Cell competition removes segmental aneuploid cells from Drosophila imaginal disc-derived tissues based on ribosomal protein gene dose
Figures
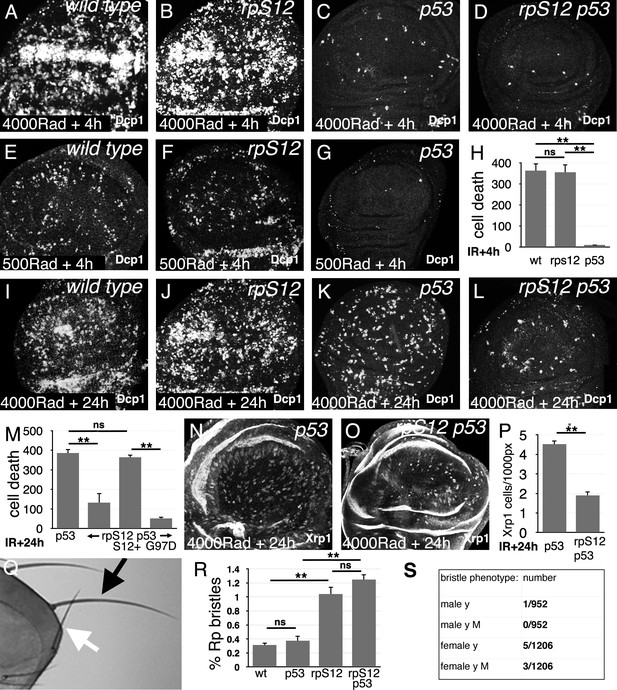
Role of cell competition after irradiation.
Panels A-G and I-L show third instar wing imaginal discs labeled to detect the activated Dcp1 caspase in apoptotic cells at the indicated times post-irradiation. (A) Extensive cell death follows within 4 hr after gamma-irradiation (4000 Rads). (B) Little change is seen in the rpS12G97D mutant. (C) Most of the acute cell death is p53-dependent, consistent with a DNA damage response. (D) A small amount of cell death persists in the rpS12G97D p53- double mutant. (E) Cell death is reduced to more quantifiable levels 4 hr after a lower radiation dose (500 Rad). (F) This DNA damage-induced cell death is not significantly affected by the rpS12G97D mutation. (G) Most cell death 4 hr after irradiation is p53-dependent. (H) Quantification of cell death (numbers of cells per wing pouch) 4 hr following irradiation with 500 Rad. ns – difference not statistically significant (p>0.05). ** - difference highly significant (p<0.01). N = 6 for each genotype. (I) Cell death 24 hr after irradiation (4000 Rad). (J) Comparable levels of cell death in rpS12G97D mutants. (K) Although cell death appears reduced in p53 mutant wing discs 24 hr after irradiation compared to the wild type control (4000 Rad), this p53-independent cell death is substantially increased compared to 4 hr after irradiation (compare panel C). (L) p53-independent cell death is reduced in the rpS12G97D p53- double mutant compared to the p53- mutant. (M) Cell death quantification in p53 and rpS12G97D p53- wing discs, and in wing discs from rpS12G97D p53- larvae carrying genomic transgenes encoding either wild type rpS12 or rpS12G97D. The results show that between 66% and 86% of p53-independent cell death was RpS12-dependent. We did not quantify cell death in wild-type and rpS12G97D wing discs because of the large number and aggregation of dead cells. ns – difference not statistically significant (p>0.05). ** - difference highly significant (p<0.01). N: 10 (p53-); 13 (rpS12G97D p53-); 9 (P{rpS12+} rpS12G97D p53-); 8 (P{rpS12G97D} rpS12G97D p53-). (N) Twenty-four hr after irradiation, p53 mutant wing discs exhibit cells expressing nuclear Xrp1 in a pattern similar to that of dying cells. We were unable to double-label with anti-Xrp1 and anti-active Dcp1 simultaneously because both are rabbit antisera. n: 33 (p53-); 26 (rpS12G97D p53-). (O) Fewer Xrp1-expressing cells were seen in rpS12G97D p53- double mutant wing discs. (P) Quantification of Xrp1 expression. Most (58%) of the p53-independent Xrp1 expression 24 hr post-irradiation was RpS12-dependent. ** - difference highly significant (p<0.01). (Q) Irradiated adult flies with a short, thin scutellar bristle (white arrow, compare normal contralateral bristle – black arrow) like those typical of Rp+/- mutant flies. (R) The frequency of sporadic Rp-like bristles increases 3.33x and 4x on the thoraces of rpS12G97D and rpS12G97D p53- flies (respectively) where cell competition is inhibited. ns – difference not statistically significant (p>0.05). ** - difference highly significant (p<0.01). N = 3 sets of 100 flies for each genotype. (S) Frequencies of y bristles found on the thoraces of ~2000 y/+; rpS12G97D female and +/Y; rpS12G97D male flies following irradiation (1000 rad) in the mid-third larval instar. The preponderance of y bristles in females suggests that induced y mutations typically affect other genes including essential genes. The occurrence of phenotypically y M bristles in females is consistent with deletions extending at least from the y locus to the nearest Rp locus, RpL36. Statistics: Significance was assessed using t-test (panel P) or one-way ANOVA with the Holm procedure for multiple comparisons (panels H,M,R). ** = p<0.01. ns = not significant. Genotypes: A, (E, I) w11-18 B, (F, J) rpS12G97D (C,G,K,N) p535A-1-4 D, (L, O) rpS12G97D p535A-1-4.
-
Figure 1—source data 1
Source Data for Figure 1H,M,P,R.
- https://cdn.elifesciences.org/articles/61172/elife-61172-fig1-data1-v1.xlsx
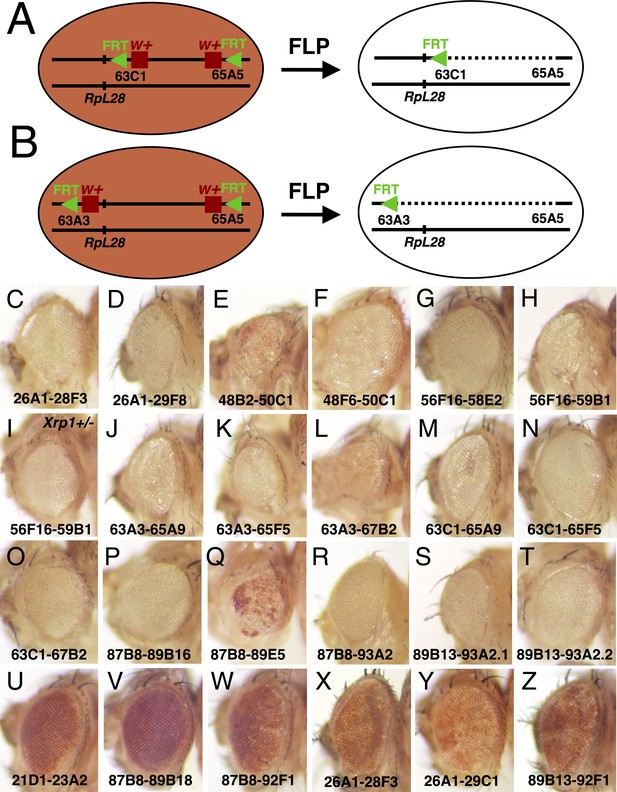
Generating segmental aneuploidy with Flp-FRT recombination.
(A) Segmental aneuploidy can be generated in a mosaic fashion using FLP-FRT. At left is a cell carrying two transposable elements encoding w+ and FRT, arranged in cis on one chromosome arm, in this case at chromosome bands 63C1 and 65A9, respectiely. (B) FLP-mediated recombination between FRT sites excising the intervening sequences. In this example, where the w+ genes lie between the FRT sites, both are excised resulting in a loss of eye pigmentation. The RpL28 gene at chromosome band 63B14 lies outside the deletion and is unaffected. (B) A comparable recombination between elements at chromosome bands 63A3-65A9 also deletes the RpL28 locus, so that the resulting segmentally aneuploid cells are heterozygously deleted for this gene. These cartoons show recombination in G1-phase of the cell cycle. In the G2-phase configuration, recombination between non-homologous FRT sites on the chromatids can occur leading to a deleted chromatid and a chromatid bearing 3 FRT insertion elements and a duplication of the intervening region. Such genotypes are substrates for further FLP recombination to the parental or deleted state, but sometimes we see them persist in adults and an example is shown in Figure 3D (Titen et al., 2020). Panels C-Z show adult eyes in the presence of eyFlp, which drives recombination close to completion in the eye and head, with the chromosome positions of parental w+ FRT insertions indicated. Panels C-T show genotypes where eyFlp recombined most eye cells, Panels U-Z illustrate genotypes where it did not. Strains that were poor substrates for Flp, either retained the parental eye color in the presence of eyFlp, or produce a salt and pepper pattern of very small clones that is indicative of excision occurring only late in development once large cell numbers are present. See Supplementary file 1 for more details and further genotypic information. (C) y w eyFlp; P{XP}d08241 PBac{WH}f04888/+; (D) y w eyFlp; P{XP}d08241 PBac{WH}f00857/+. This recombination deletes the RpL36A and RpS13 genes; (E) y w eyFlp; P{XP}d09761 PBac{WH}f00157/+. This recombination deletes the RpS11 gene. Note the particularly small size of the recombinant heads; (F) y w eyFlp; P{XP}d09417 PBac{WH}f00157/+. (G) y w eyFlp; P{XP}d02302 PBac{WH}f04349/+; (H) y w eyFlp; P{XP}d02302 PBac{WH}f00464/+. This recombination deletes the RpS16 and RpS24 genes. Note the particularly small size of the recombinant heads; (I) y w eyFlp; P{XP}d02302 PBac{WH}f00464/Xrp1m2-73. Eye size is partially rescued by heterozygosity for Xrp1. (J) y w eyF; PBac{WH}f01922 P{XP}d02570 /+. This recombination deletes the RpL28 gene. (K) y w eyF; PBac{WH}f01922 P{XP}d02813 /+. This recombination deletes the RpL28 and RpL18 genes; (L) y w eyF; PBac{WH}f01922 P{XP}d07256 /+. This recombination deletes the RpL28, RpL18, and RpL14 genes; Note the small eye size. (M) y w eyF; PBac{WH}f05041 P{XP}d02570 /+; (N) y w eyF; PBac{WH}f05041 P{XP}d02813 /+. This recombination deletes the RpL18 gene; (O) y w eyF; PBac{WH}f05041 P{XP}d07256 /+. This recombination deletes the RpL18 and RpL14 genes; (P) y w eyF; P{XP}d06796 PBac{WH}f04937 /+. This recombination deletes the eIF2γ gene. (Q) y w eyF; P{XP}d06796 PBac{WH}f00971 /+. This recombination deletes the eIF2γ gene. Note the particularly small size of the recombinant heads, which also retain an unusual amount of unrecombined cells; (R) y w eyF; P{XP}d06796 PBac{WH}f03502 /+. This recombination deletes the eIF2γ, Xrp1, RpS20 and RpS30 genes. (S) y w eyF; P{XP}d06928 PBac{WH}f03502 /+. This recombination deletes the Xrp1, RpS20 and RpS30 genes. (T) y w eyF; P{XP}d06928 PBac{WH}f01700 /+. This recombination deletes the Xrp1, RpS20, and RpS30 genes. (U) y w eyF; PBac{WH}f04180 P{XP}d07944 /+. The whole eye resembles the parental genotype lacking eyFlp; (V) y w eyF; P{XP}d06796 PBac{RB}e03186 /+. The whole eye resembles the parental genotype lacking eyFlp; (W) y w eyF; P{XP}d06796 PBac{RB}e03144 /+. The whole eye resembles the parental genotype lacking eyFlp. (X) y w eyF; P{XP}d08241 PBac{RB}e02272 /+. The eye has a mottled appearance indicative of small recombinant clones generated late in development; (Y) y w eyF; P{XP}d08241 PBac{RB}e03937 /+. The eye has a mottled appearance indicative of small recombinant clones generated late in development; (Z) y w eyF; P{XP}d06928 PBac{RB}e03144 /+. The eye has a mottled appearance indicative of small recombinant clones generated late in development.
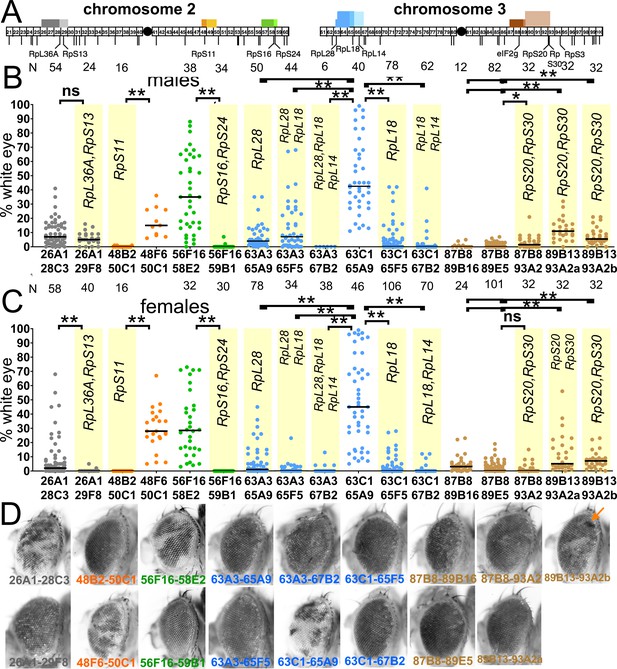
Segmental-aneuploid clones in the eye.
(A) Cartoon of the major autosomes that contain chromosome band intervals 21–100 out of 102 total. Lines below indicate the position of all the Rp gene loci thought to be haploinsufficient (Marygold et al., 2007), as well as the eIF2γ locus. Identities are shown only for the loci discussed in this paper. Blocks above the chromosomes show the extent of 17 segmentally aneuploid deletions reported in this figure, color-coded according to genomic region. (B) Graphs show the % of the male adult eye comprising w-, segmentally aneuploid cells for 17 chromosome regions. Each data-point represents the extent of w- territory of an individual eye, median values shown as black bars, N as indicated for each genotype. Geneotypes from overlapping genomic regions are shown in a common color. Yellow background indicates deletions that encompass one or more Rp loci. Deletion of Rp loci reduces contribution to the adult eye for all regions except the 87–93 region (brown). (C) As for panel A except data from females is shown. (D) Examples of typical eyes for each of the genotypes analyzed (males shown). Arrow on the Df(3R)89B13-93A2.2/+ eye indicates a small clone of darker cells reflecting four w+ elements associated with tandem duplication of the 89B13-93A2 region due to a FLP-FRT recombination event in G2 that was not resolved by further recombination (see Figure 2 legend). Statistics. Pairwise comparisons using the Mann-Whitney procedure with the Benjamini-Hochberg correction for multiple testing (see Supplementary file 2). ns – difference not statistically significant (p>0.05). * - difference significant (p<0.05). ** - difference highly significant (p<0.01). Genotypes: For 26A1-28C3, y w hsF; P{XP}d08241 PBac{WH}f04888/+; FRT82B/+. For 26A1-29F8, y w hsF; P{XP}d08241 PBac{WH}f00857/+; FRT82B/+. For 48B2-50C1, y w hsF; P{XP}d09761 PBac{WH}f00157/+; FRT82B/+. For 48F6-50C1, y w hsF; P{XP}d09761 PBac{WH}f00157/+; FRT82B/+. For 56F16-58E2, y w hsF; P{XP}d02302 PBac{WH}f04349/+; FRT82B/+. For 56F16-59B1, y w hsF; P{XP}d02302 PBac{WH}f00464/+; FRT82B/+. For 63A3-65A9, y w hsF; PBac{WH}f01922 P{XP}d02570 /FRT82B. For 63A3-65F5, y w hsF; PBac{WH}f01922 P{XP}d02813 /FRT82B. For 63A3-67B2, y w hsF; PBac{WH}f01922 P{XP}d07256 /FRT82B. For 63C1-65A9, y w hsF; PBac{WH}f05041 P{XP}d02570 /FRT82B. For 63C1-65F5, y w hsF; PBac{WH}f05041 P{XP}d02813 /FRT82B. For 63C1-67B2, y w hsF; PBac{WH}f05041 P{XP}d07256 /FRT82B. For 87B8-89B16, y w hsF; P{XP}d06796 PBac{WH}f04937 /FRT82B. For 87B8-89E5, y w hsF; P{XP}d06796 PBac{WH}f00971 /FRT82B. For 87B8-93A2, y w hsF; P{XP}d06796 PBac{WH}f03502 /FRT82B. For 89B13-93A2a, y w hsF; P{XP}d06928 PBac{WH}f03502 / FRT82B. For 89B13-93A2b, y w hsF; P{XP}d06928 PBac{WH}f01700 /FRT82B.
-
Figure 3—source data 1
% eye white data.
* indicates that 4x w+ cells indicative of FLP-mediated duplication were also observed in this specimen. Gaps between measurements separate replicate data.
- https://cdn.elifesciences.org/articles/61172/elife-61172-fig3-data1-v1.xlsx
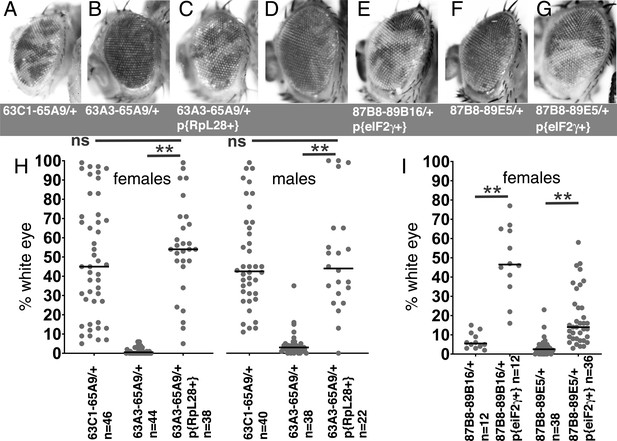
Rescue of segmental aneuploidy by single transgenes.
(A) Df63C1-65A9/+ cells contribute ~50% of the adult eye. (B) Very few cells heterozygous for the overlapping Df63A3-65A9 were recovered. (C) Df63A3-65A9/+ cells contribute ~50% of the adult eye in the presence of the RpL28+ transgene. (D). Very few cells heterozygous for Df87B8-89B16 contribute to the adult eye. (E). Df87B8-89B16/+ cells contribute ~50% of the adult eye in the presence of the eIF2γ + genomic transgene. (F). Very few cells heterozygous for Df87B8-89E5 contribute to the adult eye. (G). Df87B8-89E5/+ cells contribute ~20% of the adult eye in the presence of the eIF2γ + genomic transgene. (H) Quantification of results for the RpL28 region shown in panels A-C. Data for Df63C1-65A9 is the same as that already shown in Figure 2A,B. (I) Quantification of results for the eIF2γ region shown in panels D-G. Statistics. Pairwise comparisons used the Mann-Whitney procedure with the Benjamini-Hochberg correction for multiple testing (see Supplementary file 2). N as indicated for each genotype. ns – difference not statistically significant (p>0.05). ** - difference highly significant (p<0.01). Genotypes: For 63C1-65A9, y w hsF; PBac{WH}f05041 P{XP}d02570 /FRT82B. For 63A3-65A9, y w hsF; PBac{WH}f01922 P{XP}d02570 /FRT82B and y w hsF; {RpL28+P3-DsRed}/+; PBac{WH}f01922 P{XP}d02570 /+; FRT82B/+. For 87B8-89B16, y w hsF/+; P{XP}d06796 PBac{WH}f04937 /FRT82B and y w hsF/+; P{ry+ Su(var3-9+) eIF2γ+}/+; P{XP}d06796 PBac{WH}f04937 /+. For 87B8-89E5, y w hsF/+; P{XP}d06796 PBac{WH}f00971 /FRT82B and y w hsF/+; P{ry+ Su(var3-9+) eIF2γ+}/+; P{XP}d06796 PBac{WH}f00971 /+.
-
Figure 4—source data 1
% eye white data.
* Indicates that 4x w+ cells indicative of FLP-mediated duplication were also observed in this specimen. Gaps between measurements separate replicate data.
- https://cdn.elifesciences.org/articles/61172/elife-61172-fig4-data1-v1.xlsx
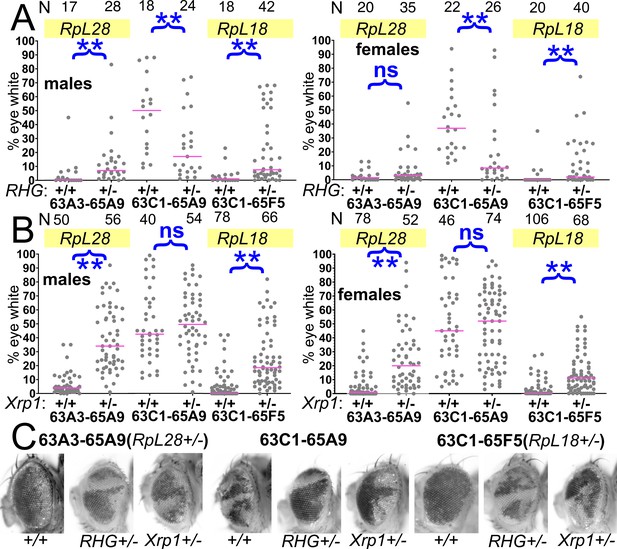
Contributions of apoptosis and Xrp1 to elimination of segmentally aneuploid cells.
Panels A-B show the contribution of w- segmentally aneuploid cells to adult eyes of indicated genotypes, N as indicated for each genotype. (A) Heterozygosity for Df(3L)H99, which deletes the proapoptotic genes rpr, hid, and grim (‘RHG’) significantly increases contribution of Df63A3-65A9/+ and Df63C1-65F5/+ cells to adult eyes. These genotypes approach the contribution level of Df63C1-65A9/Df(3L)H99 cells which do not affect any Rp locus the results for 63A3-65A9/Df(3L)H99, 63C1-65A9/Df(3L)H99, and 63C1-65F5/Df(3L)H99 cannot be distinguished in males: KruskalWallis test, p=0.18; in females, this hypothesis is rejected and post-hoc testing identified the 63C1-65A9/Df(3L)H99 data as different from the other genotypes (p=0.018 for the comparison to 63A3-65A9/H99, p=0.027 for the comparison to 63C1-65F5/H99). (B) Heterozygosity for Xrp1, which is required for the slow growth and cell competition of Rp+/-point-mutant cells, significantly increases contribution of Df63A3-65A9/+ and Df63C1-65F5/+ cells to adult eyes. Xrp1 did not affect the contribution of Df63C1-65A9/+ cells that do not affect any Rp locus. We also tested the hypothesis that Df(3L)H99, Xrp1 mutation, and an RpS3/+ genetic background (see Figure 8) suppressed cell competition to an equal degree. This hypothesis was rejected for both males and females of Df63A3-65A9/+ and Df(3L)63C1-65F5/+ (Kruskal Wallis test p=2.9×10−17, 9.7 × 10−21, 2.1 × 10−5, and 1.8 × 10−7, respectively). For each deletion, all the genotypes were individually significantly different except Df(3L)63C1-65F5/H99 and Df(3L)63C1-65F5/Xrp1 males, which were not significantly different (p=0.06, Conover post-hoc test with Holm correction). (C) Representative examples of these genotypes. Statistics. Pairwise comparisons using the Mann-Whitney procedure with the Benjamini-Hochberg correction for multiple testing (see Supplementary file 2). ns – difference not statistically significant (p>0.05). ** - difference highly significant (p<0.01). Multiple comparisons using the Kruskal Wallis test as described for panel B. Genotypes. For 63A3-65A9 in panel A: y w hsF; PBac{WH}f01922 P{XP}d02570 /FRT80B and y w hsF; PBac{WH}f01922 P{XP}d02570 /Df(3L)H99 FRT80B. For 63C1-65A9 in panel A: y w hsF; PBac{WH}f05041 P{XP}d02570 /FRT80B and y w hsF; PBac{WH}f05041 P{XP}d02570 /Df(3L)H99 FRT80B. For 63C1-65F5 in panel A: y w hsF; PBac{WH}f05041 P{XP}d02813 /FRT80B and y w hsF; PBac{WH}f05041 P{XP}d02813 /Df(3L)H99 FRT80B. For 63A3-65A9 in panel B: y w hsF; PBac{WH}f01922 P{XP}d02570 /FRT82B and y w hsF; PBac{WH}f01922 P{XP}d02570 /FRT82B Xrp1m2-73. For 63C1-65A9 in panel B: y w hsF; PBac{WH}f05041 P{XP}d02570 /FRT82B and y w hsF; PBac{WH}f05041 P{XP}d02570 /FRT82B Xrp1m2-73. For 63C1-65F5 in panel B: y w hsF; PBac{WH}f05041 P{XP}d02813 /FRT82B and y w hsF; PBac{WH}f05041 P{XP}d02813 /FRT82B Xrp1m2-73.
-
Figure 5—source data 1
% eye white data.
* Indicates that 4x w+ cells indicative of FLP-mediated duplication were also observed in this specimen. Gaps between measurements separate replicate data.
- https://cdn.elifesciences.org/articles/61172/elife-61172-fig5-data1-v1.xlsx
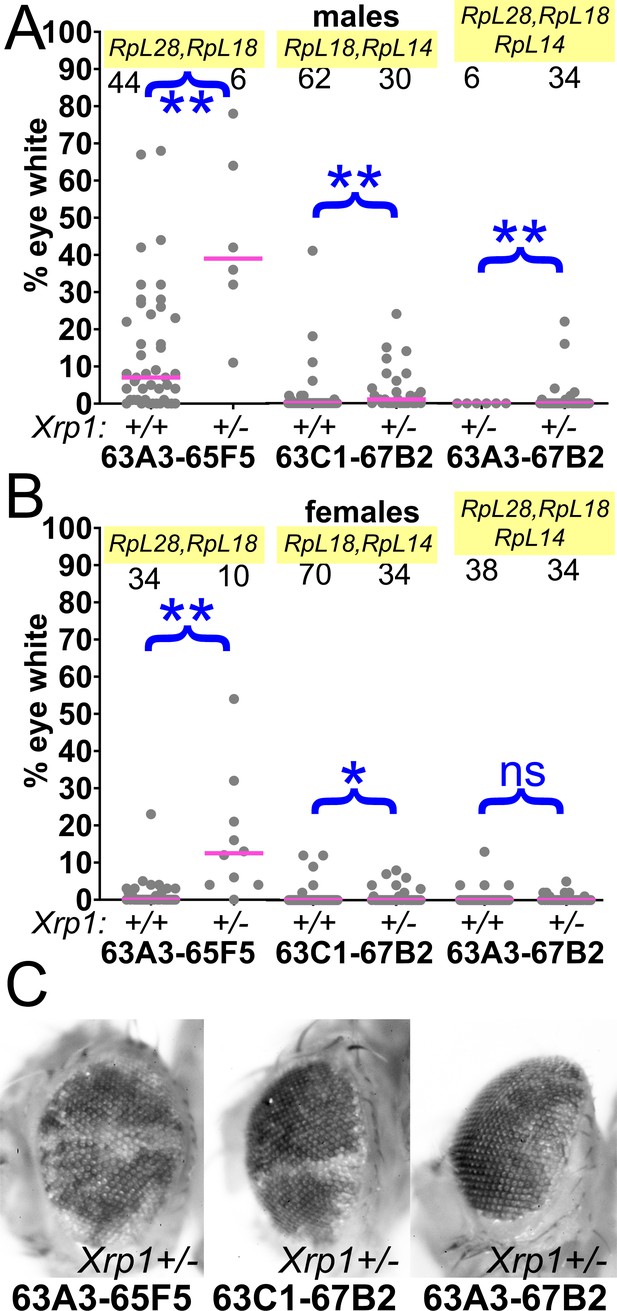
Contributions of Xrp1 to competition of larger segmental aneuploidies.
(A) Heterozygosity for Xrp1 has progressively less effect on the contributions of larger segmental-aneuploid genotypes in males, although still significant statistically. (B) In females, the effect of Xrp1 on Df(3L)63A3-67B2/+ cells, haploinsufficient for RpL14, RpL18, and RpL28, in no longer significant statistically. Xrp1 heterozygosity does affect Df(3L)63C1-67B2 females, although many zero values are superimposed so that this is hard to appreciate on the Df(3L)63C1-67B2/+ Xrp1+/+ graph. (C). Representative examples of these phenotypes (males are shown). Statistics. Pairwise comparisons used the Mann-Whitney procedure with the Benjamini-Hochberg correction for multiple testing (see Supplementary file 2). N as indicated for each genotype. ns – difference not statistically significant (p>0.05). * - difference significant (p<0.05). ** - difference highly significant (p<0.01). Genotypes. For 63A3-65F5: y w hsF; PBac{WH}f01922 P{XP}d02813 /FRT82B and y w hsF; PBac{WH}f01922 P{XP}d02813 /FRT82B Xrp1m2-73. For 63C1-67B2: y w hsF; PBac{WH}f05041 P{XP}d07256 /FRT82B and y w hsF; PBac{WH}f05041 P{XP}d07256 /FRT82B Xrp1m2-73. For 63A3-67B2: y w hsF; PBac{WH}f01922 P{XP}d07256 /FRT82B and y w hsF; PBac{WH}f01922 P{XP}d07256 /FRT82B Xrp1m2-73.
-
Figure 6—source data 1
% eye white data.
* Indicates that 4x w+ cells indicative of FLP-mediated duplication were also observed in this specimen. Gaps between measurements separate replicate data.
- https://cdn.elifesciences.org/articles/61172/elife-61172-fig6-data1-v1.xlsx
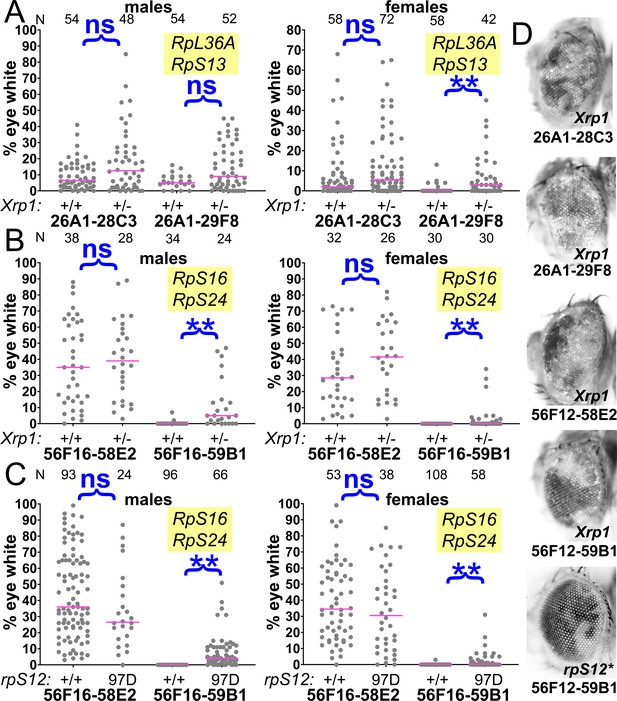
Contributions of Xrp1 and RpS12 to competition of chromosome two genotypes.
(A) Xrp1 mutation improved the eye contributions of the Df(26A1-29F8)/+ genotype. Data for Df(26A1-28C3)/+ are the same as shown in Figure 2. (B) Xrp1 mutation improved the eye contribution of Df(56F16-59B1)/+, heterozygous for RpS16 and RpS24, but had no effect on the contribution of Df(56F16-58E2)/+ cells that affect no Rp loci. (C) Homozygosity for the rpS12G97D mutation improved the eye contribution of Df(56F16-59B1)/+, heterozygous for RpS16 and RpS24, but had no effect on the contribution of Df(56F16-58E2)/+ cells that affect no Rp loci. (D) Representative examples of these phenotypes (males shown). Statistics. Pairwise comparisons used the Mann-Whitney procedure with the Benjamini-Hochberg correction for multiple testing (see Supplementary file 2). N as indicated for each genotype. ns – difference not statistically significant (p>0.05). ** - difference highly significant (p<0.01). Genotypes. For 26A1-28C3: y w hsF; P{XP}d08241 PBac{WH}f04888/+; FRT82B/+ and y w hsF; P{XP}d08241 PBac{WH}f04888/+; FRT82B Xrp1m2-73/+. For 26A1-29F8: y w hsF; P{XP}d08241 PBac{WH}f00857/+; FRT82B/+ and y w hsF; P{XP}d08241 PBac{WH}f00857/+; FRT82B Xrp1m2-73/+. For 56F16-58E2 in panel B: y w hsF; P{XP}d02302 PBac{WH}f04349/+; FRT82B/+ and y w hsF; P{XP}d02302 PBac{WH}f04349/+; FRT82B Xrp1m2-73/+. For 56F16-59B1 in panel B: y w hsF; P{XP}d02302 PBac{WH}f00464/+; FRT82B/+ and y w hsF; P{XP}d02302 PBac{WH}f00464/+; Xrp1m2-73/+. For 56F16-58E2 in panel C: y w hsF; P{XP}d02302 PBac{WH}f04349/+; FRT80B/FRT80B and y w hsF; P{XP}d02302 PBac{WH}f04349/+; rpS12G97D FRT80B/rpS12G97D FRT80B. For 56F16-59B1 in panel B: y w hsF; P{XP}d02302 PBac{WH}f00464/+; FRT80B/FRT80B and y w hsF; P{XP}d02302 PBac{WH}f00464/+; rpS12G97D FRT80B/rpS12G97D FRT80B.
-
Figure 7—source data 1
% eye white data.
* Indicates that 4x w+ cells indicative of FLP-mediated duplication were also observed in this specimen. Gaps between measurements separate replicate data.
- https://cdn.elifesciences.org/articles/61172/elife-61172-fig7-data1-v1.xlsx
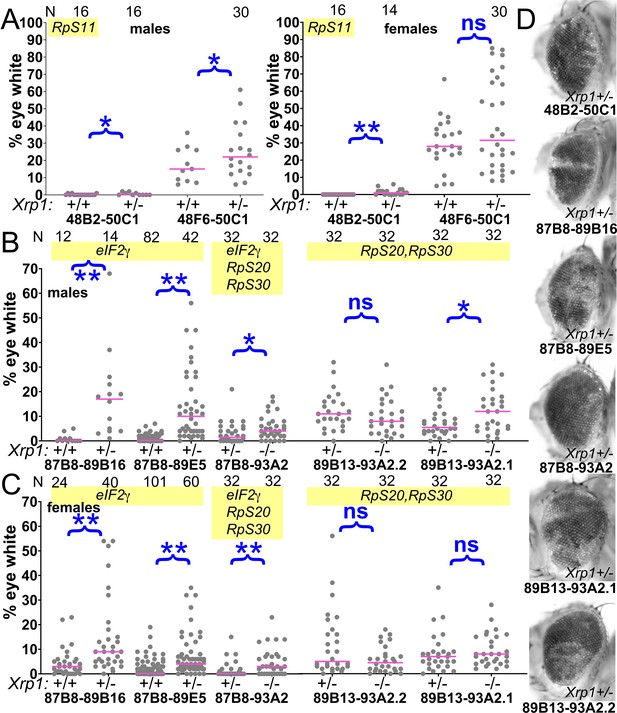
Contribution of Xrp1 to competition of further genotypes.
(A) Xrp1 mutation made a small but significant improvement to the contribution of Df(48B2-50C1)/+ cells, heterozygous for RpS11, but its effect on Df048F6-50C1/+ cells, which have no affected Rp genes, was only significant in males. (B,C) Xrp1 mutation significantly rescued the contribution of all segmental aneuploid genotypes affecting the eIF2γ locus. There was little effect of mutating the second Xrp1 copy on Df(89B13-93A2.1)/+ and Df(89B13-93A2.2)/+ cells, both of which are haplo-insufficient for RpS20 and RpS30 as well as Xrp1. (D) Representative examples of these genotypes (males shown). For panels A–C, the control data (Xrp1+/+ for A and Xrp1+/- for (B,C) were also shown in Figure 2). Statistics. Pairwise comparisons used the Mann-Whitney procedure with the Benjamini-Hochberg correction for multiple testing (see Supplementary file 2). N as indicated for each genotype. ns – difference not statistically significant (p>0.05). * - difference significant (p<0.05). ** - difference highly significant (p<0.01). Genotypes. For 48B2-50C1: y w hsF; P{XP}d09761 PBac{WH}f00157/+; FRT82B/+ and y w hsF; P{XP}d09761 PBac{WH}f00157/+; FRT82B Xrp1m2-73/+. For 48F6-50C1: y w hsF; P{XP}d09761 PBac{WH}f00157/+; FRT82B/+ and y w hsF; P{XP}d09761 PBac{WH}f00157/+; FRT82B Xrp1m2-73/+. For 87B8-89B16: y w hsF; P{XP}d06796 PBac{WH}f04937 /FRT82B and y w hsF; P{XP}d06796 PBac{WH}f04937 /FRT82B Xrp1m2-73. For 87B8-89E5: y w hsF; P{XP}d06796 PBac{WH}f00971 /FRT82B and y w hsF; P{XP}d06796 PBac{WH}f00971 /FRT82B Xrp1m2-73. For 87B8-93A2: y w hsF; P{XP}d06796 PBac{WH}f03502 /FRT82B and y w hsF; P{XP}d06796 PBac{WH}f03502 /FRT82B Xrp1m2-73. For 89B13-93A2.1: y w hsF; P{XP}d06928 PBac{WH}f03502 / FRT82B and y w hsF; P{XP}d06928 PBac{WH}f03502 /FRT82B Xrp1m2-73. For 89B13-93A2.2: y w hsF; P{XP}d06928 PBac{WH}f01700 /FRT82B and y w hsF; P{XP}d06928 PBac{WH}f01700 /FRT82B Xrp1m2-73.
-
Figure 8—source data 1
% eye white data.
* Indicates that 4x w+ cells indicative of FLP-mediated duplication were also observed in this specimen. Gaps between measurements separate replicate data.
- https://cdn.elifesciences.org/articles/61172/elife-61172-fig8-data1-v1.xlsx
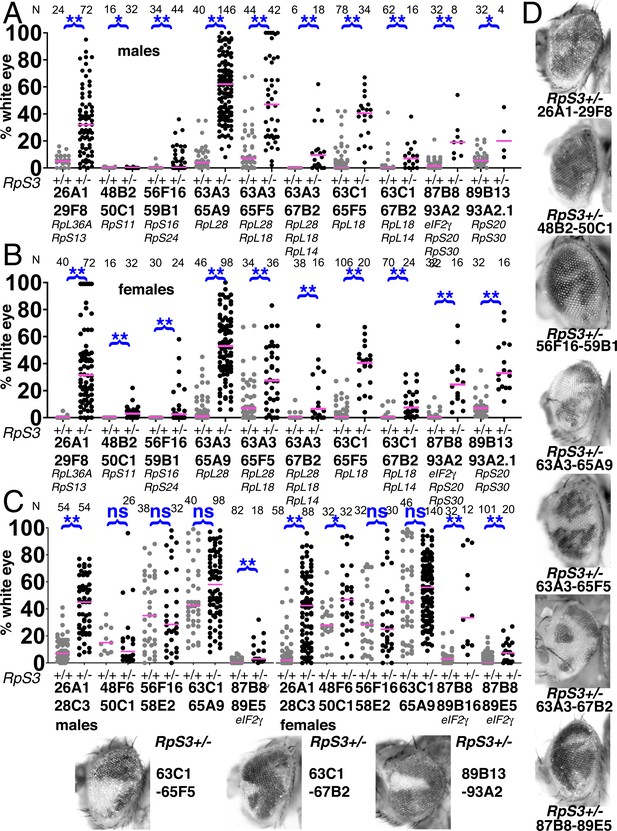
Elimination of segmentally aneuploid clones requires a Rp+/+ background.
Eye contributions of clones of segmentally aneuploid genotypes deleting the indicated Rp loci. in RpS3+/+ and RpS3+/- genetic backgrounds. Magenta bars show the median contributions, N as indicated for each genotype. (A) In males, the RpS3+/- genetic background (black datapoints) allows for significantly greater contribution in all cases except Df(48B2-50C1)/+. No Df(3R)89B13-93A2.2/RpS3 flies were obtained, this genotype is lethal due to an unidentified shared lethal outside the 89B13-93A2.2 region. (B) Comparable data from females. The RpS3+/- genetic background (black datapoints) always allows for significantly greater contribution. (C) Eye contributions of clones of segmentally aneuploid cells where no Rp loci are affected. The RpS3+/- genetic background (black datapoints) had no significant effect on many such genotypes, but did enhance the contribution of Df(26A1-28C3)/+ clones and of the Df(3R)87B8-89B16/+ and Df(3R)87B8-89E5/+ clones that were haploinsuffiicent for eIF2γ +. No Df(3R)87B8-89B16/RpS3 males were obtained. (D) Representative examples of these RpS3+/- genotypes (males shown). The RpS3+/+ control data were shown previously in Figure 2, except for 87B8-89B16 females. Statistics. Pairwise comparisons using the Mann-Whitney procedure with the Benjamini-Hochberg correction for multiple testing (see Supplementary file 2). ns – difference not statistically significant (p>0.05). * - difference significant (p<0.05). ** - difference highly significant (p<0.01). Genotypes: Same as for Figure 3, with an FRT82B RpS3 chromosome substituting for FRT82B where indicated.
-
Figure 9—source data 1
% eye white data.
* Indicates that 4x w+ cells indicative of FLP-mediated duplication were also observed in this specimen. Gaps between measurements separate replicate data.
- https://cdn.elifesciences.org/articles/61172/elife-61172-fig9-data1-v1.xlsx
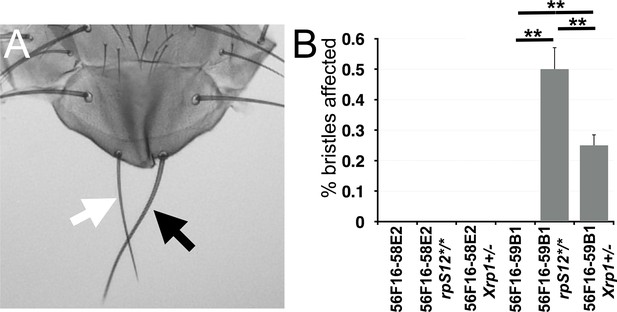
Segmentally aneuploid cells in the thorax.
(A) White arrow indicates a ‘Minute’(Rp+/-)-like short thin scutellar bristles on a fly containing clones of 56F16-59B1 cells. Compare the normal contralteral scutellar bristles (black arrow). (B) Frequency of affected bristles in genotypes as indicated. Data represent averages from three sets of 100 adults of each genotype. Minute-like bristles were not seen with the 56F16-58E2 deletion that does not affect any Rp locus. This provides as a baseline for any spontaneous loss of heterozygosity for Rp gene that might occur unrelated to the FLP-FRT excision, and which is evidently rare. Minute-like bristles were also not seen on cell competition-competent flies where 56F16-59B1 excisions would create heterozygosity for RpS16 and RpS24. These bristles only appeared in the rpS12 and Xrp1 mutant backgrounds where cell competition was compromised. Statistics. Three sets of 100 flies analyzed for each genotype. One-way ANOVA rejects the hypothesis that the six datasets are indistinguishable (p=4.28×10−7). The Holm procedure for multiple comparisons showed that results for 56F16-59B1 in the rpS12 and Xrp1 backgrounds were different from all others and from one another (adjusted p<0.05). For simplicity, significance is only indicated for 56F16-59B1 genotypes. ns – difference not statistically significant (p>0.05). * - difference significant (p<0.05). ** - difference highly significant (p<0.01).
-
Figure 10—source data 1
Source data for Figure 10B.
- https://cdn.elifesciences.org/articles/61172/elife-61172-fig10-data1-v1.xlsx
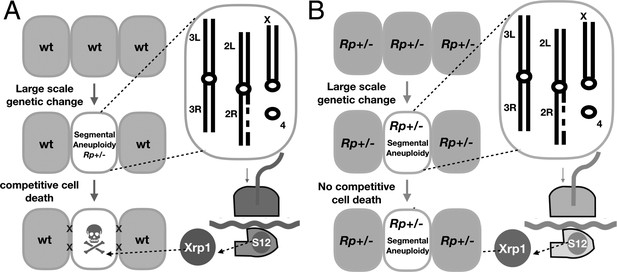
Models.
(A) Model for the elimination of segmentally aneuploid cells from imaginal discs. Cells that lose part of one chromosome may become haploinsufficient for one or more Rp loci, affecting ribosome assembly and triggering RpS12 and Xrp1 activities that lead to cell elimination by competition with unaffected neighboring cells. (B) Our studies show that segmentally aneuploid cells proliferate and contribute to adult tissues in animals where all cells are heterozygous for an Rp mutation, reducing the difference between aneuploid and diploid cells. A similar situation might apply in Diamond-Blackfan patients, many of whom are haploinsufficient for Rp loci. If Rp loci are also indicators of chromosome rearrangements in mammalian cells, such patients might face a greater accumulation of aneuploid cells.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | RpL28 | Flybase: FBgn0035422 | ||
| Gene (Drosophila melanogaster) | eIF2γ | Flybase: FBgn0263740 | ||
| Genetic reagent (D. melanogaster) | w11-18 | Hazelrigg et al., 1984; Lee et al., 2016 | FLYBASE: FBal0018186 | Bloomington Drosophila Stock Center #3605 |
| Genetic reagent (D. melanogaster) | Xrp1m2-73 | Lee et al., 2016 | FLYBASE:FBal0346068 | Bloomington Drosophila Stock Center #81270 |
| Genetic reagent (D. melanogaster) | rpS12G97D | Tyler et al., 2007 | FLYBASE:FBal0193403 | |
| Genetic reagent (D. melanogaster) | (Df3L)H99 | Abbott and Lengyel, 1991 | FLYBASE:FBab0022359 | Bloomington Drosophila Stock Center #1576 |
| Genetic reagent (D. melanogaster) | (M3R)w124 aka RpS32 | Ferrus, 1975; Abbott and Lengyel, 1991 | FLYBASE: FBal0011951 | |
| Genetic reagent (D. melanogaster) | hs-FLP | Struhl and Basler, 1993 | FLYBASE:FBtp0001101 | |
| Genetic reagent (D. melanogaster) | ey-FLP | Newsome et al., 2000 | FLYBASE:FBal0098303 | |
| Genetic reagent (D. melanogaster) | P{neoFRT}80B | Xu and Rubin, 1993 | FLYBASE:FBti0002073 | Bloomington Drosophila Stock Center #1988 |
| Genetic reagent (D. melanogaster) | P{neoFRT}82B | Xu and Rubin, 1993 | FLYBASE:FBti0002074 | Bloomington Drosophila Stock Center #2050, 2051 |
| Genetic reagent (D. melanogaster) | P{rpS12+8 kb} | Xu and Rubin, 1993; Kale et al., 2018 | FLYBASE:FBal0337985 | |
| Genetic reagent (D. melanogaster) | P{rpS12-G97D8kb} | Xu and Rubin, 1993; Kale et al., 2018 | FLYBASE:FBal0337986 | |
| Genetic reagent (D. melanogaster) | P{ry+ Su(var)3–9+ eIF2γ +} | Xu and Rubin, 1993; Tschiersch et al., 1994; Kale et al., 2018 | ||
| Genetic reagent (D. melanogaster) | P{DsRed; RpL28+} | This study | See Materials and methods; Dr. Nicholas Baker’s lab. | |
| Genetic reagent (D. melanogaster) | P535A-1-4 | Xie and Golic, 2004 | FLYBASE:FBal0138188 | Bloomington Drosophila Stock Center #6815 |
| Genetic reagent (D. melanogaster) | PBac{RB}CG11617e00462 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0162546 | Bloomington Drosophila Stock Center #17859 |
| Genetic reagent (D. melanogaster) | PBac{WH}MED15f04180 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBti0042319 | Bloomington Drosophila Stock Center #18739 |
| Genetic reagent (D. melanogaster) | P{XP}CG9016d08241 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0160858 | Bloomington Drosophila Stock Center #19290 |
| Genetic reagent (D. melanogaster) | P{XP}CG9003d09761 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0160860 | Bloomington Drosophila Stock Center #19321 |
| Genetic reagent (D. melanogaster) | P{XP}saltod09417 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0159854 | Bloomington Drosophila Stock Center #19315 |
| Genetic reagent (D. melanogaster) | P{XP}CG11200d02302 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0162606 | Bloomington Drosophila Stock Center #19173 |
| Genetic reagent (D. melanogaster) | P{XP}sobd06074 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0158622 | Bloomington Drosophila Stock Center #19230 |
| Genetic reagent (D. melanogaster) | PBac{WH}Urof04888 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0159557 | Bloomington Drosophila Stock Center #18814 |
| Genetic reagent (D. melanogaster) | PBac{RB}Ssb-c31ae02272 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0159728 | Bloomington Drosophila Stock Center #18032 |
| Genetic reagent (D. melanogaster) | PBac{RB}CG31898e03937 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0161732 | Bloomington Drosophila Stock Center #18211 |
| Genetic reagent (D. melanogaster) | PBac{WH}CG9582f00857 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0160790 | Bloomington Drosophila Stock Center #18378 |
| Genetic reagent (D. melanogaster) | PBac{WH}teif00157 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0159247 | Bloomington Drosophila Stock Center #18299 |
| Genetic reagent (D. melanogaster) | PBac{RB}CG13018e00535 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0162408 | Bloomington Drosophila Stock Center #17863 |
| Genetic reagent (D. melanogaster) | PBac{RB}Cpr51Ae03998 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0162723 | Bloomington Drosophila Stock Center #18221 |
| Genetic reagent (D. melanogaster) | PBac{WH}CG10384f04349 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0162694 | Bloomington Drosophila Stock Center #18762 |
| Genetic reagent (D. melanogaster) | PBac{WH}CG42260f00464 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0225307 | |
| Genetic reagent (D. melanogaster) | PBac{WH}Jafrac2f01922 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0160273 | Bloomington Drosophila Stock Center #18489 |
| Genetic reagent (D. melanogaster) | PBac{WH}CG17746f05041 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0162020 | Bloomington Drosophila Stock Center #18834 |
| Genetic reagent (D. melanogaster) | P{XP}Leashd06455 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0161383 | Bloomington Drosophila Stock Center #19240 |
| Genetic reagent (D. melanogaster) | P{XP}cud05983 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0158886 | Bloomington Drosophila Stock Center #19225 |
| Genetic reagent (D. melanogaster) | w1118; P{XP}d06796/TM6B, Tb1 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBti0042862 | Bloomington Drosophila Stock Center #19250 |
| Genetic reagent (D. melanogaster) | P{XP}CG10311d06928 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0162706 | Bloomington Drosophila Stock Center #19255 |
| Genetic reagent (D. melanogaster) | P{XP}d02570 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBti0054904 | |
| Genetic reagent (D. melanogaster) | P{XP}wrm1d02813 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0160902 | Bloomington Drosophila Stock Center #19182 |
| Genetic reagent (D. melanogaster) | P{XP}UGPd07256 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0159573 | Bloomington Drosophila Stock Center #19267 |
| Genetic reagent (D. melanogaster) | PBac{WH}CG14894f04937 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0162215 | Bloomington Drosophila Stock Center #18821 |
| Genetic reagent (D. melanogaster) | PBac{RB}Cad89De03186 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0160726 | Bloomington Drosophila Stock Center #18129 |
| Genetic reagent (D. melanogaster) | PBac{WH}Actn3f00971 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0162831 | Bloomington Drosophila Stock Center #18397 |
| Genetic reagent (D. melanogaster) | PBac{RB}qine03728 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0162275 | Bloomington Drosophila Stock Center #18186 |
| Genetic reagent (D. melanogaster) | PBac{RB}DPCoACe03144 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0175762 | Bloomington Drosophila Stock Center #18121 |
| Genetic reagent (D. melanogaster) | PBac{WH}KaiR1Df03502 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0161451 | Bloomington Drosophila Stock Center #18663 |
| Genetic reagent (D. melanogaster) | PBac{WH}TotCf01700 | Xu and Rubin, 1993; Thibault et al., 2004 | FLYBASE:FBal0159656 | Bloomington Drosophila Stock Center #18460 |
| Antibody | polyclonalRabbit anti-XRP1(short) | Francis et al., 2016 | (1:200) dilution | |
| Antibody | Polyclonal Rabbit anti-active-Dcp1 | Cell Signalling Technology | Cat #9578 | (1:50) dilution |
| Antibody | Polyclonal Donkey anti-Rabbit IgG, Cy3 conjugate | Jackson Immunoresearch | Cat # 711-165-152 | (1:200) dilution |
| Recombinant DNA reagent | P{DsRed; RpL28+} | This study | See Materials and methods; Dr. Nicholas Baker’s lab. | |
| Recombinant DNA reagent | pGE-attBTT-loxP-DsRed (pTL780) | Blanco-Redondo and Langenhan, 2018 | Addgene Plasmid #115160 |
Additional files
-
Supplementary file 1
FRT insertions used in this study and their combinations.
Columns indicate the specific insertion, genome location, and cytological position of the elements used as the left FRT site. Rows indicate the same information for the right FRT site. Insertions in the ‘d’ family are of the P{XP} element, the ‘e’ family the PBac{RB} element, and ‘f’ family PBac{WH} (Thibault et al., 2004). For clarity, in the main text we refer to genetic strains by the cytological insertion point eg ‘63A3-65A9’ is the shorthand descriptive name for the PBac{WH}f01922 P{XP}d02570 chromosome. Which Rp genes are included between FRT sites is shown, as is eIF2γ. Some FRT combinations were poor substrates for Flp, either retained the parental eye color in the presence of eyFlp, or produce a salt and pepper pattern of very small clones that is indicative of excision occurring only late in development once large cell numbers are present (Figure 2U–Z). These results are summarized by shading FRT combinations tested as follows: Green – eyFlp recombination in essentially all cells; Blue – eyFlp recombination in most cells, associated with small eye size (≤0.5 linear dimensions); magenta – eyFlp recombination not detected; Orange – eyFlp recombination only late in development gives a mottled eye. The interpretation that recombination is reduced or absent is preferred to the alternative possibility that excision results in a cell-lethal genotype that later disappears, in part because results correlated with individual FRT elements and not with the genetic material between them. For example, recombination between 26A1 and 28F3 or 29C1, revealed only small, late recombination, but the 26A1-29F8 recombination that deletes all the same sequences was completely excised from EyFlp eyes and developed normal eye size with entirely Df(2L)26A1-29F8/+ cells (Figure 2D). Other examples of recombinations that could not readily be obtained were 87B8-89B18, 87B8-91B8, 87B8-92F1 and 89B13-92F1, although the larger 89B8-93A2 and 89B13-93A2 recombinations were readily obtained (Figure 2R–T). In contrast to the lack of correlation with deleted chromosome regions, when an element was not recombined by eyFLP this was the case with all the partner elements tested, so each FRT element could be designated as green or orange/magenta without ambiguity (for the 21-23/4 region elements there is insufficient information to identify the particular non-recombining elements). These data suggest that some FRT-containing Exelixis elements are poor substrates for cis-recombination in the head. Interestingly, all 7 insertions of the PBac (Adams and Cory, 1998) element tested belong in this category, although this element has previously been recombined successfully in the germline (Parks et al., 2004).
- https://cdn.elifesciences.org/articles/61172/elife-61172-supp1-v1.xlsx
-
Supplementary file 2
Statistical comparisons of segmental-aneuploid cell contribution to adult eyes.
109 pairwise comparison between segmental aneuploid genotypes were performed in this study, which requires multiple testing correction. The table shows each comparison ranked according to raw p-value (Mann-Whitney), Benjamini-Hochberg critical value for FDR ≤ 0.05, adjusted p-value, and significance.
- https://cdn.elifesciences.org/articles/61172/elife-61172-supp2-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61172/elife-61172-transrepform-v1.pdf





