The half-life of the bone-derived hormone osteocalcin is regulated through O-glycosylation in mice, but not in humans
Figures
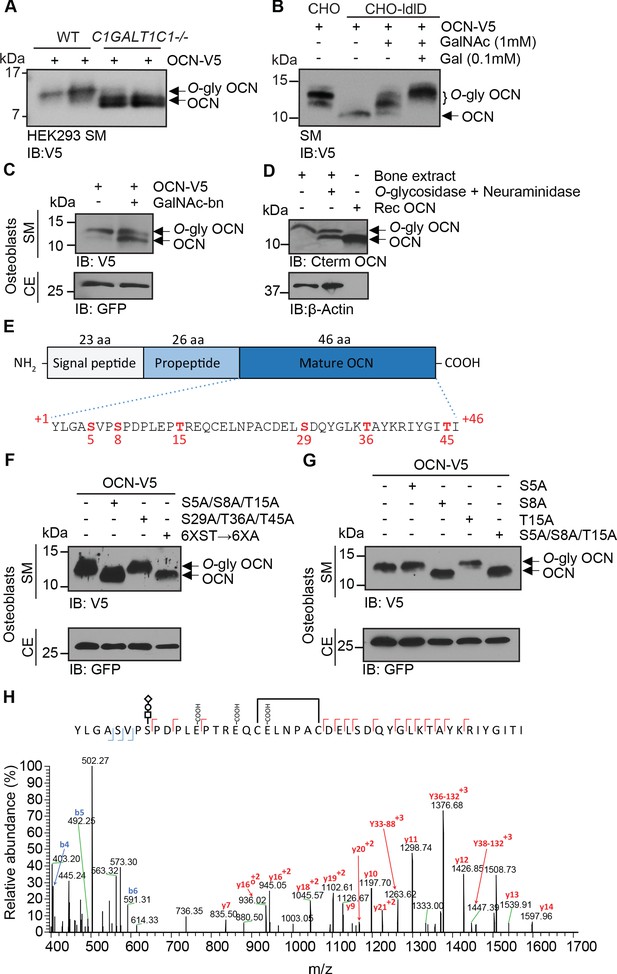
OCN is O-glycosylated in vitro and in vivo on serine 8.
(A) Western blot analysis on the secretion medium (SM) of HEK293 (WT) and HEK293 lacking COSMC (C1GALT1C1-/-) transfected with mouse OCN-V5. (B) Western blot analysis on the SM of CHO and CHO-ldlD cells transfected with mouse OCN-V5. CHO-ldlD cells were treated or not with 0.1 mM Galactose (Gal) and/or 1 mM N-acetylgalactosamine (GalNAc). (C) Effect of N-acetylgalactosaminyltransferase (GalNAc-Ts) inhibition on mouse OCN O-glycosylation in osteoblasts. Western blot analysis on the SM and cell extract (CE) of primary osteoblasts transfected with mouse OCN-V5 and treated or not with 2 mM of GalNAc-bn. (D) OCN deglycosylation assay. Bone extracts of C57BL/6J mice were treated or not with O-glycosidase and neuraminidase for 4 hr at 37°C and analyzed by western blot using anti-C-termimal OCN antibody (Cterm OCN). β-actin was used as a loading control. Rec OCN: Non-glycosylated OCN produced in bacteria. (E) Structure of mouse pre-pro-OCN and amino acid sequence of mature mouse OCN. The serine (S) and threonine (S) residues are in red. (F) Western blot analysis on the SM and cell extract (CE) of primary osteoblasts transfected with OCN-V5 containing or not the indicated mutations. In the 6XST→6XA mutant, all six serine and threonine residues from OCN were mutated to alanine. (G) Western blot analysis on the SM and cell extract (CE) of primary osteoblasts transfected with OCN-V5 containing or not the indicated mutations. (H) Annotated HCD MS/MS spectrum of a modified form of OCN (HexNAc-Hex-NANA + 3 Gla + S-S) pulled down from the bone homogenate of C57BL/6J mice. The precursor m/z value is 1180.95003 (M+5H)+5 and mass accuracy with the annotated OCN modified form is 4.6 ppm. In C, F and G, GFP co-expressed from OCN-V5 expression vector, was used as a loading control.
-
Figure 1—source data 1
Original western blot image from Figure 1A.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Original western blot image from Figure 1B.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig1-data2-v2.xlsx
-
Figure 1—source data 3
Original western blot image from Figure 1C.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig1-data3-v2.xlsx
-
Figure 1—source data 4
Original western blot image from Figure 1D.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig1-data4-v2.xlsx
-
Figure 1—source data 5
Original western blot image from Figure 1F.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig1-data5-v2.xlsx
-
Figure 1—source data 6
Original western blot image from Figure 1G.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig1-data6-v2.xlsx
-
Figure 1—source data 7
Raw proteomic data from Figure 1H.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig1-data7-v2.xlsx
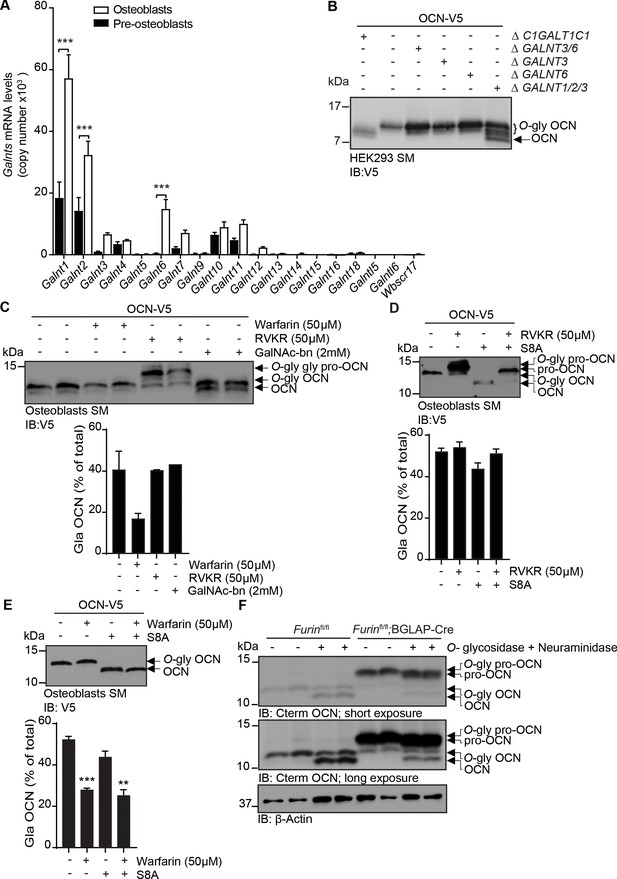
OCN O-glycosylation by N-acetylgalactosaminyltransferase (GalNAc-Ts) is independent of its processing and γ-carboxylation.
(A) Galnts expression in pre-osteoblasts (undifferentiated) and osteoblasts (differentiated) by quantitative PCR (n = 3 per condition). Results are represented as copy number of Galnts normalized to Actb. (B) Western blot analysis of OCN in the secretion media (SM) of HEK293 cells deficient for specific GalNAc-Ts. OCN-V5 was transfected in parental, C1GALT1C1-/- (Δ C1GALT1C1), or GALNTs deficient (Δ) HEK293 cells and analysed by western blot using anti-V5 antibody. (C) Western blot analysis on the SM of osteoblasts transfected with mouse OCN-V5 and treated or not with 2 mM of GalNAc-bn, 50 μM warfarin or 50 μM Dec-RVKR-CMK (RVKR) (upper panel), and percentage of carboxylated OCN (Gla-OCN) over total OCN measured by ELISA (lower panel; n = 2 per condition). (D) Western blot analysis on the SM of osteoblasts transfected with mouse OCN-V5 containing or not the S8A mutation and treated with 50 μM Dec-RVKR-CMK (RVKR) (upper panel), and percentage of carboxylated OCN (Gla-OCN) over total OCN measured by ELISA (lower panel; n = 3 per condition). (E) Western blot analysis on the SM of osteoblasts transfected with mouse OCN-V5 containing or not the S8A mutation and treated with 50 μM warfarin (upper panel), and percentage of carboxylated OCN over total OCN measured by ELISA (lower panel; n = 3 per condition). (F) Western blot analysis of OCN deglycosylation assay on bone extracts from Furinfl/fl and Furinfl/fl;BGLAP-Cre mice (n = 2 independent mice per genotype). Bone extracts were treated or not with O-glycosidase and neuraminidase for 4 hr at 37°C and analyzed by western blot using anti-C-termimal OCN antibody (Cterm OCN). **p<0.01; ***p<0.001 using one-way ANOVA with Bonferroni multiple comparisons test.
-
Figure 2—source data 1
Numerical data from the graph in Figure 2A.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Original western blot image from Figure 2B.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Original western blot image from Figure 2C.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig2-data3-v2.xlsx
-
Figure 2—source data 4
Numerical data from the graph in Figure 2C.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig2-data4-v2.xlsx
-
Figure 2—source data 5
Original western blot image from Figure 2D.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig2-data5-v2.xlsx
-
Figure 2—source data 6
Numerical data from the graph in Figure 2D.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig2-data6-v2.xlsx
-
Figure 2—source data 7
Original western blot image from Figure 2E.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig2-data7-v2.xlsx
-
Figure 2—source data 8
Numerical data from the graph in Figure 2E.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig2-data8-v2.xlsx
-
Figure 2—source data 9
Original western blot image from Figure 2F.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig2-data9-v2.xlsx

Mouse OCN O-glycosylation occurs independently of its carboxylation and processing in HEK293 cells.
Western blot analysis on the secretion media (SM) of HEK293 cells transfected with mouse OCN-V5 and treated or not with 2 mM of GalNAc-bn, 50 μM warfarin or 50 μM Dec-RVKR-CMK (RVKR).
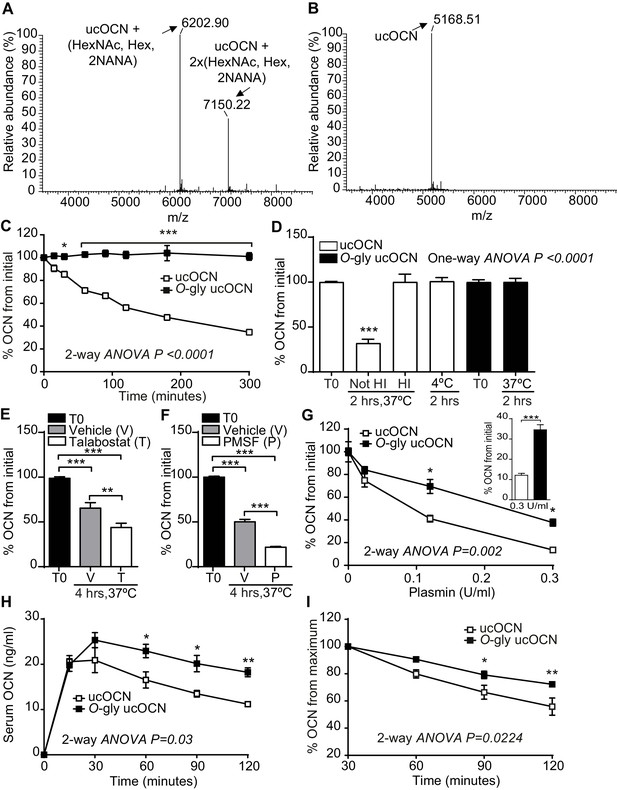
Mouse OCN O-glycosylation increases its half-life ex vivo and in vivo.
(A) Annotated and deconvoluted MS spectrum of purified glycosylated mouse OCN (O-gly ucOCN). (B) Annotated and deconvoluted MS spectrum of purified non-glycosylated mouse OCN (ucOCN). (C–D) Ex vivo half-life of O-gly ucOCN and ucOCN in OCN deficient (Bglap-/-) plasma. (C) 100 ng/ml of O-gly ucOCN or ucOCN were incubated in plasma at 37°C for 0 to 5 hr and OCN levels were measured at the indicated times (n = 4 independent plasma per condition). (D) 100 ng/ml of O-gly ucOCN or ucOCN were incubated in plasma for 2 hr at 37°C in different conditions or at 4°C. HI: heat-inactivated plasma (n = 3 independent plasma per condition). (E) 50 ng/ml of ucOCN was incubated in plasma from Bglap-/- mice for 4 hr at 37°C in the presence of vehicle (V) or 10 mM talabostat (T) (n = 3 per condition). (F) 50 ng/ml of ucOCN was incubated in plasma from Bglap-/- mice for 4 hr at 37°C in the presence of vehicle (V) or 10 mM phenylmethylsulfonyl fluoride (PMSF) (n = 3 per condition). (G) Effect of plasmin on OCN stability ex vivo. 50 ng/ml of O-gly ucOCN and ucOCN were incubated for two hours in Bglap-/- heat-inactivated plasma containing different concentration of plasmin (n = 2 per condition). Inset graph shows the stability of the O-gly ucOCN and ucOCN incubated for two hours with 0.3 U/ml of plasmin (n = 5). (H) OCN deficient male mice (Bglap-/-) were fasted for 16 hr, O-gly ucOCN (n = 5 mice) or ucOCN (n = 5 mice) were injected intraperitoneally at a dose of 40 ng/g of body weight and serum OCN levels were measured at the indicated time points. (I) Using the data in (H) the percentage (%) of OCN in the declining phase was calculated relative to the maxima of each OCN forms at 30 min. T0: start point, see Materials and methods; HexNAc: N-acetylhexosamine; Hex: Hexose; NANA: N-acetylneuraminic acid. OCN measurements were performed using total mouse OCN ELISA assay (see Materials and methods). Results are given as mean ± SEM. *p<0.05; **p<0.01; ***p<0.001 using two-way ANOVA for repeated measurements with Bonferroni multiple comparisons test.
-
Figure 3—source data 1
Raw proteomic data from Figure 3A.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Raw proteomic data from Figure 3B.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-data2-v2.xlsx
-
Figure 3—source data 3
Numerical data from the graph in Figure 3C.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-data3-v2.xlsx
-
Figure 3—source data 4
Numerical data from the graph in Figure 3D.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-data4-v2.xlsx
-
Figure 3—source data 5
Numerical data from the graph in Figure 3E.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-data5-v2.xlsx
-
Figure 3—source data 6
Numerical data from the graph in Figure 3F.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-data6-v2.xlsx
-
Figure 3—source data 7
Numerical data from the graph in Figure 3G.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-data7-v2.xlsx
-
Figure 3—source data 8
Numerical data from the graph in Figure 3H.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-data8-v2.xlsx
-
Figure 3—source data 9
Numerical data from the graph in Figure 3I.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-data9-v2.xlsx

Purification of recombinant O-glycosylated mouse ucOCN.
(A) Map of the pcDNA3.1-Fc-hinge-Thr-OCN construct used to produce and purify mouse ucOCN fusion protein. (B) Coomassie staining of purified O-glycosylated mouse ucOCN (O-gly ucOCN) compared to non- glycosylated mouse ucOCN (ucOCN) produced in bacteria.
-
Figure 3—figure supplement 1—source data 1
Original gel image from Figure 3—figure supplement 1 (panel B).
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-figsupp1-data1-v2.xlsx

Mouse OCN ELISA recognize O-glycosylated mouse ucOCN (O-gly ucOCN) and non-glycosylated mouse ucOCN (ucOCN).
Standard curve of O-gly ucOCN (n = 3–5) and ucOCN (n = 3–5) ranging from 0 to 100 ng/ml.
-
Figure 3—figure supplement 2—source data 1
Numerical data from the graph in Figure 3—figure supplement 2.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-figsupp2-data1-v2.xlsx
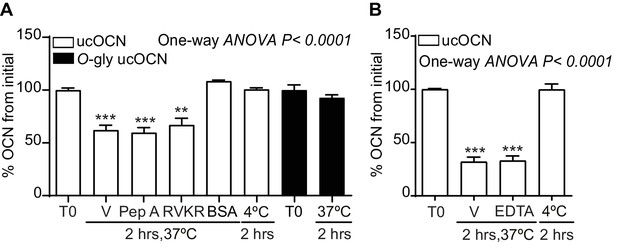
Effect of different protease inhibitors on non-glycosylated mouse ucOCN plasma half-life.
(A–B) Ex vivo half-life of O-gly ucOCN and ucOCN in OCN-deficient plasma (Bglap-/-; n = 3 plasma for each condition). (A) 100 ng/ml O-gly ucOCN (n = 3) and ucOCN (n = 3) were incubated for 2 hr in plasma at 37°C or 4°C and treated with vehicle (V) or with protease inhibitors or incubated in 3.5% BSA (bovine serum albumin prepared in saline solution). (B) 100 ng/ml O-gly ucOCN and ucOCN was incubated for 2 hr in normal plasma at 37°C or 4°C and treated with vehicle (V) or with EDTA (10 mM). T0: start point, see Materials and methods; Pep A: 10 μM Pepstatin A; RVKR: 50 μM Dec-RVKR-CMK.
-
Figure 3—figure supplement 3—source data 1
Numerical data from the graph in Figure 3—figure supplement 3 (panel A).
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-figsupp3-data1-v2.xlsx
-
Figure 3—figure supplement 3—source data 2
Numerical data from the graph in Figure 3—figure supplement 3 (panel B).
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-figsupp3-data2-v2.xlsx
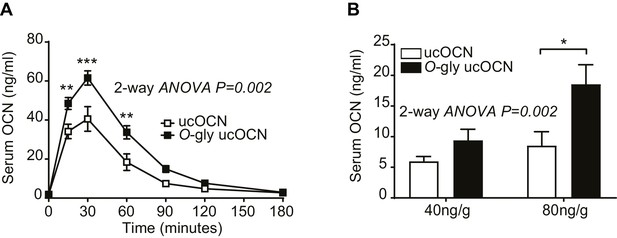
Mouse OCN O-glycosylation increases its stability in vivo in fed conditions.
(A) In vivo stability of O-gly ucOCN and ucOCN in fed condition in mice. O-gly ucOCN or ucOCN were injected intraperitoneally in OCN deficient male mice (Bglap-/-; n = 9 mice each) at a dose of 40 ng/g of body weight and serum OCN levels were measured at the indicated time points. (B) O-gly ucOCN (n = 4 mice) or ucOCN (n = 4 mice) were injected intraperitoneally in OCN deficient male mice (Bglap-/-), in fed condition, at a dose of 40 or 80 ng/g of body weight. Serum OCN level was measured two hours post-injection.
-
Figure 3—figure supplement 4—source data 1
Numerical data from the graph in Figure 3—figure supplement 4 (panel A).
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-figsupp4-data1-v2.xlsx
-
Figure 3—figure supplement 4—source data 2
Numerical data from the graph in Figure 3—figure supplement 4 (panel B).
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig3-figsupp4-data2-v2.xlsx
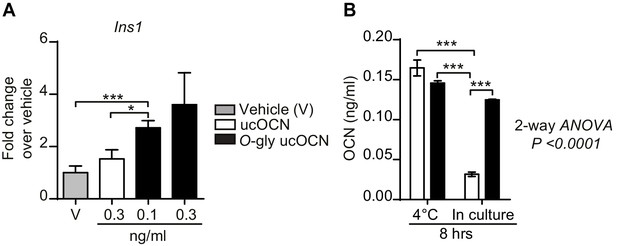
Effect of O-glycosylation on mouse OCN bioactivity and stability in culture.
(A) Insulin gene expression (Ins1) in INS-1 832/3 cells following an 8 hr treatment with vehicle (n = 6), non-glycosylated mouse OCN (ucOCN) (n = 10) or glycosylated mouse ucOCN (O-gly ucOCN) (n = 8–10) at the indicated concentrations. (B) Concentration of OCN in the media incubated for 8 hr at 4°C without cells or for 8 hr in culture with INS-1 832/3 cells at 37°C (n = 3 for each condition). Results are shown as mean ± SEM. *p<0.05; ***p<0.001 using Student t-test or ordinary two-way ANOVA with Bonferroni multiple comparisons test.
-
Figure 4—source data 1
Numerical data from the graph in Figure 4A.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Numerical data from the graph in Figure 4B.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig4-data2-v2.xlsx
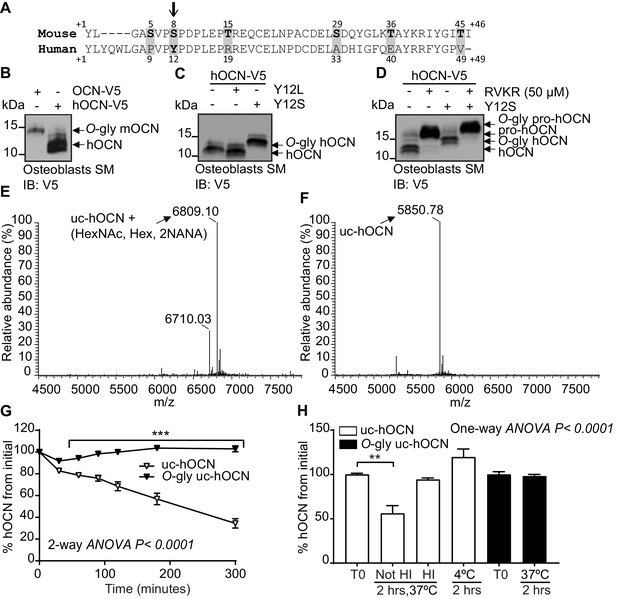
Human OCN O-glycosylation increases its half-life ex vivo.
(A) Amino acid alignment of mouse and human OCN. The six serine and threonine residues present in the mouse protein and their corresponding amino acids in human OCN are highlighted in gray. The site of O-glycosylation in mouse OCN (S8) is indicated by an arrow. (B) Western Blot analysis on the secretion media (SM) of primary osteoblasts transfected with human OCN-V5 (hOCN) and mouse OCN-V5 (OCN). (C) Western blot analysis on the SM of primary osteoblasts transfected with hOCN-V5 containing or not the indicated mutations. (D) Western Blot analysis on the SM of primary osteoblasts transfected with hOCN-V5 containing or not the Y12S mutations and treated or not with 50 μM Dec-RVKR-CMK (RVKR). (E) Annotated and deconvoluted MS spectrum of purified O-glycosylated uncarboxylated human OCN (O-gly uc-hOCN). (F) Annotated and deconvoluted MS spectrum of and purified non-glycosylated uncarboxylated human OCN (uc-hOCN). (G–H) Ex vivo half-life of O-gly uc-hOCN and uc-hOCN in mouse plasma. (G) 60 ng/ml of O-gly uc-hOCN and uc-hOCN were incubated at 37°C in plasma of OCN deficient mice (Bglap-/-) (n = 4) for 0 to 5 hr and hOCN levels were measured at the indicated time. (H) O-gly uc-hOCN and uc-hOCN were incubated in Bglap-/- plasma for 2 hr at 37°C in different conditions or at 4°C (n = 4 independent plasma per condition). T0: start point, see Materials and methods; HI: heat-inactivated plasma; HexNAc: N-acetylhexosamine; Hex: Hexose; NANA: N-acetylneuraminic acid. Uc-hOCN levels were measured at the indicated time points using an uc-hOCN ELISA assay. Results are given as mean ± SEM. **p<0.01; ***p<0.001 using two-way ANOVA for repeated measurements with Bonferroni multiple comparisons test.
-
Figure 5—source data 1
Original western blot image from Figure 5B.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Original western blot image from Figure 5C.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig5-data2-v2.xlsx
-
Figure 5—source data 3
Original western blot image from Figure 5D.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig5-data3-v2.xlsx
-
Figure 5—source data 4
Raw proteomic data from Figure 5E.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig5-data4-v2.xlsx
-
Figure 5—source data 5
Raw proteomic data from Figure 5F.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig5-data5-v2.xlsx
-
Figure 5—source data 6
Numerical data from the graph in Figure 5G.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig5-data6-v2.xlsx
-
Figure 5—source data 7
Numerical data from the graph in Figure 5H.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig5-data7-v2.xlsx
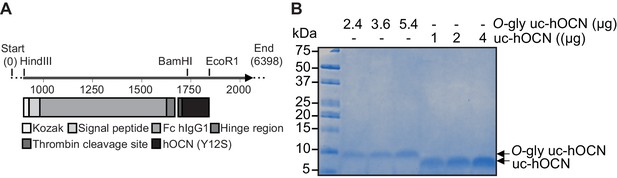
Purification of recombinant O-glycosylated human ucOCN.
(A) Map of pcDNA3.1-Fc-hinge-Thr-hOCN (Y12S) construct used to produce and purify O-glycosylated human ucOCN fusion protein. (B) Coomassie staining of purified O-glycosylated human ucOCN (O-gly uchOCN) compared to non-glycosylated human ucOCN (uc-hOCN) produced in bacteria.
-
Figure 5—figure supplement 1—source data 1
Original gel image from Figure 5—figure supplement 1 (panel B).
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig5-figsupp1-data1-v2.xlsx
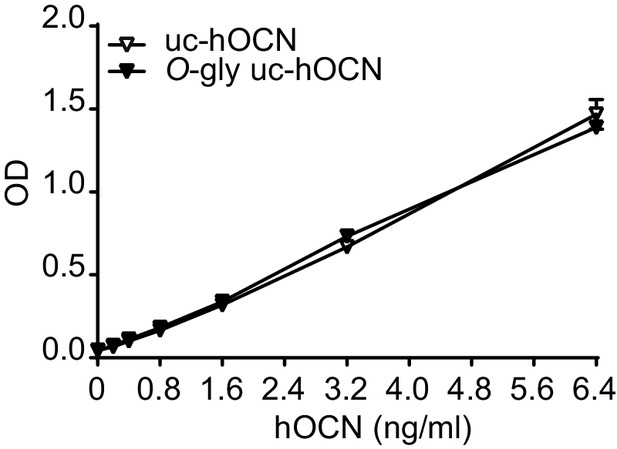
Human ucOCN ELISA equally recognize O-glycosylated human ucOCN (O-gly uc-hOCN) and non-glycosylated human ucOCN (uc-hOCN).
Standard curve of O-gly uc-hOCN (n = 2) and uc-hOCN (n = 2) ranging from 0 to 100 ng/ml.
-
Figure 5—figure supplement 2—source data 1
Numerical data from the graph in Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig5-figsupp2-data1-v2.xlsx
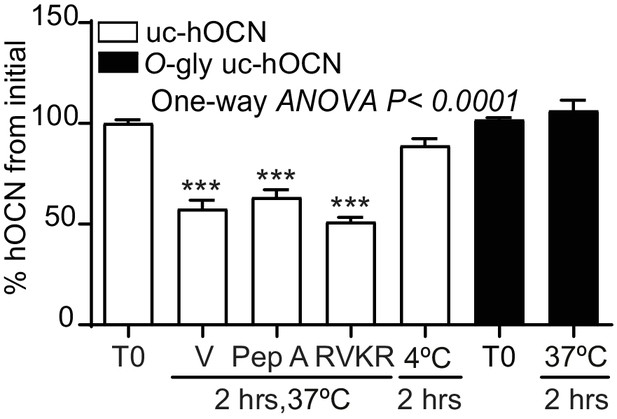
Effect of different protease inhibitors on non-glycosylated human ucOCN plasma half-life.
Ex vivo half-life of O-gly uc-hOCN and uc-hOCN in OCN deficient plasma (Bglap-/-; n = 3–4 plasma). 650 ng/ml O-gly uc-hOCN (n = 3) and uc-hOCN (n = 3) were incubated for 2 hr in normal plasma at 37°C or 4°C and treated with vehicle (V) or with protease inhibitors. T0: start point, see Materials and methods; Pep A: 10 μM Pepstatin A; RVKR: 50 μM Dec-RVKR-CMK.
-
Figure 5—figure supplement 3—source data 1
Numerical data from the graph in Figure 5—figure supplement 3.
- https://cdn.elifesciences.org/articles/61174/elife-61174-fig5-figsupp3-data1-v2.xlsx
Tables
OCN serum levels in mouse and human at different ages.
| OCN serum levels (ng/ml) | ||
|---|---|---|
| Age (mice/human) | Mouse [mean ± SD (n)] | Human* [mean ± SD (n)] |
| 2 weeks/ 1 year old | 1369.7 ± 146.7 (8) | 62.9 ± 8.1 (43) |
| 4 weeks/ 11–13 years old | 617.2 ± 192.5 (5) | 74.1 ± 8.9 (41) |
| 13 weeks/ 25–29 years old | 252.2 ± 8.0 (4) | 21.0 ± 6.3 (49) |
| 60 weeks/ 50–54 years old | 50.0 ± 7.2 (4) | 13.5 ± 6.3 (127) |
-
Table 1—source data 1
Numerical data from Table 1.
- https://cdn.elifesciences.org/articles/61174/elife-61174-table1-data1-v2.xlsx
The monoisotopic mass and relative abundance of the different OCN forms detected in the supernatant of differentiated osteoblasts.
| Monoisotopic mass range (Da) | Relative abundance (%) | Most probable modification | Most probable oligosaccharide |
|---|---|---|---|
| O-glycosylated OCN | 83.88 | ||
| 5767.6961 | 4.80 | Glycosylation | HexNAc, Hex, NANA |
| 5783.6801 | 0.25 | Glycosylation + oxidation | HexNAc, Hex, NANA |
| 5855.6676 | 4.60 | Glycosylation + 2x Gla | HexNAc, Hex, NANA |
| 5899.7161 | 3.16 | Glycosylation + 3x Gla | HexNAc, Hex, NANA |
| 5915.6386–5968.5796 | 5.51 | Glycosylation + 3x Gla + oxidation + additional unidentified modifications or adduct ions | HexNAc, Hex, NANA |
| 6058.7916 | 24.48 | Glycosylation | HexNAc, Hex, 2x NANA |
| 6074.7766–6096.7409 | 2.48 | Glycosylation + oxidation | HexNAc, Hex, 2x NANA |
| 6102.7681 | 7.89 | Glycosylation + 1x Gla | HexNAc, Hex, 2x NANA |
| 6146.7609 | 23.97 | Glycosylation + 2x Gla | HexNAc, Hex, 2x NANA |
| 6190.8061–6214.7278 | 6.72 | Glycosylation + 3x Gla + additional unidentified modifications or adduct ions | HexNAc, Hex, 2x NANA |
| Non O-glycosylated OCN | 16.12 | ||
| 5127.4676 | 3.00 | Oxidation | NA |
| 5171.4301 | 1.85 | 1x Gla + oxidation | NA |
| 5199.4446 | 2.08 | 2x Gla | NA |
| 5215.4296 | 8.50 | 2x Gla + oxidation | NA |
| 5259.4204 | 0.66 | 3x Gla + oxidation | NA |
-
Gla: Gamma-carboxyglutamic acid residue; HexNAc: N-acetylhexosamine; Hex: Hexose; NANA: N-acetylneuraminic acid; 1x: one time; 2x: two times; 3x: three times; NA: not applicable.
-
Table 2—source data 1
Raw proteomic data from Table 2.
- https://cdn.elifesciences.org/articles/61174/elife-61174-table2-data1-v2.xlsx
The monoisotopic mass and relative abundance of the different OCN forms detected in mouse bone homogenates.
| Monoisotopic mass range | Relative abundance (%) | Most probable modification | Most probable oligosaccharide |
|---|---|---|---|
| O-glycosylated OCN | 99.07 | ||
| 5855.6676 | 0.54 | Glycosylation + 2x Gla | HexNAc, Hex, NANA |
| 5899.7161 | 7.43 | Glycosylation + 3x Gla | HexNAc, Hex, NANA |
| 5915.6386–6135.6796 | 36.07 | Glycosylation + 3x Gla + oxidation + additional unidentified modifications or adduct ions | HexNAc, Hex, NANA |
| 6146.7609–6162.7991 | 5.42 | Glycosylation + 2x Gla + additional unidentified modifications | HexNAc, Hex, 2xNANA |
| 6190.8061 | 4.49 | Glycosylation + 3x Gla | HexNAc, Hex, 2xNANA |
| 6206.8016–6441.7636 | 45.12 | Glycosylation + 3x Gla + oxidation + additional unidentified modifications or adduct ions | HexNAc, Hex, 2xNANA |
| Non O-glycosylated OCN | 0.93 | ||
| 5259.4204 | 0.93 | 3x Gla + oxidation | NA |
-
Gla: Gamma-carboxyglutamic acid residue; HexNAc: N-acetylhexosamine; Hex: Hexose; NANA: N-acetylneuraminic acid; 1x: one time; 2x: two times; 3x: three times; NA: not applicable.
-
Table 3—source data 1
Raw proteomic data from Table 3.
- https://cdn.elifesciences.org/articles/61174/elife-61174-table3-data1-v2.xlsx
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (M. musculus) | Bglap | GenBank | Gene ID: 12096 | Mouse osteocalcin gene 1 |
| Gene (M. musculus) | Bglap2 | GenBank | Gene ID: 12097 | Mouse osteocalcin gene 2 |
| Gene (Homo sapiens) | BGLAP | GenBank | Gene ID: 632 | Human osteocalcin gene |
| Genetic reagent (M. musculus) | Bglap-/- | PMID:8684484 | Bglap/Bglap2tm1Kry RRID:MGI:3837364 | Genetic background: C57BL/6J |
| Genetic reagent (M. musculus) | Furinfl/fl | PMID:15471862 | Furintm1Jwmc RRID:MGI:3700793 | Genetic background: C57BL/6J |
| Genetic reagent (M. musculus) | BGLAP-Cre | PMID:12215457 | Tg(BGLAP-cre)1Clem RRID:IMSR_JAX:019509 | Genetic background: C57BL/6J |
| Genetic reagent (M. musculus) | C57BL/6J wildtype mice | The Jackson Laboratory | Stock No: 000664 RRID:IMSR_JAX:000664 | For primary osteoblasts preparation |
| Cell line (R. norvegicus) | INS-1 832/3 | Millipore-Sigma | SCC208 RRID:CVCL_ZL55 | |
| Cell line (C. griseus) | Chinese hamster ovary (CHO-K1) cells | ATCC | CCL-61 RRID:CVCL_0214 | |
| Cell line (C. griseus) | Chinese hamster ovary ldlD cells (CHO-ldlD) | PMID:3948246 | RRID:CVCL_1V03 | Cell maintained in N. Seidah lab. |
| Cell line (M. musculus) | Primary osteoblasts | This paper | Prepared from C57BL/6J wildtype mice newborn calvaria | |
| Cell line (H. sapiens) | Human embryonic kidney cells HEK293 | ATCC | CRL-1573 RRID:CVCL_0045 | |
| Cell line (H. sapiens) | COSMC knockout HEK293 cells (C1GALT1C1-/-) | PMID:23584533 | RRID:CVCL_S025 | Cell maintained in H. Clausen lab. |
| Cell line (H. sapiens) | GALNT3/6 knockout HEK293 cells | PMID:31040225 | Cell maintained in H. Clausen lab. | |
| Cell line (H. sapiens) | GALNT3 knockout HEK293 cells | PMID:31040225 | Cell maintained in H. Clausen lab. | |
| Cell line (H. sapiens) | GALNT6 knockout HEK293 cells | PMID:31040225 | Cell maintained in H. Clausen lab. | |
| Cell line (H. sapiens) | GALNT1/2/3 knockout HEK293 cells | PMID:31040225 | Cell maintained in H. Clausen lab. | |
| Transfected construct (M. musculus) | pIRES2-EGFP-mOCN-V5 | This paper | To express mouse OCN V5 tagged in primary osteoblasts, CHO-K1, CHO-ldlD and HEK293 | |
| Transfected construct (M. musculus) | pIRES2- EGFP-mOCN (S5A/S8/AT15A) -V5 | This paper | To express (S5A/S8/AT15A) mutant mouse OCN V5 tagged in primary osteoblasts | |
| Transfected construct (M. musculus) | pIRES2- EGFP-mOCN (S29A/T36A/T45A) -V5 | This paper | To express (S29A/T36A/T45A) mutant mouse OCN V5 tagged in primary osteoblasts | |
| Transfected construct (M. musculus) | pIRES2- EGFP-mOCN (6XST→6XA)-V5 | This paper | To express (6XST→6XA) mutant mouse OCN V5 tagged in primary osteoblasts | |
| Transfected construct (M. musculus) | pIRES2- EGFP-mOCN (S5A)-V5 | This paper | To express (S5A) mutant mouse OCN V5 tagged in primary osteoblasts | |
| Transfected construct (M. musculus) | pIRES2- EGFP-mOCN (S8A)-V5 | This paper | To express (S8A) mutant mouse OCN V5 tagged in primary osteoblasts | |
| Transfected construct (M. musculus) | pIRES2- EGFP-mOCN (T15A)-V5 | This paper | To express (T15A) mutant mouse OCN V5 tagged in primary osteoblasts | |
| Transfected construct (H. sapiens) | pIRES2-EGFP-hOCN-V5 | This paper | To express human OCN V5 tagged in primary osteoblasts | |
| Transfected construct (H. sapiens) | pIRES2-EGFP-hOCN (Y12S)-V5 | This paper | To express (Y12S) mutant human OCN V5 tagged in primary osteoblasts | |
| Transfected construct (H. sapiens) | pIRES2-EGFP-hOCN (Y12L)-V5 | This paper | To express (Y12L) human OCN V5 tagged in primary osteoblasts | |
| Transfected construct (H. sapiens) | pcDNA3.1-Fc-hinge-Thr-mOCN | This paper | Used to produce O-gly ucOCN in HEK293 | |
| Transfected construct (H. sapiens) | pcDNA3.1-Fc-hinge-Thr-hOCN (Y12S) | This paper | Used to produce O-gly uc-hOCN in HEK293 | |
| Antibody | Anti-GFP, mouse monoclonal, clones 7.1 and 13.1 | Sigma-Aldrich | 11814460001 RRID:AB_390913 | WB (1:1000) |
| Antibody | Anti-V5, mouse monoclonal, clone V5-10 | Sigma-Aldrich | V8012 RRID:AB_261888 | WB (1:3000) |
| Antibody | Anti–β-actin, mouse monoclonal, clone AC-15 | Sigma-Aldrich | A5441 RRID:AB_476744 | WB (1:7000) |
| Antibody | Anti-Gla-OCN goat polyclonal antibody (recognize amino acids 11–26 of carboxylated mature mouse OCN) | PMID:20570657 | WB (1:3000) ELISA (2 μg/ml) | |
| Antibody | Anti-CTERM OCN goat polyclonal antibody recognize amino acids26–46 of mature mouse OCN | PMID:20570657 | WB (1:3000) ELISA (1:600) IP (1:100) | |
| Antibody | Anti-MID OCN goat polyclonal antibody recognize amino acids11 to 26 of mature mouse OCN | PMID:20570657 | ELISA (1.5 μg/ml) | |
| Recombinant DNA reagent | pTT5-Fc1_CTL | PMID:23951290 | Used as PCR template to amplify Fc and hinge region | |
| Peptide, recombinant protein | Collagenase type 2 | Worthington Biochemical Corporation | LS004176 | For primary osteoblasts preparation |
| Peptide, recombinant protein | O-Glycosidase and Neuraminidase Bundle | NEB | E0540S | Deglycosylation assay |
| Peptide, recombinant protein | Thrombin | GE Healthcare Life Sciences | 27-0846-01 | Protein purification |
| Peptide, recombinant protein | Human plasmin | Sigma | P1867 | |
| Chemical compound, drug | Warfarin | Santa Cruz Biotechnology | sc-205888 | VKORC1 inhibitor |
| Chemical compound, drug | Decamoyl-RVKR-CMK | Tocris | 3501/1 | Furin inhibitor |
| Chemical compound, drug | N-acetylgalactosaminyltransferase inhibitor (GalNAc-bn) | Sigma | 200100 | GalNAc-Ts inhibitor |
| Chemical compound, drug | Benzamidine sepharose | GE healthcare | 17-5123-10 | Protein purification |
| Chemical compound, drug | Pepstatin A | Sigma | P5318 | Aspartyl proteases inhibitor |
| Chemical compound, drug | Talabostat | Tocris, | 3719/10 | FAP inhibitor |
| Chemical compound, drug | Phenylmethylsulfonyl fluoride (PMSF) | Amresco | 329-98-6 | Serine proteases inhibitor |
| Chemical compound, drug | Vitamin K1 | Sigma | V3501 | Cofactor for gamma carboxylation |
| Commercial assay or kit | HiTrap protein A high performance | GE Healthcare Life Sciences | GE29-0485-76 | Protein purification |
| Commercial assay or kit | Human ucOCN ELISA | BioLegend (PMID:31935114) | 446707 | |
| Commercial assay, kit | JetPrime | Polypus transfection | 114–15 | |
| Commercial assay, kit | Lipofectamine 2000 | Thermo Fisher | 11668019 | |
| Software, algorithm | Prism version 7.03 | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | Xcalibur 4.0 | Thermo Fisher Scientific | RRID:SCR_014593 |
Additional key resources.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Sequence-based reagent | mOCN-For-EcoRI | This paper | PCR primers (Cloning of mOCN in pIRES2-EGFP-V5) | AATTGAATTCgCcaccatgaggaccctctctc |
| Sequence-based reagent | mOCN-For-EcoRI | This paper | PCR primer (cloning of mOCN in pIRES2-EGFP-V5) | AATTGAATTCGCCACCATGAGGACCCTCTCTC |
| Sequence-based reagent | mOCN-Rev-Stop-AgeI | This paper | PCR primer (cloning of mOCN in pIRES2-EGFP-V5, No V5 tagged protein) | AATTACCGGTCTAAATAGTGATACCGTAGATG |
| Sequence-based reagent | mOCN-Rev-AgeI | This paper | PCR primer (cloning of mOCN in pIRES2-EGFP-V5) | AATTACCGGTAATAGTGATACCGTAGATGCG |
| Sequence-based reagent | mOCNSTT-stop-Age1-Rev | This paper | PCR primer (cloning of S29A/T36A/T45A mOCN in pIRES2-EGFP-V5, No V5 tagged protein) | AATTACCGGTCTAAATAGCGATACCGTAGATG |
| Sequence-based reagent | mOCNSTT-Age1-Rev | This paper | PCR primer (cloning of S29A/T36A/T45A mOCN in pIRES2-EGFP-V5) | AATTACCGGTAATAGCGATACCGTAGATGCG |
| Sequence-based reagent | mOCN-S5A-For | This paper | PCR primer (mutagenesis of Serine five to Alanine in mOCN) | TACCTTGGAGCCGCCGTCCCCAGCCCA |
| Sequence-based reagent | mOCN-S5A-Rev | This paper | PCR primer (mutagenesis of Serine five to Alanine in mOCN) | TGGGCTGGGGACGGCGGCTCCAAGGTA |
| Sequence-based reagent | mOCN-S8A-For | This paper | PCR primer (mutagenesis of Serine eight to Alanine in mOCN) | GCCTCAGTCCCCGCCCCAGATCCCCTG |
| Sequence-based reagent | mOCN-S8A-Rev | This paper | PCR primer (mutagenesis of Serine eight to Alanine in mOCN) | CAGGGGATCTGGGGCGGGGACTGAGGC |
| Sequence-based reagent | mOCN-T15A-For | This paper | PCR primer (mutagenesis of Threonine 15 to Alanine in mOCN) | CTGGAGCCCGCCCGGGAGCAG |
| Sequence-based reagent | mOCN-T15A-Rev | This paper | PCR primer (mutagenesis of Threonine 15 to Alanine in mOCN) | CTGCTCCCGGGC GGGCTCCAG |
| Sequence-based reagent | hOCN-EcoRI-For | This paper | PCR primer (cloning human osteocalcin in pIRES2-EGFP-V5) | AATTGAATTCGCCACCATGAGAGCCCTCACACTCCT |
| Sequence-based reagent | hOCN-AgeI-Rev | This paper | PCR primer (cloning human osteocalcin in pIRES2-EGFP-V5) | AATT ACCGGT GACCGGGCCGTAGAAGCG |
| Sequence-based reagent | hOCN-Y12S-For | This paper | PCR primer (mutagenesis of Tyrosine 12 to Serine in hOCN) | GCCCCAGTCCCCAGCCCGGATCCCCTG |
| Sequence-based reagent | hOCN-Y12S-Rev | This paper | PCR primer (mutagenesis of Tyrosine 12 to Serine in hOCN) | CAGGGGATCCGGGCTGGGGACTGGGGC |
| Sequence-based reagent | hOCN-Y12L-For | This paper | PCR primer (mutagenesis of Tyrosine 12 to Leucine in hOCN) | GCCCCAGTCCCCCTACCGGATCCCCTG |
| Sequence-based reagent | hOCN-Y12L-Rev | This paper | PCR primer (mutagenesis of Tyrosine 12 to Leucine in hOCN) | CAGGGGATCCGGTAGGGGGACTGGGGC |
| Sequence-based reagent | HindIII-FchIgG1 -For | This paper | PCR primer (amplification of FC fragment+ hinge region in pTT5FC-CTL plasmid, and cloning in pcDNA3 in HindIII-BamHI) | AATTAAGCTTGCCACCATGGAGTTTGGGCTG |
| Sequence-based reagent | BamHI-FchIgG1-Rev | This paper | PCR primer (amplification of FC fragment+ hinge region in pTT5FC-CTL plasmid, and cloning in pcDNA3 in HindIII-BamHI) | AATTGGATCCTGGGCACGGTGGGCATGTG |
| Sequence-based reagent | BamHI-Thrombin-mOCN-For | This paper | PCR primer (cloning of Thrombin mOCN in pcDNA3 FchIgG1 using BamHI-EcoRI) | AATTGGATCCCTGGTTCCGCGTGGATCTTACCTTGGAGCCTCAGTCC |
| Sequence-based reagent | EcoRI-mOCN-Rev | This paper | PCR primer (cloning of Thrombin mOCN in pcDNA3 FchIgG1 using BamHI-EcoRI) | AATTGAATTCCTAAATAGTGATACCGTAGATG |
| Sequence-based reagent | BglII-Thrombin-hOCN- For | This paper | PCR primer (cloning of Thrombin hOCN (Y12S) in pcDNA3 FchIgG1 using BglII-EcoRI) | AATTAGATCTCTGGTTCCGCGTGGATCTTACCTGTATCAATGGCTGG |
| Sequence-based reagent | EcoRI-hOCN-Rev | This paper | PCR primer (cloning of Thrombin hOCN (Y12S) in pcDNA3 FchIgG1 using BglII-EcoRI) | AATTGAATTCCTAGACCGGGCCGTAGAAGCGC |
| Sequence-based reagent | GalnT1-For | This paper | QPCR primer (amplify Galnt1, M. musculus) | GCAGCATGTGAACAGCAATCA |
| Sequence-based reagent | GalnT1-Rev | This paper | QPCR primer (amplify Galnt1, M. musculus) | GCTGAGGTAGCCCAGTCAATC |
| Sequence-based reagent | GalnT2-For | This paper | QPCR primer (amplify Galnt2, M. musculus) | GGCAACTCCAAACTGCGACA |
| Sequence-based reagent | GalnT2-Rev | This paper | QPCR primer (amplify Galnt2, M. musculus) | TCAACAAACTGGGCCGGTG |
| Sequence-based reagent | GalnT3-For | This paper | QPCR primer (amplify Galnt3, M. musculus) | ACTTAGTGCCATGTGACGCA |
| Sequence-based reagent | GalnT3-Rev | This paper | QPCR primer (amplify Galnt3, M. musculus) | GGGTTTCTGCAGCGGTTCTA |
| Sequence-based reagent | GalnT4-For | This paper | QPCR primer (amplify Galnt4, M. musculus) | CAAAACTGCCCCAAAGACGG |
| Sequence-based reagent | GalnT4-Rev | This paper | QPCR primer (amplify Galnt4, M. musculus) | CGCTCTGCTGCTAGCCTATT |
| Sequence-based reagent | GalnT5-For | This paper | QPCR primer (amplify Galnt5, M. musculus) | CCCTGAAACTGGCTGCTTGT |
| Sequence-based reagent | GalnT5-Rev | This paper | QPCR primer (amplify Galnt5, M. musculus) | ATGGAGAGAAATTCAGTCAGCAA |
| Sequence-based reagent | GalnT6-For | This paper | QPCR primer (amplify Galnt6, M. musculus) | CCAGCTCTGGCTGTTTGTCTA |
| Sequence-based reagent | GalnT6-Rev | This paper | QPCR primer (amplify Galnt6, M. musculus) | TTGGGCCAAGTAGCATGTGA |
| Sequence-based reagent | GalnT7-For | This paper | QPCR primer (amplify Galnt7, M. musculus) | GCACAGGTTTACGCACATCA |
| Sequence-based reagent | GalnT7-Rev | This paper | QPCR primer (amplify Galnt7, M. musculus) | TTCCAGGCGGTTTTCAGTCC |
| Sequence-based reagent | GalnT9-For | This paper | QPCR primer (amplify Galnt9, M. musculus) | CAACTTTGGGCTGCGGTTAG |
| Sequence-based reagent | GalnT9-Rev | This paper | QPCR primer (amplify Galnt9, M. musculus) | CCCACATTGCTCTTGGGTCT |
| Sequence-based reagent | GalnT10-For | This paper | QPCR primer (amplify Galnt10, M. musculus) | GGAGTACCGCCACCTCTCAG |
| Sequence-based reagent | GalnT10-Rev | This paper | QPCR primer (amplify Galnt10, M. musculus) | AGGTCCCAGGCAATTTTGGT |
| Sequence-based reagent | GalnT11-For | This paper | QPCR primer (amplify Galnt11, M. musculus) | GGCTGTACCAAGTGTCCGTT |
| Sequence-based reagent | GalnT11-Rev | This paper | QPCR primer (amplify Galnt11, M. musculus) | GCAGGCATGACAAAACCAGG |
| Sequence-based reagent | GalnT12-For | This paper | QPCR primer (amplify Galnt12, M. musculus) | ACAACGGCTTTGCACCATAC |
| Sequence-based reagent | GalnT12-Rev | This paper | QPCR primer (amplify Galnt12, M. musculus) | ACACTCTTGTGACACCCAGC |
| Sequence-based reagent | GalnT13-For | This paper | QPCR primer (amplify Galnt13, M. musculus) | CTGGCAATGTGGAGGTTCTT |
| Sequence-based reagent | GalnT13-Rev | This paper | QPCR primer (amplify Galnt13, M. musculus) | AATTCATCCATCCACACTTCTGC |
| Sequence-based reagent | GalnT14-For | This paper | QPCR primer (amplify Galnt14, M. musculus) | TCTTTCCGAGTGTGGATGTGT |
| Sequence-based reagent | GalnT14-Rev | This paper | QPCR primer (amplify Galnt14, M. musculus) | CCCATCGGGGAAAACATAAGGA |
| Sequence-based reagent | GalnT15-For | This paper | QPCR primer (amplify Galnt15, M. musculus) | CTGCGGTGGCTCTGTTGAAA |
| Sequence-based reagent | GalnT15-Rev | This paper | QPCR primer (amplify Galnt15, M. musculus) | CTGGGATGTGCCTGTAGAAGG |
| Sequence-based reagent | GalnT16-For | This paper | QPCR primer (amplify Galnt16, M. musculus) | TGGTGACCAGCAAATGTCAGA |
| Sequence-based reagent | GalnT16-Rev | This paper | QPCR primer (amplify Galnt16, M. musculus) | TCCGGTCGAAATGTGAGGAG |
| Sequence-based reagent | GalnT18-For | This paper | QPCR primer (amplify Galnt18, M. musculus) | CAGAAGTGCTCGGGACAACA |
| Sequence-based reagent | GalnT18-Rev | This paper | QPCR primer (amplify Galnt18, M. musculus) | TTGGCTCTCCCTCTCAGACT |
| Sequence-based reagent | Galntl5-For | This paper | QPCR primer (amplify Galntl5, M. musculus) | AGTGAGCGCGTGGAATTAAG |
| Sequence-based reagent | Galntl5-Rev | This paper | QPCR primer (amplify Galntl5, M. musculus) | AGATTTGTCCTGTGGTGCGA |
| Sequence-based reagent | Wbscr17-For | This paper | QPCR primer (amplify Wbscr17, M. musculus) | CTTAGGTGCTCTGGGGACCA |
| Sequence-based reagent | Wbscr17-Rev | This paper | QPCR primer (amplify Wbscr17, M. musculus) | TGTACAAGCTGCTCTTGACCT |
| Sequence-based reagent | Galntl6-For | This paper | QPCR primer (amplify Galntl6, M. musculus) | ACCGAGACTAGCAGTTCCCT |
| Sequence-based reagent | Galntl6-Rev | This paper | QPCR primer (amplify Galntl6, M. musculus) | GTCATGCGCTCTGTTTCCAC |
| Sequence-based reagent | Actin beta- For | This paper | QPCR primer (amplify Actb, M. musculus) | GACCTCTAT GCCAACACAGT |
| Sequence-based reagent | Actin beta- Rev | This paper | QPCR primer (amplify Actb, M. musculus) | AGTACTTGC GCTCAGGAGGA |
| Sequence-based reagent | Ins1- For | This paper | QPCR primer (amplify Ins1, R. Norvegicus) | ACCCTAAGTGACCAGCTACA |
| Sequence-based reagent | Ins1-Rev | This paper | QPCR primer (amplify Ins1, R. Norvegicus) | TTCACGACGGGACTTGGG |
| Sequence-based reagent | Gapdh-For | This paper | QPCR primer (amplify Gapdh, R. Norvegicus) | AGTGCCAGCCTCGTCTCATA |
| Sequence-based reagent | Gapdh-Rev | This paper | QPCR primer (amplify Gapdh, R. Norvegicus) | GATGGTGATGGGTTTCCCGT |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61174/elife-61174-transrepform-v2.docx
-
Reporting standard 1
Check list for the "Reporting guidelines for mass spectrometry".
- https://cdn.elifesciences.org/articles/61174/elife-61174-repstand1-v2.docx





