The Nesprin-1/-2 ortholog ANC-1 regulates organelle positioning in C. elegans independently from its KASH or actin-binding domains
Figures
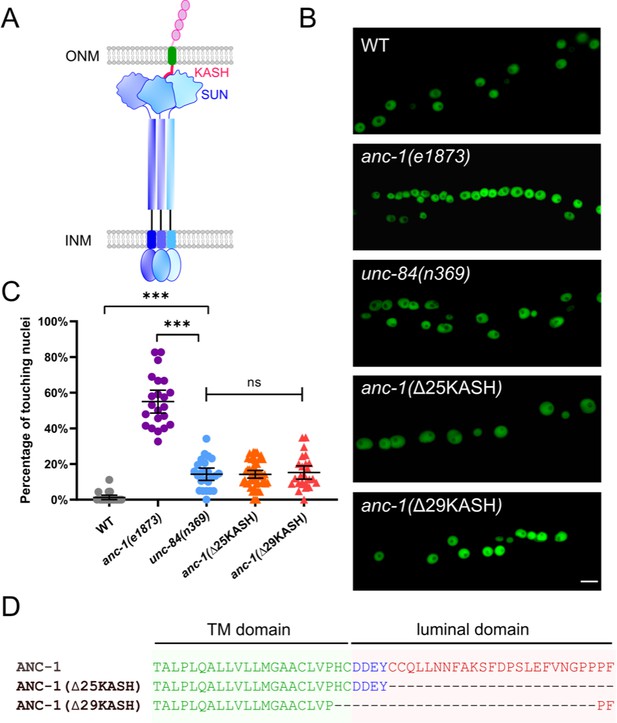
ANC-1 has a linker of nucleoskeleton and cytoskeleton (LINC) complex-independent role in anchoring nuclei.
(A) Model of the LINC complex. Trimers of the SUN protein UNC-84 (purple and blue) and the KASH protein ANC-1 (red, green, and pink, only one of the trimers is shown) form the LINC complex, which spans the outer nuclear membrane (ONM) and inner nuclear membrane (INM). (B) Lateral views of adult C. elegans expressing hypodermal nuclear GFP in wild type (WT) or indicated mutants. Scale bar, 10 µm. (C) Quantification of nuclear anchorage defects. Each point represents the percentage of touching nuclei on one side of a young adult animal. Means with 95% CI error bars are shown. ANOVA and Tukey’s multiple comparisons tests were used for statistical analysis; ns means not significant, p>0.05; ***p≤0.001. n ≥ 20 for each strain. (D) Sequences of the transmembrane (TM) domain and the luminal domain of ANC-1 showing the deletions analyzed.
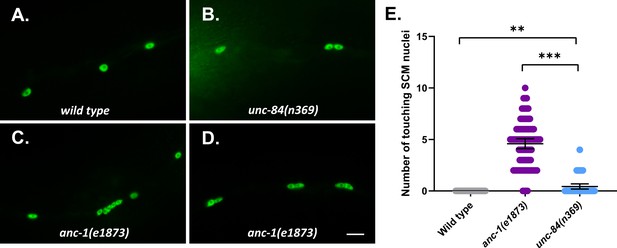
Nuclear anchorage defects in seam cell syncytia.
(A–D) Lateral views of C. elegans wild type (WT) (A), unc-84(n369) (B), and anc-1(e1873) (C–D) mutants expressing wIs54[scm::gfp]. (E) Quantification of the number of touching seam cell nuclei. Each point represents the number of touching nuclei in the seam cell of a young adult animal. Means with 95% CI error bars are shown. Unpaired student two-tail t-test was used for statistical analysis. **p≤0.005. ***p≤0.001. n ≥ 42 for each strain. Scale bar, 10 µm.
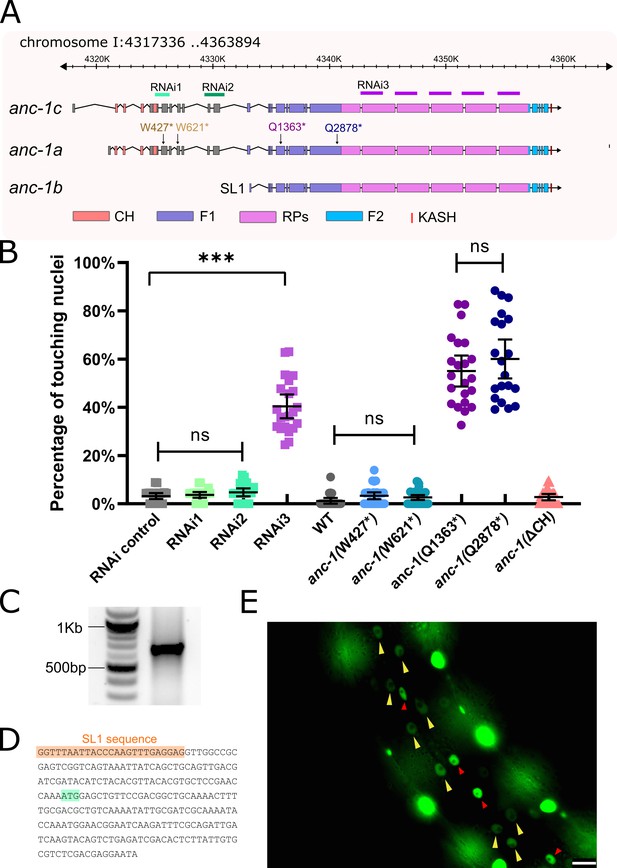
anc-1b is the major isoform in hyp7 nuclear anchorage.
(A) Schematic gene structure of anc-1a, b, and c isoforms (modified from the J-Brower in Wormbase). Domains are color-coded. CH = calponin homology; F1 = fragment one from the ATG of ANC-1b to the beginning of the repeats; RPs = the six exact repeats of about 900 residues each; F2 = fragment two from the end of the repeats to the transmembrane (TM) span; WT = wild type; and KASH = Klarsicht, ANC-1, Syne homology, in this case referring to the residues in the lumen of the nuclear envelope. The target regions of RNAi constructs are labeled. Premature stop mutations are indicated using the numbering the anc-1a isoform. (B) Quantification of nuclear anchorage defects in anc-1 mutant and RNAi animals. Means with 95% CI are shown in the graph. ANOVA and Tukey’s multiple comparisons tests were used for statistical analysis. ns, not significant, p>0.05; ***p≤0.001. n ≥ 20 for each strain. (C) An agarose gel showing the 5’-RACE products on the right lane. (D) Partial sequence of the 5’-RACE product. An SL1 sequence (orange) adjacent to the 5’ end of the anc-1b transcript was identified. The predicted start codon is in light green. (E) Lateral view of a worm showing the expression of nls::GFP driven by anc-1b promoter. Yellow arrows mark hyp7 nuclei; the red arrows mark seam cell nuclei; and the bright, unmarked nuclei are in muscle cells. Scale bar, 10 µm.
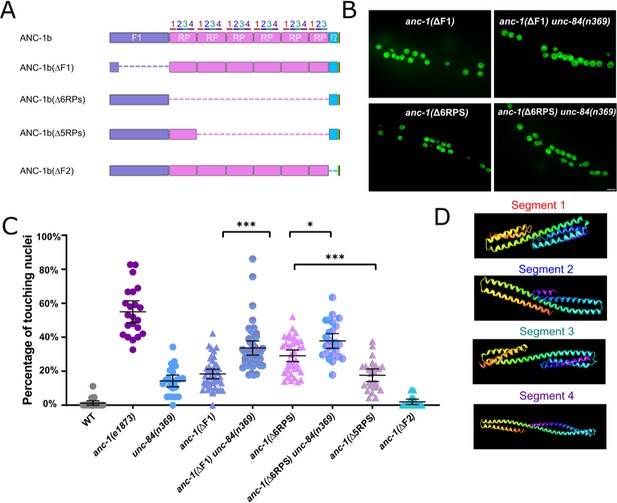
Cytoplasmic domain deletion analysis of ANC-1b.
(A) Schematics of the ANC-1b cytoplasmic domain deletions. (B) Lateral views are shown of young adult C. elegans expressing hypodermal nuclear GFP in the indicated genotypes. Scale bar, 10 µm. (C) Quantification of nuclear anchorage in the wild type (WT) as well as anc-1b domain deletion mutants. Each point represents the percentage of touching nuclei on one side of a young adult animal. Means with 95% CI error bars are shown. ANOVA and Tukey’s multiple comparisons tests were used for statistical analysis. *p≤0.05; **p≤0.01; ***p≤0.001. n ≥ 20 for each strain. The data in the first three columns, WT, anc-1(e1873), and unc-84(n369) are duplicated from Figure 1C and copied here for easy reference. (D) QUARK result for three fragments in the tandem repeats (RPs). The positions of the fragments are indicated in 3A.
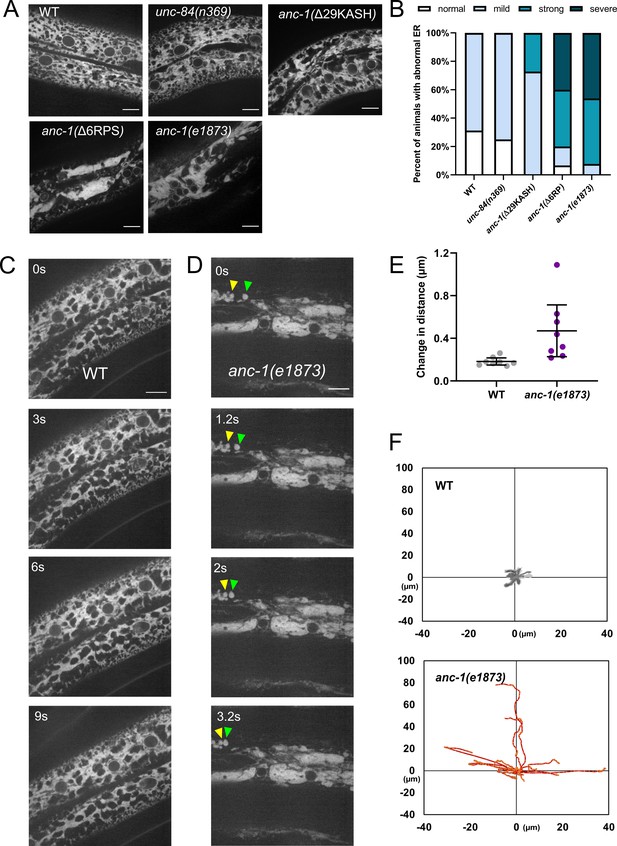
The ER is mispositioned in anc-1 mutants.
(A) Representative images of the hyp7 ER labeled with the GFP::KDEL marker in the young adult animals. (B) Scoring of the ER positioning defects. ER images of the listed strains were mixed and randomized for blind analysis by multiple researchers. n ≥ 11 for each strain. (C–D) Time-lapse images of hyp7 GFP::KDEL marker (C) over 9 s in wild type (WT) or (D) over 3.2 s in anc-1 null. Arrowheads show two fragments of ER that changed their relative distance from one another over a short period of time. (E–F) To quantify ER displacement, three spots on each WT and anc-1(e1873) movie were tracked. The average change in distance between two points in 200 ms intervals is plotted in (E). The trajectories of the relative movements of each spot apart from the others are shown in (F). Eight movies of each strain were analyzed. Scale bar, 10 µm for all the images.
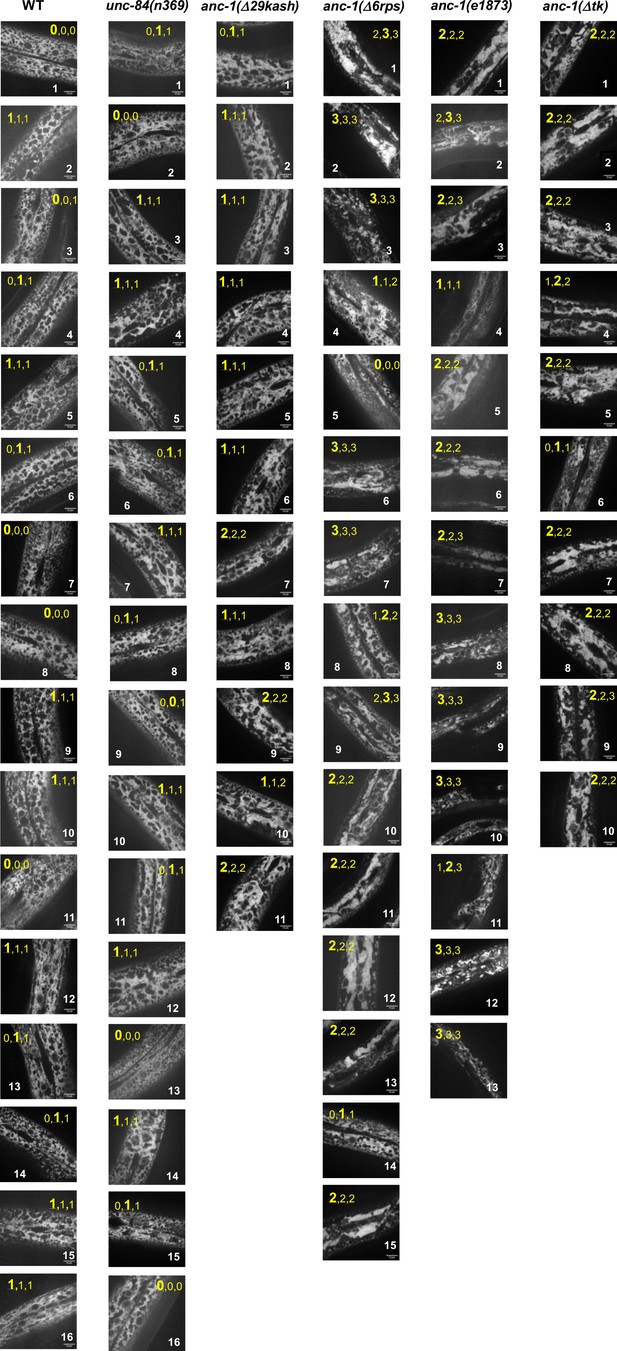
Raw data for ER morphology assays.
The ER in hyp7 was labeled with a GFP::KDEL marker. Young adult animals are shown. Each column shows all the images scored from a single genetic strain (labeled on the top). The three numbers in yellow at the top of each image are the scores of three independent investigators giving them bind scores. 0, 1, 2, and 3 represent normal, mild, strong, and severe ER anchorage defects, respectively. The score given by more than two people was chosen as the final score and is in a larger font. Scale bars, 10 µm. All the images are at the same scale. WT = wild type.
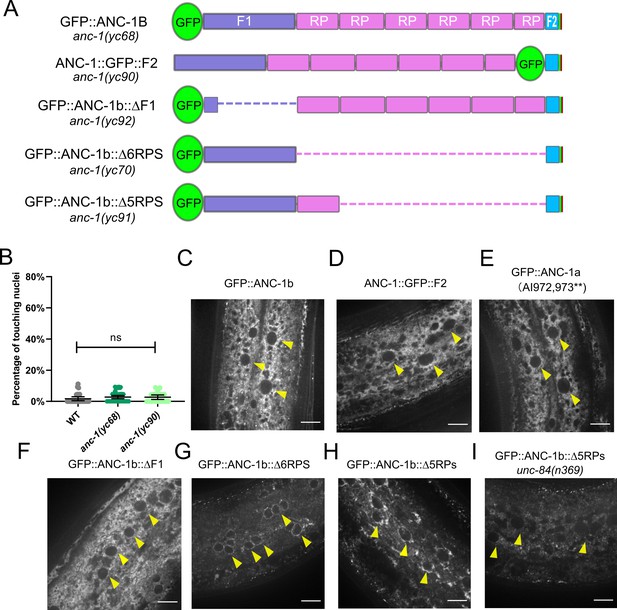
The subcellular localization of ANC-1.
(A) Schematic depicting the ANC-1 GFP knock-in constructs with or without deletion of ANC-1 cytoplasmic domains. (B) Nuclear positioning in GFP::ANC-1b is wild type (WT). Each point represents the percentage of touching nuclei on one side of a young adult animal. Means with 95% CI error bars are shown. ns, not significant (p>0.05). n ≥ 20. (C–I) Confocal images of hyp7 subcellular localization in the indicated strains. Yellow arrowheads point to nuclei. Scale bar, 10 µm.
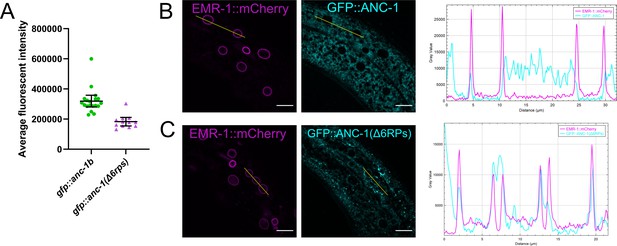
GFP::ANC-1(Δ6RPS) is expressed at a lower level and is enriched at the nuclear envelope.
(A) Average fluorescent intensity of GFP::ANC-1b and GFP::ANC-1(Δ6RPS) in the hypodermal tissue. Images of young adults were taken under the same conditions to calculate the average fluorescent intensity. (B–C) EMR-1::mCherry (magenta) and GFP::ANC-1b or GFP::ANC-1(Δ6RPS) (cyan) from the same animal are shown. Line scans are shown on the far right. Scale bar, 10 µm.
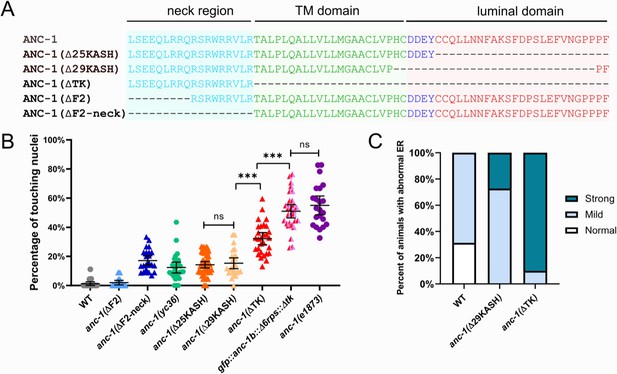
The transmembrane domain of ANC-1 is necessary for nuclear and ER positioning.
(A) Sequence of deletion in the neck region (blue), transmembrane (TM) span (green), or the luminal domain (red). (B) Quantification of nuclear anchorage defects in anc-1 mutants. Each point represents the percentage of touching nuclei on one side of a young adult animal. Means with 95% CI error bars are shown. n ≥ 20. ANOVA and Tukey’s multiple comparisons tests were used in comparisons. ***p≤0.001; ns means p>0.05. The data of wild type (WT), anc-1(ΔF2), anc-1(Δ25KASH), anc-1(Δ29KASH), and anc-1(e1873) are duplicated from Figures 1C and 3C and copied here for easy reference. (C) Qualitative analysis of the ER anchorage defects as in Figure 4. Significantly more anc-1(ΔTK) animals show strong ER anchorage defects than anc-1(Δ29KASH) mutants (p≤0.01 by Fisher’s exact test). Sample sizes were all >10.
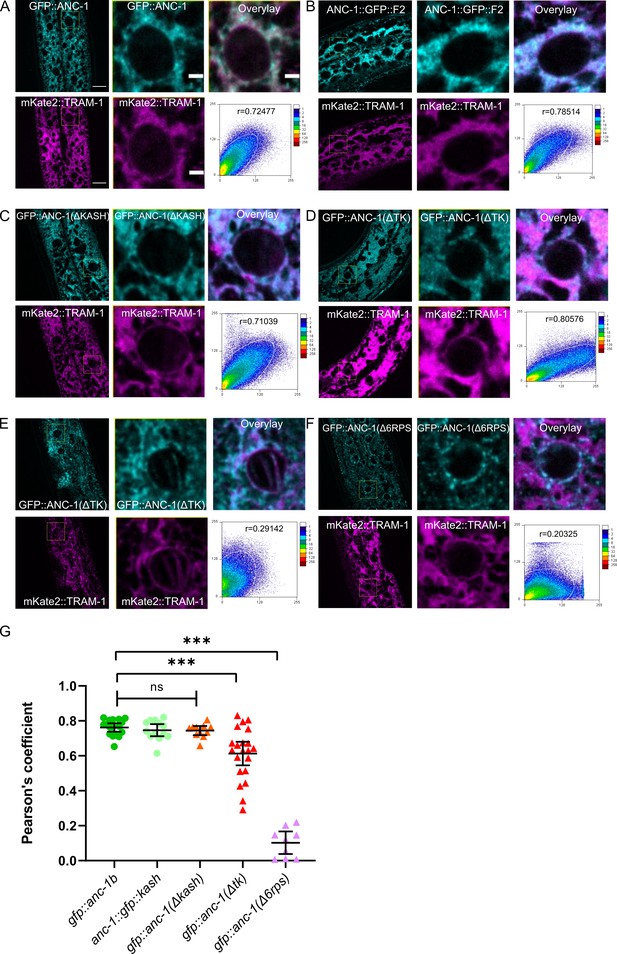
ANC-1 localizes to the ER.
ANC-1-GFP fusion protein (cyan) localization with respect to the ER membrane as marked by a mKate::TRAM-1 (magenta) is shown. (A) Wild type (WT) GFP::ANC-1b. (B) Wild type ANC-1::GFP::F2. (C) GFP::ANC-1b::ΔKASH. (D) An example of GFP::ANC-1b::ΔTK with good overlap with the ER and (E) a GFP::ANC-1b::ΔTK with poor overlap. (F) GFP::ANC-1b::Δ6RPS. (A–F) For each section, the left two panels are low magnification of the young adult hypodermis (scale bar, 10 µm) and the middle two panels are a zoom in of the boxed part of the left panels (scale bar, 2 µm). The top right shows the merge of the two channels. The bottom right uses the used ImageJ plug-in ScatterJ to quantify the co-localization and the Pearson's correlation coefficient for overlap is shown as an r value. (G) A scatter plot of Pearson’s coefficients showing overlap between the indicated GFP and the mKate::TRAM-1 ER membrane marker. Mean ±95% CI are shown. ANOVA and Tukey’s multiple comparisons tests were used in comparisons. ***p≤0.001; ns means not significant.
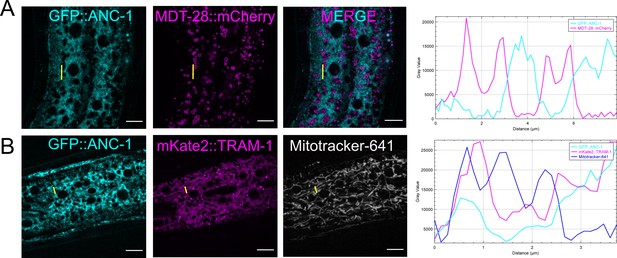
GFP::ANC-1b does not co-localize with lipid droplets or mitochondria.
(A) GFP::ANC-1b (cyan) and MDT-28::mCherry (magenta) are merged on the tight. (B) GFP::ANC-1b (cyan), mKate2::TRAM-1 (cyan), and mitotacker-641 (gray) are shown from the same animal. (A–B) Line scans are shown on the far right. Scale bar, 10 µm.
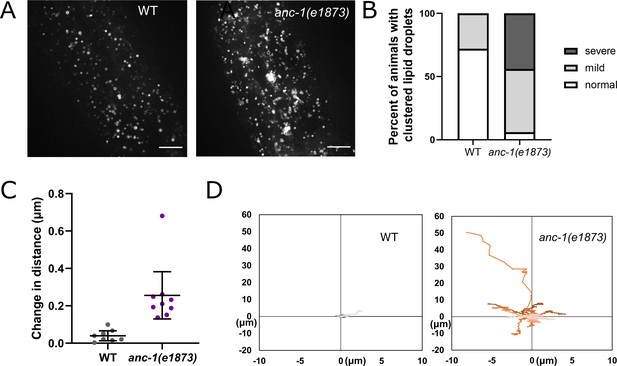
Lipid droplets are mispositioned in anc-1 mutants.
(A) Representative images of hyp7 lipid droplets labeled with the Pmdt-28::mdt-28::mCherry marker in the young adult animals. (B) Scoring of the lipid droplet positioning defects. Images were randomized for blind analysis by multiple researchers. n ≥ 11. (C–D) Time-lapse images of hyp7 lipid droplets. (C–D) The lipid droplet displacement phenotype was quantified as described in Figure 4E–F. The average change in distance between two points in 242 ms intervals is plotted in (C). The trajectories of the relative movements of each spot apart from the others are shown in (D). Eight movies of each strain were analyzed. Scale bar, 10 µm for all the images. Also see Videos 4–5. WT = wild type.
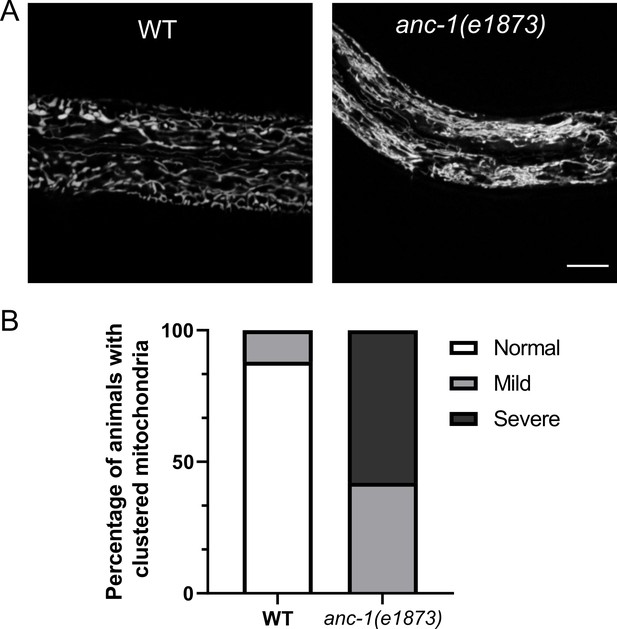
Mitochondria are mispositioned in anc-1 mutants.
(A) The hyp7 mitochondria were labeled with the Pcol-10::mito::GFP marker in the L3 animals. (B) The mitochondria positioning defect was scored. Images were randomized for blind analysis by multiple researchers. n ≥ 18 for each strain. Scale bar, 10 µm for all the images. Also see Videos 6–7. WT = wild type.
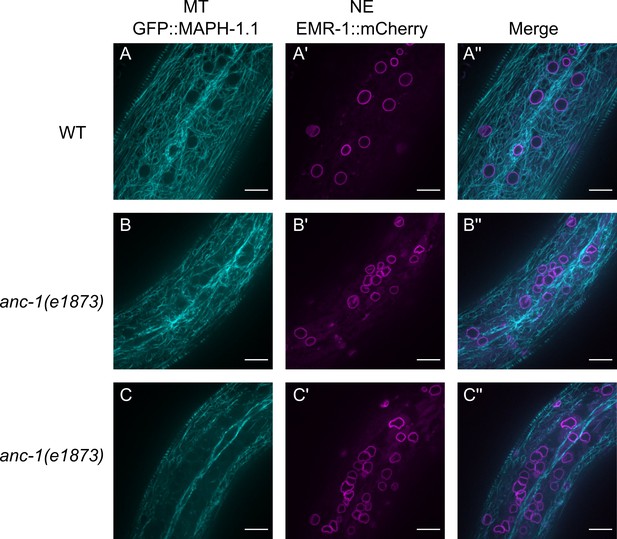
Microtubule organization in anc-1 mutants.
Representative images of microtubules (cyan) and nuclear envelope (NE) (magenta) from the same young adult animals are shown. (A–A’’) wild type (WT), (B–B’’) anc-1(e1873) mutant animal where the microtubules are mostly normal. (C–C’’) In other anc-1(e1873) animals, microtubule organization is disrupted in the channels where nuclei move. Microtubules are labeled with GFP::MAPH-1.1 and the nuclear envelope is labeled with EMR-1::mCherry. Scale bar, 10 µm. Also see Videos 8–10.
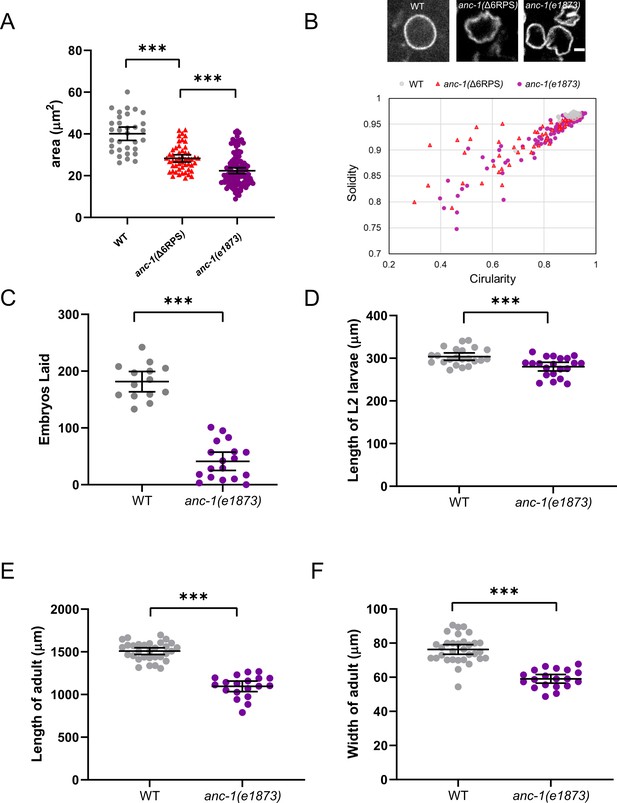
anc-1 mutants have developmental defects.
(A) The area of cross-sections of hyp7 nuclei is shown. Each dot represents the area of a single nucleus. n = 32 for wild type (WT), n = 50 for anc-1(Δ6RPS), n = 111 for anc-1(e1873). (B) Top panel: Representative images of hyp7 nuclei marked by EMR-1::mCherry of WT, anc-1(Δ6RPS), and anc-1(e1873) mutants. Scale bar, 2 µm. Bottom panel: Plot of the solidity and the circularity. (C–F) The brood size (C), length of the L2 larvae (D), adult length (E) and width (F) are significantly reduced in anc-1(e1873) mutants. Each dot represents a single animal. n ≥ 14 for (C) and n ≥ 19 for (D–F). Means with 95% CI error bars are shown. Unpaired student two-tail t-test was used for statistical analysis. **p≤0.01; ***p≤0.001.
Videos
The ER is anchored in wild type (WT) hyp7.
An example video of the hyp7 ER in young adult wild type C. elegans expressing pwSi83[phyp7gfp::kdel]. Images were captured at the interval of 0.2 s for 10 s. Scale bar, 10 µm.
The ER is unanchored in anc-1(e1873) mutant hyp7.
An example video of the hyp7 ER in the young adult anc-1(e1873) mutant C. elegans expressing pwSi83[phyp7gfp::kdel]. Images were captured at the interval of 0.2 s for 10 s. Scale bar, 10 µm.
ER positioning in unc-84(null) mutant hyp7.
An example of one of the most severe ER positioning defects observed is shown in a video of the hyp7 ER in the young adult unc-84(n369) mutant C. elegans expressing pwSi83[phyp7gfp::kdel]. Images were captured at the interval of 0.25 s for 4 s. Scale bar, 10 µm.
Lipid droplets are anchored in wild type (WT) hyp7.
An example video of the hyp7 lipid droplets in young adult wild type C. elegans expressing ldrIs2 [mdt-28p::mdt-28::mCherry +unc-76(+)]. Images were captured at the interval of 0.2 s for 10 s. Scale bar, 10 µm.
Lipid droplets are unanchored in anc-1(e1873) mutant hyp7.
An example video of the hyp7 lipid droplets in the young adult anc-1(e1873) mutant C. elegans expressing ldrIs2 [mdt-28p::mdt-28::mCherry +unc-76(+)]. Images were captured at the interval of 0.24 s for 10 s. Scale bar, 10 µm.
Mitochondria are anchored in wild type (WT) hyp7.
An example video of hyp7 mitochondria in young adult wild type C. elegans as followed by the Pcol-10::mitoLS::GFP marker. Images were captured at the interval of 0.1 s for 10 s. Scale bar, 10 µm.
Mitochondria are unanchored in anc-1(e1873) mutant hyp7.
An example video of hyp7 mitochondria in the young adult anc-1(e1873) mutant C. elegans as followed by the Pcol-10::mitoLS::GFP marker. Images were captured at the interval of 0.1 s for 10 s. Scale bar, 10 µm.
Microtubules in wild type (WT) hypodermal syncytia.
A representative clip of the hyp7 in a wild type young adult expressing GFP:: MAPH-1.1 to mark microtubules in cyan and EMR-1::mCherry to mark nuclear envelopes in magenta. Images were at the interval of 1 s for 20 s. Scale bar, 10 µm.
Microtubules in anc-1(null) hypodermal syncytia.
A representative clip of the hyp7 in a anc-1(e1783) mutant young adult expressing GFP:: MAPH-1.1 to mark microtubules in cyan and EMR-1::mCherry to mark nuclear envelopes in magenta. Images were at the interval of 0.845 s for 40 s. Scale bar, 10 µm.
Microtubules in anc-1(null) hypodermal syncytia.
A representative clip of the hyp7 in a anc-1(e1783) mutant young adult expressing GFP::MAPH-1.1 to mark microtubules in cyan and EMR-1::mCherry to mark nuclear envelopes in magenta. Images were at the interval of 0.638 s for 32 s. Scale bar, 10 µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia. coli) | OP50 | Caenorhabditis Genetics Center (CGC) | OP50 | https://cgc.umn.edu/strain/OP50 |
| Strain, strain background (E. coli) | DH10B | New England Biolabs (NEB) | C3019H | |
| Recombinant DNA reagent | pLF3FShC | Nonet, 2020 | addgene: #153083 | https://www.addgene.org/153083/ |
| Recombinant DNA reagent | pSL845 | This paper | Py37a1b.5::mKate2::tram-1::tram-1 3’UTR plasmid to generate the ycSi2 transgenic C. elegans strain | |
| Recombinant DNA reagent | pSL835 | This paper | Panc-1b::nls::gfp::lacZ Plasmid to generate ycEx260 | |
| Recombinant DNA reagent | pSL289 | This paper | Pcol-10::mitoLS::gfp Plasmid to generate ycEx217 |
C. elegans strains in this study.
| Strain | Genotype | Reference |
|---|---|---|
| N2 | Wild type | |
| UD522 | ycEx249[pcol-19::gfp::lacZ, pmyo-2::mCherry] | Cain et al., 2018 |
| UD532 | unc-84(n369) X; ycEx249 | Cain et al., 2018 |
| UD538 | anc-1(e1873) I; ycEx249 | Cain et al., 2018 |
| UD578 | anc-1(yc52[∆25KASH]) I; ycEx249 | This study |
| UD615 | anc-1(yc69[∆29KASH]) I; ycEx249 | This study |
| JR672 | wIs54[scm::gfp] V | Terns et al., 1997 |
| UD457 | unc-84(n369) X; wIs54 V | This study |
| UD451 | anc-1(e1873) I; wIs54 V | This study |
| VC20178 | anc-1(gk109010[W427*]) I | Thompson et al., 2013 |
| UD737 | anc-1(gk109010[W427*]) I; ycEx265[pcol-19::gfp::lacZ, pmyo-2::mCherry] | This study |
| VC40007 | anc-1(gk109018[W621*]) I | Thompson et al., 2013 |
| UD736 | anc-1(gk109018[W621*]) I; ycEx266[pcol-19::gfp::lacZ, pmyo-2::mCherry] | This study |
| VC40614 | anc-1(gk722608[Q2878*]) I | Thompson et al., 2013 |
| UD565 | anc-1(gk722608[Q2878*]) I; ycEx249 | This study |
| UD535 | anc-1(yc41[anc-1::gfp3Xflag::kash,Δch]) I; ycEx249 | This study |
| UD608 | ycEx260[panc-1b::nls::gfp::lacZ] | This study |
| UD599 | anc-1(yc62[ΔF1]) I; ycEx249 | This study |
| UD591 | anc-1(yc61[Δ6RPS]) I; ycEx249 | This study |
| UD668 | anc-1(yc80[ΔF2]) I; ycEx249 | This study |
| UD669 | anc-1(yc81[ΔF2-neck]) I; ycEx249 | This study |
| UD695 | anc-1(yc61[Δ6RPS]) I; unc-84(n369) X; ycEx249 | This study |
| UD696 | anc-1(yc62[ΔF1]) I; unc-84(n369) X; ycEx249 | This study |
| UD612 | anc-1(yc68[gfp::anc-1b]) I | This study |
| UD655 | anc-1(yc78[Δ5RPS]) I; ycEx249 | This study |
| UD619 | anc-1(yc68[gfp::anc-1b]) I; ycEx249 | This study |
| UD694 | anc-1(yc90[anc-1::gfp::F2]) I | This study |
| UD697 | anc-1(yc90[anc-1::gfp::F2]) I; ycEx249 | This study |
| UD618 | anc-1(yc70[gfp::anc-1b::Δ6rps]) I | This study |
| UD698 | anc-1(yc91[gfp::anc-1b::Δ5rps]) I | This study |
| UD701 | anc-1(yc92[gfp::anc-1b::Δf1]) I | This study |
| UD702 | anc-1(yc93[anc-1AI972,973**::gfp]) I | This study |
| UD625 | anc-1(yc71[ΔTK]) I; ycEx249 | This study |
| UD645 | anc-1(yc73[gfp::anc-1b::Δ6rps::Δtk]) I; ycEx249 | This study |
| RT3739 | pwSi83[phyp7gfp::kdel] | A gift from Barth Grant |
| UD652 | anc-1(e1873) I; pwSi83 | This study |
| UD679 | unc-84(n369) X; pwSi83 | This study |
| UD707 | anc-1(yc71[∆TK]) I; pwSi83 | This study |
| UD651 | anc-1(yc61[∆6RPS]) I; pwSi83 | This study |
| UD672 | anc-1(yc69[∆29KASH]) I; pwSi83 | This study |
| UD520 | anc-1(yc36[anc-1::gfp3Xflag::kash]) I | This study |
| BN147 | bqSi142 [pemr-1::emr-1::mCherry + unc-119(+)] II | Morales-Martínez et al., 2015 |
| BOX188 | maph-1.1(mib12[gfp::maph-1.1]) I | Waaijers et al., 2016 |
| UD649 | maph-1.1(mib12[gfp::maph-1.1]) I; bqSi142 II | This study |
| UD650 | anc-1(e1873) I; maph-1.1(mib12[gfp::maph-1.1]) I; bqSi142 II | This study |
| UD677 | anc-1(yc61[∆6RPS]) I; maph-1.1(mib12[gfp::maph-1.1]) I; bqSi142 II | This study |
| UD3 | ycEx217[Pcol-10::mitoLS::GFP, Podr-1::RFP] | This study |
| UD676 | anc-1(e1873) I; [Pcol-10::mitoLS::GFP, Podr-1::RFP] | This study |
| LIU2 | ldrIs2 [mdt-28p::mdt-28::mCherry + unc-76(+)]. | Na et al., 2015 |
| UD681 | anc-1(e1873) I; ldrIs2 | This study |
| UD728 | anc-1(yc94[gfp::anc-1b::Δtk]) I | This study |
| UD789 | anc-1(yc106[gfp::anc-1b::Δkash]) I | This study |
| UD756 | ycSi2[pSL845 Py37a1b.5::mKate2::tram-1::tram-1 3’UTR] IV | This study |
| NM5179 | jsTi1493[LoxP::mex-5p::FLP:SL2::mNeonGreen::rpl-28p::FRT::GFP::his-58::FRT3] IV | Nonet, 2020 |
| UD778 | anc-1(yc68[gfp::anc-1b]) I; ycSi2 | This study |
| UD779 | anc-1(yc90[anc-1::gfp::F2]) I; ycSi2 | This study |
| UD780 | anc-1(yc70[gfp::anc-1b::Δ6rps]) I; ycSi2 | This study |
| UD781 | anc-1(yc106[gfp::anc-1b::Δkash]) I; ycSi2 | This study |
| UD782 | anc-1(yc94[gfp::anc-1b::Δtk]) I; ycSi2 | This study |
| UD783 | anc-1(yc68[gfp::anc-1b]) I; bqSi142 | This study |
| UD785 | anc-1(yc70[gfp::anc-1b::Δ6rps]) I; bqSi142 | This study |
| UD790 | anc-1(yc68[gfp::anc-1b]) I; ldrIs2 | This study |
crRNA and repair templates used in this study.
| New alleles | Strain | crRNA * | DNA repair template * †,§, ‡ |
|---|---|---|---|
| anc-1(yc52[∆25KASH]) I | N2 | CAGUACUCGUCGUCGCAAUG | GCACTGCTTGTTCTACTTATGGGAGCCGCTTGTTTGGTTCCACAcTGtGAtGAtGAaTAtTAATCTTTAATTTTTTATTTTCATTACTATTCACTATTGTTTCATTCATCATGAACCTG |
| anc-1(yc69[∆29KASH]) I | N2 | CAGUACUCGUCGUCGCAAUG | NHEJ ‡ |
| anc-1(yc41[anc-1::gfp3Xflag::kash,Δch]) I | N2 | UUUCAUCUUGAAGAGGUUCG | --AAAATCTATTTTGAAAATTTTCAGATGAGGACGAG<EGFP-3xFLAG > AGATCAGGAGCTAGCGGAGCCATGTTCGGAGAAAGATCACCAATG-- |
| anc-1(yc62[ΔF1]) I | N2 | ACUUGAUCAAUCUAUAAUAA | GAAACATGAAAGCAAAGTACATTTTTTTAAAAATCGATTATTTCagATcGAcCAgGTACAGTCTGAGATCGACACTCTTTCAGACTTCGAGGAGATCGAGCGTGAAATAAACGGCTCACTCGAAGCTTTCGAAGCCGAG |
| anc-1(yc61[Δ6RPS]) I | N2 | GCGUUCAAUUUCUUCAAAAU UGUUAGUAUUGGCGGCGAGU GGAGCGUUUUGUAAAAGCAA | AAGGTACAAAACATTGGAAAAACATCGATTGACGACGTGAATGTATCTGACTTCGAGGAGATCGAGCGTGAGATCAATGGCTCCCTTGAGGCTTTCTCTATTTGGGAACGCTTCGTCAAGGCTAAAGATGATTTGTACGATTATTTGGAGAAATTAGAGAACAATGTAAGC |
| anc-1(yc80[ΔF2]) I | N2 | GGAGCGUUUUGUAAAAGCAA UCCAACGGGAUCUUUGUCGU | ACTCTTATTCCGGACCTTGAAGAAAGAGCTTCTATTTGGGAGCGTACTGCTTTGCCACTTCAGGTTTGTTTATATTTTTTAATATTAATA |
| anc-1(yc36[anc-1::gfp3Xflag::kash]) I | N2 | GACAAAGATCCCGTTGGAGA | TCCGACGACAgAGATCtCGcTGGcGcCGcGTACTCAGAACTTCAGGAGCTAGCGGAGCC < EGFP-3xFLAG > tcaggagctagcggagccGCTTTGCCACTTCAGgtttgtttatatttttt |
| anc-1([yc68[gfp::anc-1b]]) I | N2 | CCGUCGGAACAGCUCCAUUU | TCTTAACCTTTTGTTCCATTCACTAATTATTTTCAATTACAGGAGGTTGGCCGCGAGTCGGTCAGTAAATTATCAGCTGCAGTTGACGATCGATACATCTACACGTTACACGTGCTCCGAACTAAG < EGFP > GGAGGTTCCGGAGGTGGATCTGGAGGTGAaCTcTTtCGtCGtCTGCAAAACTTTTGCGACGCTGTCAAAATATTGCGATCGCAAAATACCAAATGGAACGGAATCAAGATTTCGCAGGTTTGTTTCAAAAGCATCACAAATTAGCGG |
| anc-1(yc78[Δ5RPS]) I | N2 | GGAGCGUUUUGUAAAAGCAA UUCCUCUGGCUUCAACGAGU | GATCAGCTCAAGTCGGACGATTTGAAGACGGCAGAAAAGGAAATCACTAAtagccTcAAaCCcGAaTCTATTTGGGAaaGaTTcGTtAAgGCtAAAGATGATTTGTACGATTATTTGGAGAAATTAGAGAACAAT |
| anc-1(yc90[anc-1::gfp::F2]) I | N2 | GGAGCGUUUUGUAAAAGCAA | CCGGACCTTGAAGAAAGAGCTTCTATTTGGGAGCGTGGTGGAAGTGGTGGAGGAAGCGGTGGA < EGFP > GCATGGATGAACTATACAAAGGAGGTTCCGGAGGTGGATCTGGAGGTTTcGTcAAgGCtAAAGATGATTTGTACGATTATTTGGAGAAATTAGAGAACA |
| anc-1(yc70[gfp::anc-1b::Δ6rps]) I | UD612 | GCGUUCAAUUUCUUCAAAAU UGUUAGUAUUGGCGGCGAGU GGAGCGUUUUGUAAAAGCAA | AAGGTACAAAACATTGGAAAAACATCGATTGACGACGTGAATGTATCTGACTTCGAGGAGATCGAGCGTGAGATCAATGGCTCCCTTGAGGCTTTCTCTATTTGGGAACGCTTcGTcAAgGCtAAAGATGATTTGTACGATTATTTGGAGAAATTAGAGAACAATGTAAGC |
| anc-1(yc91[gfp::anc-1b::Δ5rps]) I | UD612 | GGAGCGUUUUGUAAAAGCAA UUCCUCUGGCUUCAACGAGU | GATCAGCTCAAGTCGGACGATTTGAAGACGGCAGAAAAGGAAATCACTAAtagccTcAAaCCcGAaTCTATTTGGGAaaGaTTcGTtAAgGCtAAAGATGATTTGTACGATTATTTGGAGAAATTAGAGAACAAT |
| anc-1(yc92[gfp::anc-1b::Δf1]) I | UD612 | ACUUGAUCAAUCUAUAAUAA | GAAACATGAAAGCAAAGTACATTTTTTTAAAAATCGATTATTTCagATcGAcCAgGTACAGTCTGAGATCGACACTCTTTCAGACTTCGAGGAGATCGAGCGTGAAATAAACGGCTCACTCGAAGCTTTCGAAGCCGAG |
| anc-1(yc93[anc-1AI972,973**::gfp]) I | UD612 | ACUCACCUCUAGAAAUUCGA | CAAAATTTAGAGCTCAGCAATGAGCAGGACTGTCCAGATtaatgaGgtaccCTAGAGGTGAGTATAGTCATTTTCCGCTCATTACACTCTT |
| anc-1(yc71[∆TK]) I | N2 | CAGAACUGCUUUGCCACUUC AUUAAAGAUUAAAAUGGUGG | GAACAACTCCGACGACAAAGATCCCGTTGGAGACGGGTACTCAGATAATCTTTAATTTTTTATTTTCATTACTATTCACTATTGTTTCATTCATC |
| anc-1(yc106[gfp::anc-1b::Δkash]) I | UD612 | CAGUACUCGUCGUCGCAAUG | GCACTGCTTGTTCTACTTATGGGAGCCGCTTGTTTGGTTCCACAcTGtGAtGAtGAaTAtTAATCTTTAATTTTTTATTTTCATTACTATTCACTATTGTTTCATTCATCATGAACCTG |
| anc-1(yc94[gfp::anc-1b::Δtk]) | UD612 | CAGAACUGCUUUGCCACUUC AUUAAAGAUUAAAAUGGUGG | GAACAACTCCGACGACAAAGATCCCGTTGGAGACGGGTACTCAGATAATCTTTAATTTTTTATTTTCATTACTATTCACTATTGTTTCATTCATC |
| anc-1(yc73[gfp::anc-1b::Δ6rps::Δtk]) I | UD618 | CAGAACUGCUUUGCCACUUC AUUAAAGAUUAAAAUGGUGG | GAACAACTCCGACGACAAAGATCCCGTTGGAGACGGGTACTCAGATAATCTTTAATTTTTTATTTTCATTACTATTCACTATTGTTTCATTCATC |
| dpy-10(cn64) | Co-CRISPR | GCUACCAUAGGCACCACGAG Arribere et al., 2014 | CACTTGAACTTCAATACGGCAAGATGAGAATGACTGGAAACCGTACCGCATGCGGTGCCTATGGTAGCGGAGCTTCACATGGCTTCAGACCAACAGCCTAT (Arribere et al., 2014) |
-
*all nucleotide sequences are displayed as single strand in the 5’ to 3’ orientation.
†In many cases a ssDNA oligonucleotide was used. For larger inserts, a PCR product was used.
-
§An imprecise NHEJ event led to an in-frame deletion without using the repair template.
¶Underlined sequences introduce silent mutations so the repair template is not cut by Cas9.
-
‡Underline indicates the silent mutation in the repair template.





