405306 | ELISA(1:2800) |
| Antibody | Anti-Mouse IgG-HRP (Goat polyclonal) | Bethyl Laboratories INC | Cat#A90-116P | ELISA(1:7000) |
| Antibody | Biotinylated Anti-Rabbit (Goat Polyclonal) | Vector Laboratories | Cat#BA-1000 | IHC(1:500) |
| Other | Mouse Polyomavirus (Strain A2) | N/A | N/A | |
| Strain, strain background (Escherichia coli) | BL21 | Agilent | Cat#200133 | |
| Recombinant DNA reagent | PyVP1-pGEX-4T-2 (plasmid) | Provided by Robert Garcea | N/A | |
| Recombinant DNA reagent | H2B-GFP (plasmid) | Kanda et al., 1998, Addgene | Plasmid #11680 | |
| Peptide, recombinant protein | Benzonase Nuclease | Sigma | Cat#E1014 | Virus Purification (1:3333) |
| Other | Db-LT359 Tetramer | NIH Tetramer Core | N/A | FC(1:400) |
| Peptide, recombinant protein | Neuraminidase from Vibrio cholerae (Type II) | Sigma | Cat#N6514 | Virus Purification (1:2000) |
| Peptide, recombinant protein | RevertAid H Minus Reverse Transcriptase | ThermoFisher | Cat#EP0451 | |
| Chemical compound, drug | OptiPrep | STEMCELL Technologies | Cat#07820 | |
| Chemical compound, drug | Glutathione Sepharose 4B | GE Healthcare | Cat#17075601 | |
| Chemical compound, drug | TRIzol Reagant | ThermoFisher | Ref#15596018 | |
| Chemical compound, drug | Lipofectamine 2000 Transfection Reagent | ThermoFisher | Cat#11668030 | |
| Commercial assay, kit | TBP PrimeTime XL qPCR Assay | IDT | Mm.PT.39a.22214839 | |
| Other | ProLong Gold antifade reagent with DAPI | ThermoFisher | Ref#P36931 | |
| Other | Fixable Viability Dye eFluor780 | ThermoFisher | Cat# 65-0865-14 | FC(1:1000) |
| Commercial assay, kit | Avidin/Biotin Blocking Kit | Vector Laboratories | Cat#SP-2001 | |
| Commercial assay, kit | VECTASTAIN Elite ABC-HRP Kit | Vector Laboratories | Cat#PK-6100 | |
| Commercial assay, kit | NovaRED Substrate Kit | Vector Laboratories | Cat#SK-4800 | |
| Commercial assay, kit | 1-Step Ultra TMB-ELISA | ThermoFisher | Ref#34028 | |
| Commercial assay, kit | PerfectCTa SYBR Green FastMix | Quantabio | P/N 84069 | |
| Commercial assay, kit | PerfectCTa FastMix II ROX | Quantabio | P/N 84210 | |
| Commercial assay, kit | PureLink Viral RNA/DNA mini Kit | ThermoFisher | Ref#12280–050 | |
| Commercial assay, kit | Pierce Fab Micro Preparation Kit | ThermoFisher | Ref#44685 | |
| Commercial assay, kit | Nab Protein G Spin Columns | ThermoFisher | Ref#89953 | |
| Commercial assay, kit | Wizard Genomic DNA Purification Kit | Promega | Ref#A1120 | |
| Commercial assay, kit | QuikChange II Site-Directed Mutagenesis Kit | Agilent | Cat#200523 | |
| Commercial assay, kit | QIAquick PCR Purification Kit | Qiagen | Cat#28104 | |
| Cell Line (Mus musculus) | BALB/3T3 Clone A31 | ATCC | CCL-163, RRID:CVCL_0184 | |
| Cell Line (M. musculus) | NMuMG | ATCC | CRL-1636, RRID:CVCL_0075 | |
| Cell Line (M. musculus) | mIMCD-3 | ATCC | CRL-2123, RRID:CVCL_0429 | |
| Cell Line (M. musculus) | C57BL/6 MEF | This paper | Primary murine embryonic fibroblasts | |
| Strain, strain background (M. musculus) | C57BL/6 | National Cancer Institute | Cat#OIC55 | |
| Genetic reagent (M. musculus) | Stat1–/– | Jackson Laboratory | Cat#012606 | |
| Genetic reagent (M. musculus) | μMT | Jackson Laboratory | Cat#002288 | |
| Software, algorithm | Prism | Graphpad | RRID:SCR_002798 | |
| Software, algorithm | FlowJo | BD | RRID:SCR_008520 | |
| Software, algorithm | ImageJ | NIH | RRID:SCR_003070 | |
| Software, algorithm | Leica LAS X | Leica | RRID:SCR_013673 | |
| Software, algorithm | Photoshop | Adobe | RRID:SCR_014199 | |
| Software, algorithm | Relion | Scheres et al., 2009 | RRID:SCR_016274 | |
| Software, algorithm | cryoSPARC | Structura Biotechnology | RRID:SCR_016501 | |
| Software, algorithm | ISECC | See Data and code availability | v 2019.09 | |
| Software, algorithm | PHENIX | phenix-online.org | RRID:SCR_014224 | |
| Software, algorithm | Coot | Emsley et al., 2010 | RRID:SCR_014222 |
Additional files
-
Supplementary file 1
Collection statistics.
Independent datasets were collected for MuPyV and the MuPyV-Fab complex.
- https://cdn.elifesciences.org/articles/61056/elife-61056-supp1-v2.docx
-
Supplementary file 2
Refinement statistics.
Local refinement allowed models to be built into the higher resolution capsomer maps.
- https://cdn.elifesciences.org/articles/61056/elife-61056-supp2-v2.docx
-
Supplementary file 3
Fab contact residues.
The majority of Fab contacts are through the heavy chain, with minor contributions from the light chain.
- https://cdn.elifesciences.org/articles/61056/elife-61056-supp3-v2.docx
-
Supplementary file 4
Statistics of existing and new cryo-EM polyomavirus maps.
Capsomer-based local refinement allowed rapid refinement of polyomavirus to high resolution, using only a modest particle number.
- https://cdn.elifesciences.org/articles/61056/elife-61056-supp4-v2.docx
-
Supplementary file 5
ISECC source code.
Archive of the ISECC source code used in sub-particle generation. The most current version of the software is maintained at https://github.com/goetschius/isecc.
- https://cdn.elifesciences.org/articles/61056/elife-61056-supp5-v2.zip
-
Supplementary file 6
Oligonucleotide sequences.
Sequences of oligonucleotides used for site-directed mutagenesis, qPCR, sequencing.
- https://cdn.elifesciences.org/articles/61056/elife-61056-supp6-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61056/elife-61056-transrepform-v2.docx





