Concerted action of kinesins KIF5B and KIF13B promotes efficient secretory vesicle transport to microtubule plus ends
Figures
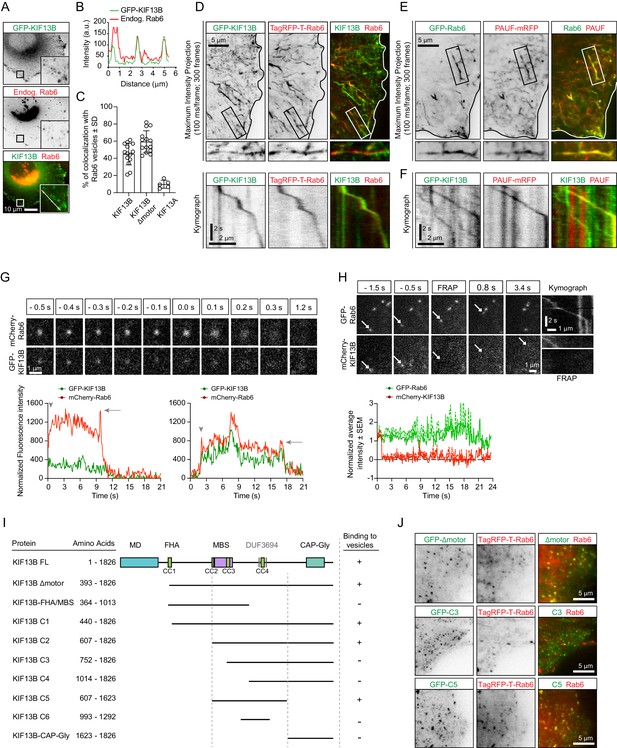
KIF13B localizes to Rab6-positive secretory carriers.
(A) Immunofluorescence images of a HeLa cell expressing GFP-KIF13B and stained for endogenous Rab6. The insets correspond to magnified views of the boxed areas. (B) Signal intensity profile of GFP-KIF13B (green) and endogenous Rab6 (red) along the white line in panel A. (C) Quantification of the percentage of TagRFP-T labeled Rab6A vesicles colocalizing with GFP-KIF13B, GFP-KIF13B Δmotor and GFP-KIF13A. n = 14 (GFP-KIF13B, GFP-KIF13B Δmotor) and n = 5 cells (GFP-KIF13A). (D) Maximum intensity projections (300 consecutive frames, 100 ms interval) illustrating imaging of live HeLa cells expressing GFP-KIF13B and TagRFP-T-Rab6A using TIRFM. Magnifications of the boxed area and kymographs illustrating the movement of co-labeled vesicle are shown below. (E) Maximum intensity projections with magnified views of the boxed areas illustrating TIRFM imaging of live HeLa cells transfected with GFP-Rab6A and PAUF-mRFP. (F) Kymographs illustrating the movement of vesicles labeled with GFP-KIF13B and PAUF-mRFP. (G) (Top) Frames from live TIRFM imaging showing the behavior of GFP-KIF13B and mcherry-Rab6A vesicles before and during fusion. 0.0 s corresponds to the sharp increase of fluorescent signal associated with vesicle docking at the plasma membrane. (Bottom) Two examples showing the average fluorescence intensity of a single vesicle labeled with GFP-KIF13B and mCherry-Rab6A plotted versus time. Vesicle appearance in the focal plane is indicated by an arrowhead. Arrow points to the peaks of fluorescence intensity prior to vesicle fusion with the plasma membrane. (H) (Top) TIRFM imaging combined with FRAP was performed in live HeLa cells containing exocytotic vesicles labeled for GFP-Rab6A and mCherry-KIF13B. The mCherry signal was photobleached (frames labeled FRAP) on moving vesicles labeled with GFP-Rab6A. Arrows indicate the same vesicle over time. Kymographs are shown to illustrate the absence of fluorescence recovery of mCherry-KIF13B on the vesicle. (Bottom) Quantification of the signal intensity of mCherry-KIF13B (red) on moving GFP-Rab6A vesicle (green) over time after FRAP. n = 6 vesicles in 4 cells. (I) Scheme of the GFP-KIF13B deletion constructs used in this study. The constructs were transfected in HeLa cells and colocalization with TagRFP-T-Rab6A-positive vesicles was determined by live cell imaging. The amino acid positions in KIF13B are indicated. MD, motor domain; FHA, forkhead-associated domain, MBS, MAGUK binding stalk; DUF, domain of unknown function; CC, coiled-coil. (J) Live images of HeLa cells expressing TagRFP-T-Rab6A and the indicated GFP-KIF13B deletion construct using TIRF microscopy.
-
Figure 1—source data 1
An Excel sheet with numerical data on the fluorescence intensity profile of Rab6 and KIF13B on vesicles, colocalization of KIF13B, KIF13B Δmotor and KIF13A with Rab6 vesicles, fluorescence intensity profile of Rab6 and KIF13B on vesicles during fusion, and FRAP dynamics of Rab6 and KIF13B on vesicles represented as plots in Figure 1B,C,G,H.
- https://cdn.elifesciences.org/articles/61302/elife-61302-fig1-data1-v2.xlsx
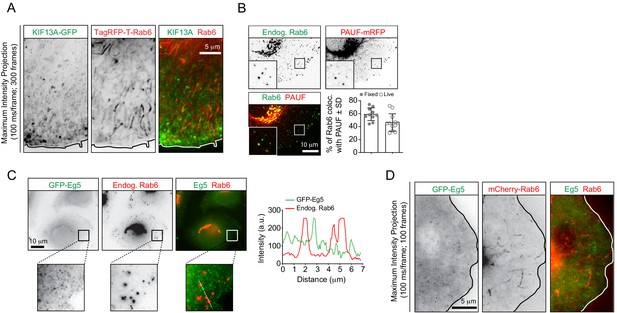
Analysis of localization of different markers to Rab6 vesicles.
(A) Maximum intensity projections (300 consecutive frames, 100 ms interval) of a HeLa cell expressing KIF13A-GFP and TagRFP-T-Rab6A imaged using TIRFM. (B) Imaging of HeLa cells expressing PAUF-mRFP and stained for endogenous Rab6. The insets correspond to magnified views of the boxed areas. The plot shows quantification of the percentage of PAUF-mRFP positive vesicles colocalizing with endogenous Rab6 or with GFP-Rab6A. n = 10 and 12 cells, respectively, in two independent experiments. (C) Imaging of a GFP-Eg5-expressing HeLa cell stained for Rab6. Panels at the bottom correspond to magnified views of the boxed areas. The graph displays the signal intensity profile of GFP-Eg5 (green) and endogenous Rab6 (red) along the white dashed line. (D) Maximum intensity projections (100 consecutive frames, 100 ms interval) of a HeLa cell expressing GFP-Eg5 and mCherry-Rab6A imaged using TIRFM.
-
Figure 1—figure supplement 1—source data 1
An Excel sheet with numerical data on the quantification of the colocalization of Rab6 with PAUF-positive vesicles, and the fluorescence intensity profile of Rab6 and Eg5 on vesicles represented as plots in Figure 1—figure supplement 1B,C.
- https://cdn.elifesciences.org/articles/61302/elife-61302-fig1-figsupp1-data1-v2.xlsx

Transport of Rab6 vesicles in HeLa cells is driven by KIF5B and KIF13B.
(A) Western blots of HeLa cell extracts showing the knockout of the different kinesins, using the indicated antibodies. Ku80 antibody was used as loading control. KIF13B-KO clone #1, KIF5B-KO clone #1, KIF5B/KIF13B-KO clone #1 and 4X-KO clone #2 have been used in latter experiments. (B) A maximum intensity projection over 500 frames (100 ms exposure with no delays) illustrating mCherry-Rab6A vesicle movement imaged by TIRFM and automated tracking using the SOS/MTrackJ plugin of mCherry-Rab6A labeled vesicles in the same HeLa cells. (C,E) Examples of automatic tracking of mCherry-Rab6A labeled vesicles using the SOS/MTrackJ plugin in the indicated knockout HeLa cells (C), or in 4X-KO cells expressing the indicated kinesins. (D) Analysis of the number of vesicle runs in the conditions depicted in (C). n = 29, 27, 30, 27, 22 and 21 cells, respectively, in three independent experiments. Mann-Whitney U test: ***p=0.0001; **p=0.0015; ns, no significant difference. (F) Analysis of the number of vesicle runs in the conditions depicted in (E). n = 40, 30, 30, 30, 30 and 30 cells in three independent experiments for each condition except for Eg5 (two independent experiments). Mann-Whitney U test: ****p<0.0001; ns, no significant difference. (G) Tracks containing processive runs as a percentage of total number of Rab6 tracks per cell. n = 29, 27, 30, 27, 22, 21, 30, 30, 30 and 30 cells, respectively, in two independent experiments for Eg5 and 3 independent experiments in all other conditions. Mann-Whitney U test: ****p<0.0001; **p=0.0011; ns, no significant difference. (H) Cumulated (total, summed) duration of processive runs as a percentage of total duration of all Rab6 tracks, per cell. The dataset is the same as in (G). Mann-Whitney U test: ****p<0.0001; *p<0.047; ns, no significant difference. (I) Distribution of the fraction of processive motion to the total duration of a track. Only the tracks that contain processive runs are included. n = 4237, 3532, 5219, 1115, 494, 556, 5289, 3811, 833 and 842 tracks. The dataset is the same as in (G).
-
Figure 2—source data 1
An Excel sheet with numerical data on the quantification of the effect of kinesin silencing and rescue on the number of vesicle runs, percentage of tracks with runs, cumulated processive run duration and the distribution of processive movements per track represented as plots in Figure 2D,F–I.
- https://cdn.elifesciences.org/articles/61302/elife-61302-fig2-data1-v2.xlsx
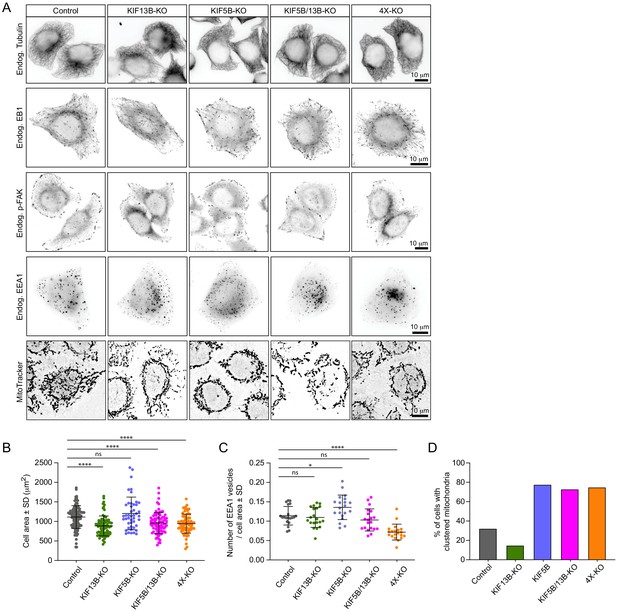
Validation of kinesin knockout cell lines.
(A) Representative immunofluorescence images showing the effect of the knockout of different kinesins on the MT cytoskeleton, EB1-positive MT plus ends and distribution of focal adhesions, early endosomes and mitochondria. (B–D) Quantification of cell area (B), number of EEA1-positive vesicles (C) and percentage of cells with clustered mitochondria (D) in the different knockout cell lines. B: n = 104, 81, 45, 81 and 81 cells, respectively. C and D: n = 20 in all conditions. Mann-Whitney U test: ****p<0.0001, ***p=0.0009, *p=0.02, ns, no significant difference.
-
Figure 2—figure supplement 1—source data 1
An Excel sheet with numerical data on the quantification of the effect of kinesin silencing on cell area, number of EEA1 vesicles and mitochondria clustering represented as plots in Figure 2—figure supplement 1B–D.
- https://cdn.elifesciences.org/articles/61302/elife-61302-fig2-figsupp1-data1-v2.xlsx
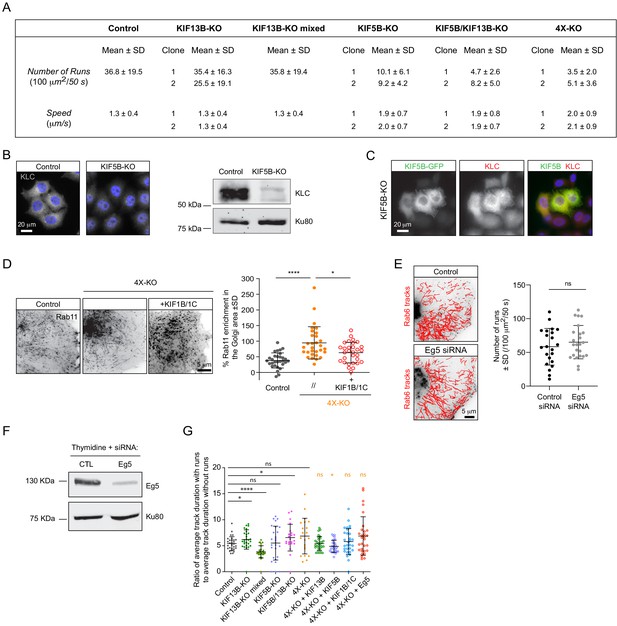
Characterization of kinesin knockout cell lines.
(A) Table summarizing parameters of Rab6 vesicle runs (number and speed) in two independent HeLa cell clones for each kinesin knockout. Data for the mixed population of KIF13B-KO cells is included for comparison. KIF13B-KO clone #1, KIF5B-KO clone #1, KIF5B/KIF13B-KO clone #1 and 4X-KO clone #2 have been used in latter experiments. (B) Impact of KIF5B-KO on the expression of kinesin light chain (KLC) assessed by immunostaining with a KLC antibody and Western blotting. (C) Representative images of KIF5B-KO HeLa cells expressing a KIF5B-GFP construct and stained for KLC. (D) Analysis of Rab11 tracks in control and 4X-KO HeLa cells or 4X-KO cells expressing KIF1B and KIF1C. Maximum intensity projections (500 consecutive frames, 100 ms interval) of GFP-Rab11 were used to calculate the enrichment of Rab11 signal in the perinuclear region as described in Methods. n = 30 cells for each condition in three independent experiments. Mann-Whitney U test: ****p<0.0001, *p=0.0225. (E) Automatic tracking with SOS/MTrackJ plugin of GFP-Rab6A labeled vesicles imaged by TIRFM and quantification of the number of Rab6 vesicle runs in HeLa cells transfected with control or Eg5 siRNA and treated with thymidine to prevent mitotic entry. Number of runs in control is a bit high compared to controls in Figure 2D,F. We attribute this difference to the transfection of control Luciferase siRNA and slightly different acquisition settings. n = 22 and 24 cells in two independent experiments. Unpaired t-test: ns, no significant difference. (F) Western blot demonstrating Eg5 knockdown efficiency in HeLa cells using an antibody against Eg5. (G) Total number of tracks per cell. n = 29, 27, 30, 27, 22, 21, 30, 30, 30 and 30 cells, respectively, in two independent experiments for Eg5 and 3 independent experiments in all other conditions. The dataset is the same as in Figure 2G. Mann-Whitney U test: ****p<0.0001; *p=0.0122; ns, no significant difference. (H) Ratio of average duration of tracks with runs to average duration of tracks without runs, per cell. The dataset is the same as in Figure 2G. Mann-Whitney U test: ****p<0.0001; *p<0.032; ns, no significant difference.
-
Figure 2—figure supplement 2—source data 1
An Excel sheet with numerical data on the quantification of Rab11 enrichment in the Golgi area in 4X-KO and in 4X-KO re-expressing KIF1B/1C, the effect of Eg5 depletion on the number of vesicle runs, and the effect of kinesin silencing and rescue on the average track duration with runs represented as plots in Figure 2—figure supplement 2D,E,G.
- https://cdn.elifesciences.org/articles/61302/elife-61302-fig2-figsupp2-data1-v2.xlsx
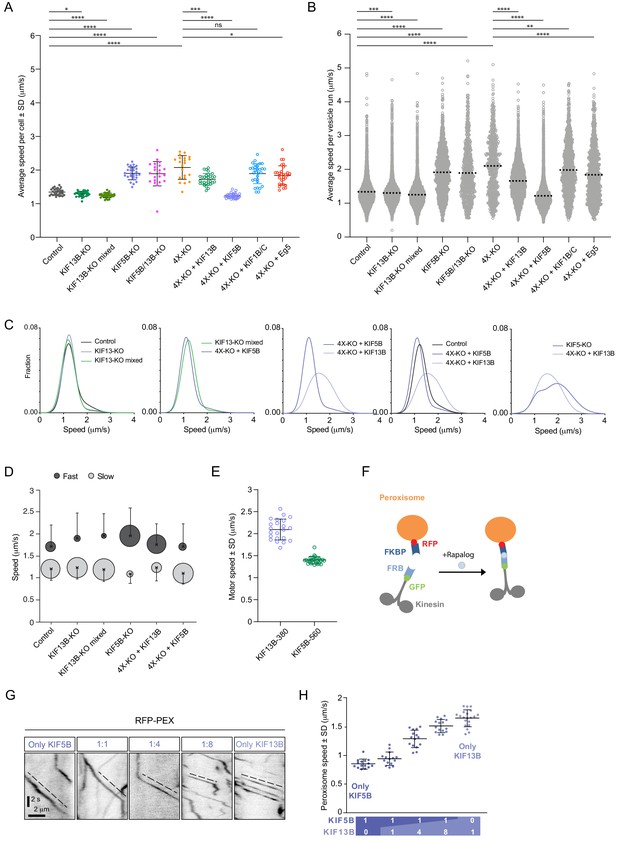
KIF5B and KIF13B have distinct speeds.
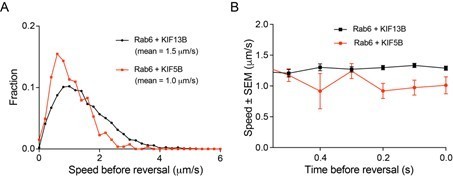
(A) Analysis of the mean speed of automatically tracked mCherry-Rab6A-positive vesicles per cell in the indicated conditions. n = 29, 27, 30, 27, 22, 21, 30, 30, 30 and 30 cells, respectively, in two independent experiments for Eg5 and 3 independent experiments in all other conditions. Unpaired t-test: ****p<0.0001; ***p=0.0002; *p<0.025, ns, no significant difference. (B) Violin plots showing the speed of individual automatically tracked mCherry-Rab6A-positive vesicles in the indicated conditions. Dotted lines represent the mean. n = 29, 27, 30, 27, 22, 21, 30, 30, 30 and 30 cells, respectively, in two independent experiments for Eg5 and 3 independent experiments in all other conditions. Same data as shown in (A), but displayed here for individual vesicle runs rather than averaged per cell. Mann-Whitney U test: ****p<0.0001; ***p=0.0002; **p=0.0064. (C) Combinations of sums of two Gaussian fits to the distribution of Rab6 vesicles run speeds for the indicated conditions (see Figure 3—figure supplement 1C). (D) Parameters of ‘slow’ (light gray) and ‘fast’ (dark gray) Gaussians components from fits in Figure 3—figure supplement 1C. Crosses correspond to the mean value and error bars to the standard deviation of fitted Gaussians. The area of the circles corresponds to the fraction of runs associated with ‘slow’ and ‘fast’ components, i.e. represents the area under the curve of each Gaussian (total sum of areas for each condition is the same). n = 7099 runs in 29 cells, 6682 runs in 27 cells, 8192 runs in 30 cells, 1772 runs in 27 cells, 7616 runs in 30 cells and 5651 runs in 30 cells, respectively. (E) Speed of kinesin-positive particles imaged with TIRF microscopy in 4X-KO HeLa cells expressing KIF5B-560-GFP (n = 30 cells, three experiments) or KIF13B-380-LZ-GFP (n = 22 cells, two experiments). (F) A scheme of inducible peroxisome trafficking assay performed by the rapalog-dependent recruitment of FRB-tagged KIF5B(1-807) or KIF13B(1-444) to FKBP-tagged PEX3, a peroxisome protein. (G) Kymographs illustrating peroxisome movements in cells transfected with the indicated FRB-tagged KIF5B(1-807):KIF13B(1-444) plasmid ratios. (H) MRC5 cells were co-transfected with PEX3-mRFP-FKBP and the indicated ratios of FRB-tagged KIF5B and KIF13B plasmids mentioned in (F) simultaneously, while the total amount of DNA was kept constant. Peroxisomes were imaged by TIRF or SD microscopy and their speeds were measured 10–40 min after rapalog addition. n = 15, 15, 15, 15 and 20 cells, respectively in three experiments (Kif13B alone - in four experiments).
-
Figure 3—source data 1
An Excel sheet with numerical data on the quantification of the effect of kinesin silencing and rescue on vesicle speed, KIF13B-380 and KIF5B-650 motor speed, and peroxisome speed in cells expressing different ratios of KIF5B and KIF13B represented as plots in Figure 3A–E,H.
- https://cdn.elifesciences.org/articles/61302/elife-61302-fig3-data1-v2.xlsx
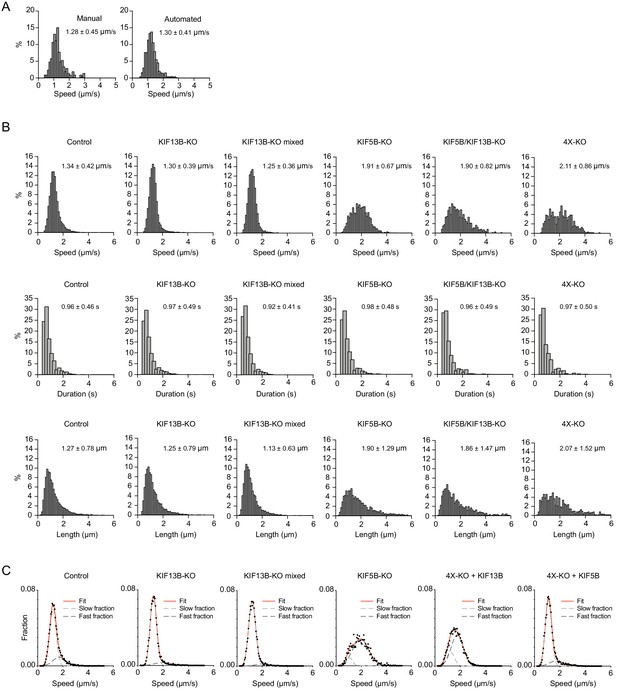
Characterization of Rab6 vesicle motility in kinesin knockout lines.
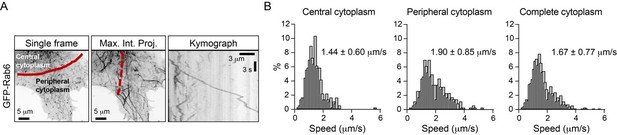
(A) Analysis of Rab6 vesicle speeds per run using manual and automatic tracking in HeLa cells expressing mCherry-Rab6A and imaged using TIRF microscopy. The speed of Rab6 vesicles was calculated either manually based on kymograph analysis or automatically using the SOS/MTrackJ plugin, as shown in Figure 2B. Plots show frequency distribution histograms annotated with the mean ± SD. n = 205 and 1213 tracks from the same 5 cells. (B) Histograms of the run speed, run duration and run length distributions of Rab6-positive vesicles in the indicated conditions. n = 7099 in 29 cells, 6682 in 27 cells, 8192 in 30 cells, 1772 in 27 cells, 707 in 22 cells and 617 in 21 cells in three independent experiments each condition. (C) Distribution of Rab6 vesicle run speeds (dots) and their fit to sum of two Gaussians model (red solid line). Light and dark gray dashed lines represent ‘slow’ and ‘fast’ Gaussian components of fits. For all condition data represents three independent experiments. n = 7099 runs in 29 cells, 6682 runs in 27 cells, 8192 runs in 30 cells, 1772 runs in 27 cells, 7616 runs in 30 cells and 5651 runs in 30 cells, respectively. For all conditions the sum of two Gaussian functions describes distributions better than one, according to extra sum-of-squares F test (all conditions p<0.0001). Same data as shown in Figure 3C,D.
-
Figure 3—figure supplement 1—source data 1
An Excel sheet with numerical data on the quantification of Rab6 vesicle speed distribution from manual and automated analysis, and the effect of kinesin silencing on the distribution of speed, duration and length of vesicle runs represented as plots in Figure 3—figure supplement 1A–C.
- https://cdn.elifesciences.org/articles/61302/elife-61302-fig3-figsupp1-data1-v2.xlsx
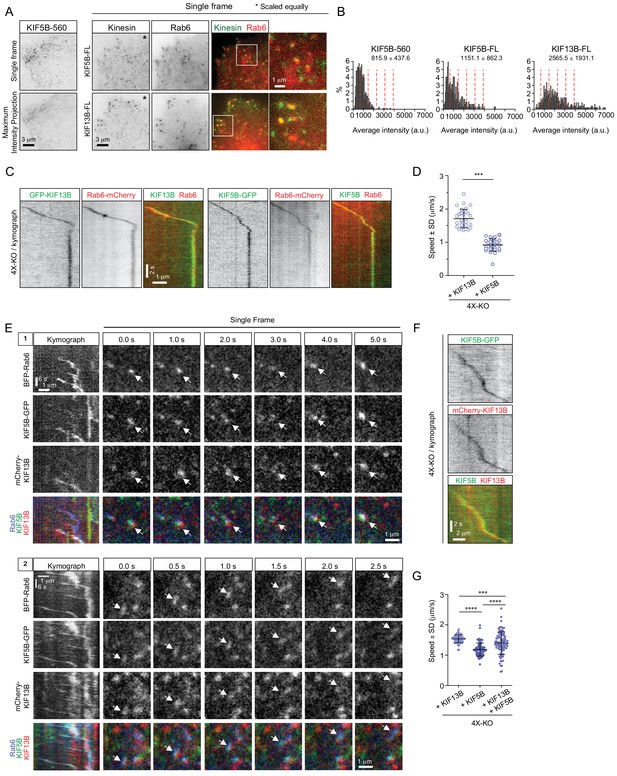
KIF5B and KIF13B colocalize on moving vesicles.
(A) Live TIRFM imaging of 4X-KO cells expressing either KIF5B-560-GFP alone or mCherry-Rab6A together with the full-length (FL) KIF5B-GFP or GFP-KIF13B. A single frame and a maximum intensity projection of a KIF5B-560-GFP movie is shown on the left, and single frames illustrating the localization of KIF5B-FL-GFP and GFP-KIF13B-FL on mCherry-Rab6A-labeled vesicles are shown on the right. Insets show enlargement of boxed areas. (B) Histograms showing the frequency distributions of the average intensity of the indicated kinesin constructs in 4X-KO cells annotated with the mean ± SD; for KIF5B-FL-GFP and GFP-KIF13B-FL, only the signals colocalizing with Rab6 vesicles were quantified. n = 660 in 17 cells, 320 in 24 cells and 495 in 19 cells in two independent experiments (KIF5B-560) and three independent experiments (KIF5B-FL and KIF13B-FL). Dashed lines mark intensity of 1, 2, 3, 4 and 5 kinesin molecules estimated from the average value of KIF5B-560 distribution. (C–D) 4X-KO HeLa cells expressing mCherry-Rab6A together with GFP-KIF13B-FL or KIF5B-FL-GFP were imaged using TIRFM. Kymographs illustrating the movement of Rab6 vesicles positive for KIF13B or KIF5B were drawn and used to manually measure the velocity of each motor. n = 30 and 28 cells in three independent experiments. Mann-Whitney U test: ***p<0.001. (E) Live confocal imaging of 4X-KO cells co-expressing TagBFP-Rab6, full-length KIF5B-GFP and full-length mCherry-KIF13B. Kymographs of two different moving vesicles and corresponding consecutive single frames (1 and 2) are shown to illustrate the simultaneous localization of KIF5B-GFP and mCherry-KIF13B on moving TagBFP-Rab6-positive vesicles. (F) A kymograph illustrating the movement of a particle co-labeled with KIF5B-GFP and mCherry-KIF13B and expressed in a 4X-KO HeLa cell. (G) Automated analysis of speeds of Rab6 vesicles colocalized with indicated full-length kinesins in 4X-KO HeLa cells expressing either KIF5B-GFP and mCherry-KIF13B alone, or the condition where two kinesins colocalize together. Dots show average speed per cell, bars represent mean and SD. n = 82, 79 and 89 cells, same data and analysis as in Figure 5G, H. Mann-Whitney U test: ****p<0.0001; ***p=0.0006.
-
Figure 4—source data 1
An Excel sheet with numerical data on the quantification, in 4X-KO cells, of the distribution of the average intensity of fluorescently tagged KIF5B-560, KIF5B-FL and KIF13B-FL, the speed of KIF13B and KIF5B, and the speed of vesicles containing KIF13B, KIF5B or both motors represented as plots in Figure 4B,D,G.
- https://cdn.elifesciences.org/articles/61302/elife-61302-fig4-data1-v2.xlsx
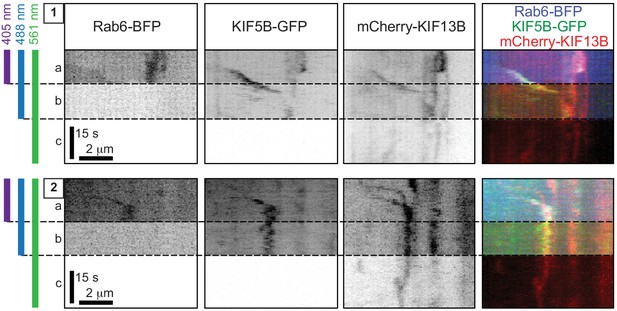
Validation of simultaneous three-color image acquisition.
Live TIRF imaging of 4X-KO cells co-expressing TagBFP-Rab6, full-length KIF5B-GFP and full-length mCherry-KIF13B. Kymographs of two different vesicles (1 and 2) from two different cells simultaneously imaged with 405 nm, 488 nm and 561 nm lasers are shown. Simultaneous three-color imaging shows that the vesicles are co-labeled with the three fluorescently tagged proteins (section (a) of each kymograph). As blue fluorescence can potentially be detected in the green channel, we confirmed that switching off the 405 nm laser (b) did not have a major impact on the green channel signal (meaning that the initial green signal was not caused by cross-talk with the blue channel). Turning off the 488 nm laser (c) also did not have a major impact on the red channel. These controls show that there is no significant crosstalk between channels in our microscope setup, and that we can reliably simultaneously detect blue, green and red channels on moving vesicles.
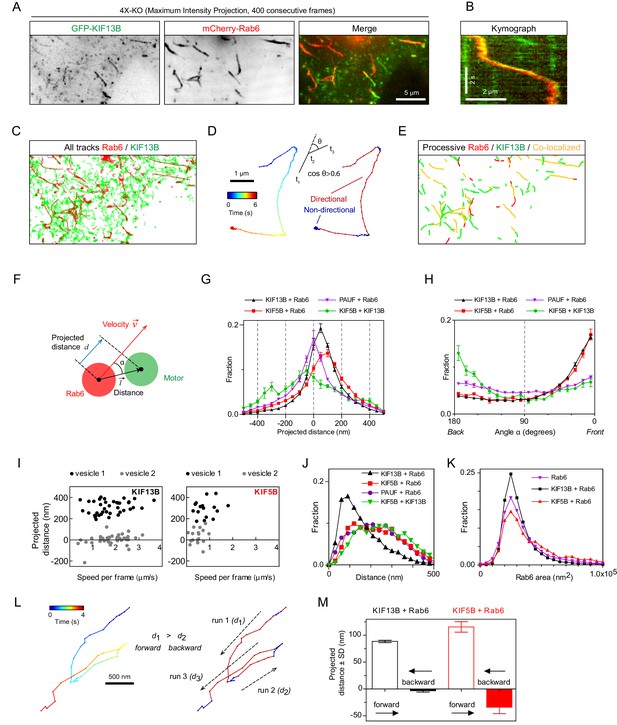
Kinesins exhibit distinct distributions on Rab6 vesicles.
(A) A representative example of maximum intensity projection (400 consecutive frames, 100 ms interval) of 4X-KO HeLa cells expressing mCherry-Rab6A and GFP-KIF13B to visualize events of Rab6-vesicle movement. Chromatic aberration of the red channel (mCherry-Rab6A) was corrected based on calibration, as illustrated in Figure 5—figure supplement 1A. (B) A kymograph from the movie shown in (A) illustrating the movement of a mCherry-Rab6A-labeled vesicle positive for GFP-KIF13B. (C) Automatically extracted trajectories of mCherry-Rab6A- and GFP-KIF13B-positive particles (detected independently) from the movie shown in (A). (D) Segmentation of trajectories into periods of random and directed motion. (Left) An example of Rab6 vesicle trajectory with color-coded time. (Middle) Definition of directional movements: Movement was classified as directional when a cosine of the angle θ between two consecutive velocity vectors (t3–t2 and t2–t1) was larger than 0.6. (Right) Final segmentation result with directional (red) and random (blue) periods of movement. (E) Directional segments of the tracks shown in (C), with colocalizing tracks labeled in yellow. (F) Schematics of the parameters used to characterize the distribution of two markers on the same vesicle. The projected distance d is calculated as a projection of distance between the centers of motor and cargo fluorescent signals () onto the axis defined by the instant velocity vector () of the cargo. The angle α is defined as the angle between the distance and velocity vectors. (G,H) The averaged histograms of the instantaneous projected distance (G) and the angle α (H) for GFP-KIF13B (black), KIF5B-GFP (red) with respect to mCherry-Rab6A, PAUF-mRFP with respect to GFP-Rab6A (purple) and for KIF5B-GFP with respect to mCherry-KIF13B (green). Each dot and bar represent the average and SEM over several independent experiments, each including 8–20 cells. KIF13B (N = 6 independent experiments, 82 cells, 11333 runs, 55129 time points), KIF5B (N = 6, 79 cells, 2826 runs, 10023 time points), KIF5B and KIF13B (N = 7, 89 cells, 1558 runs, 4371 time points) and PAUF (N = 2, 20 cells, 5807 runs, 21359 time points). (I) Plots of projected distance between Rab6A and KIF13B (left) or KIF5B (right) signals against speed for four different vesicles/runs (two different vesicles with distinct maximum projected distances, likely reflecting different vesicle sizes, are shown for each kinesin). (J,K) Histograms of the distance between the indicated markers (J) and Rab6A area (K) averaged per run and pooled together for all experiments. Same statistics as in (G,H). (L) Extraction of opposite polarity runs from Rab6 vesicle trajectories. On the left, an example of a trajectory with color-coded time; on the right, the same trajectory where the color denotes movement characteristics, directed (red) or random (blue). For each processive segment (run) the average direction of the velocity vector (dashed arrows) and average projected distance value (di) are calculated. Within one trajectory, the algorithm searches for all possible pair combination and keeps only those where the average movement direction is opposite. Within each pair a run with the higher average projected displacement is assigned to be the ‘forward’ run and the other one the ‘backward’ run. (M) Instantaneous (per frame) projected displacements for pairs of opposite runs, average ± SEM for the denoted conditions. The data are the same as in (G–J); for KIF13B, 664 opposite run pairs found, 3262 forward and 3122 backward time points; for KIF5B, 83 run pairs, 289 forward and 274 backward time points.
-
Figure 5—source data 1
An Excel sheet with numerical data on the quantification of the distribution and projected displacement of KIF13B and KIF5B signals on moving vesicles represented as plots in Figure 5G–K,M.
- https://cdn.elifesciences.org/articles/61302/elife-61302-fig5-data1-v2.xlsx
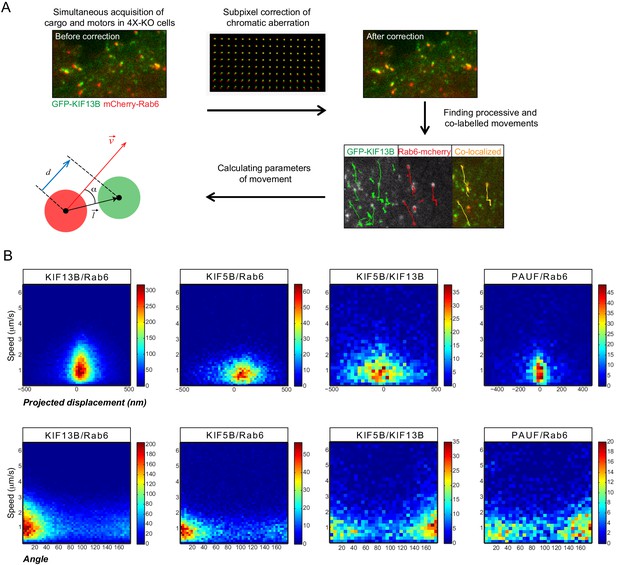
Analysis of kinesin distribution on Rab6 vesicles.
(A) Scheme depicting the workflow for subpixel localization of kinesins. Pairs of fluorescent markers were expressed in 4X-KO cells (for all kinesin constructs) or control HeLa cells (PAUF) and imaged using TIRFM. The acquired signals were corrected for chromatic aberrations using a grid of fluorescent beads as described in Materials and methods, and vesicle runs were automatically tracked and processed. Processive runs with both markers present were used to calculate the parameters of movement: the distance vector between the center of the two fluorescent signals (mCherry and GFP spots) was projected onto the Rab6 instant velocity vector to determine the projected displacement (d) (blue segment). α is the angle between the distance and velocity vector. (B) The projected displacement and the angle as described in (A) plotted as heat maps against speed. Same data as shown in Figure 5G,H.
-
Figure 5—figure supplement 1—source data 1
An Excel sheet with numerical data on the quantification of the projected displacement and angle relative to speed of KIF13B and KIF5B signals plotted as heat maps in Figure 5—figure supplement 1B.
- https://cdn.elifesciences.org/articles/61302/elife-61302-fig5-figsupp1-data1-v2.xlsx
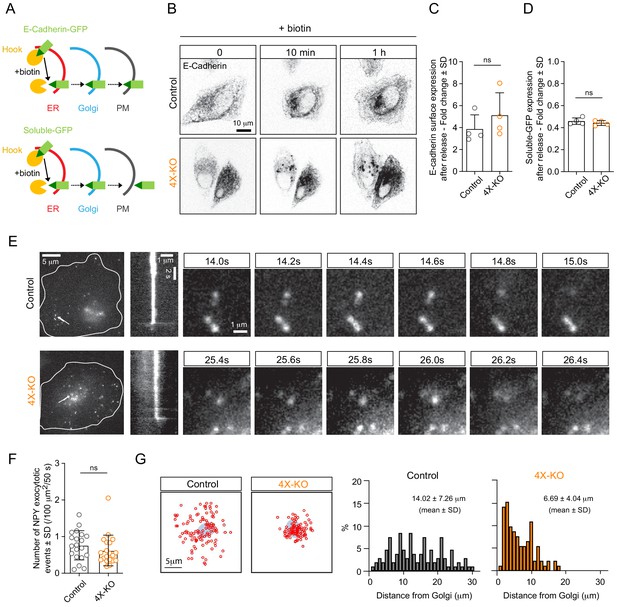
Kinesins control the spatial distribution but not the efficiency of exocytosis.
(A) A scheme depicting the RUSH assay used in this study. The interaction of SBP-GFP-E-Cadherin or soluble-GFP-SBP with streptavidin-KDEL (Hook) allows for the retention of E-Cadherin-GFP or soluble-GFP in the ER and their release for transport to the Golgi and the plasma membrane (PM) upon the addition of biotin, which competes with SBP for streptavidin binding. (B,C) RUSH assay was performed by expressing SBP-GFP-E-Cadherin and streptavidin-KDEL from the same bicistronic expression plasmid in control or 4X-KO HeLa cells. Cells were treated with biotin and imaged using time-lapse spinning-disk confocal microscopy (B, GFP-E-Cadherin signal) or subjected to surface staining with anti-GFP antibody to specifically label plasma membrane-exposed E-Cadherin followed by flow cytometry analysis (C). Plot shows the fold change of Alexa641 mean intensity (surface staining) in E-Cadherin expressing cells before and after biotin addition (1 hr). n = 4 independent experiments. Mann-Whitney U test: ns, no significant difference. (D) Control or 4X-KO HeLa cells expressing soluble-GFP-SBP and streptavidin-KDEL were treated with biotin and analyzed by flow cytometry to quantify the fold change of GFP mean intensity after biotin addition. n = 4 independent experiments. Mann-Whitney U test: ns, no significant difference. (E,F) TIRF microscopy was used to visualize and analyze exocytosis events in control or 4X-KO HeLa cells expressing NPY-GFP. Exocytotic events, defined by a fast burst of fluorescence followed by the disappearance of the signal were visually identified, confirmed by kymograph analysis and counted per cell and per surface area and per duration of movie (50 s) (F) n = 20 cells in two independent experiments. Mann-Whitney U test: ns, no significant difference. Individual time frames in (E) illustrate representative exocytotic events; their localization is indicated by white arrows, and the corresponding kymographs are shown. (G) Analysis of the spatial distribution of the NPY exocytotic events shown in (E). Schematized positions of NPY exocytosis events (red circles) compared to the position of the Golgi (blue) are shown on the left (sum of 20 cells) and frequency distributions of the distance between the center of the Golgi and the sites of exocytosis in control and 4X-KO HeLa cells are shown on the right. n = 109 and 93 tracks from 20 cells in two independent experiments.
-
Figure 6—source data 1
An Excel sheet with numerical data on the quantification of the effect of kinesin silencing on E-cadherin surface expression and soluble-GFP expression, the number of NPY exocytotic events and the distribution of the distance between the center of the Golgi and the sites of NPY exocytotic events represented as plots in Figure 6C,D,F,G.
- https://cdn.elifesciences.org/articles/61302/elife-61302-fig6-data1-v2.xlsx
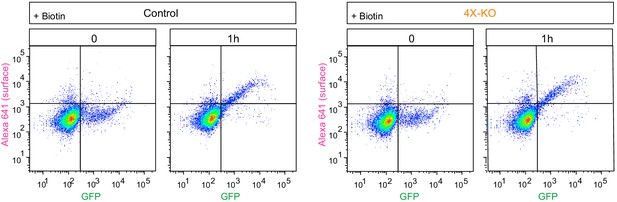
Quantification of secretion using flow cytometry.
RUSH assay was performed in control or 4X-KO HeLa cells expressing SBP-GFP-E-Cadherin and streptavidin-KDEL and analyzed by flow cytometry as shown in Figure 6A, C. The dot plots show the intensity of the signal of GFP-E-Cadherin (GFP) plotted against the signal of the surface staining (Alexa 641) per cell. Scale is logarithmic.
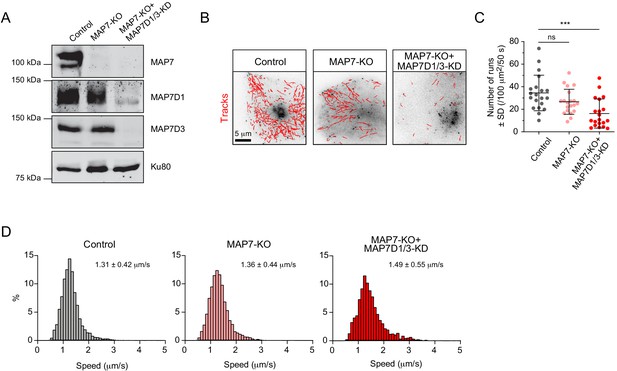
MAP7 family proteins are required for the transport of Rab6 vesicles.
(A) Western blot analysis of the extracts of control or MAP7 knockout (MAP7-KO) HeLa cells or MAP7-KO cells transfected with siRNAs against MAPD1 and MAP7D3 (MAP7-KO+MAP7D1/3-KO) with the indicated antibodies. (B–D) GFP-Rab6A was expressed and imaged using TIRFM in Hela cells described in (A). Automatic tracking using the SOS/MTrackJ plugin (500 consecutive frames, 100 ms interval) of the Rab6A signal (B), number of Rab6 vesicle runs per cell, n = 20 cells in two experiment in each condition (C) and the frequency distributions of Rab6 vesicle speeds after automatic tracking annotated with the mean ± SD (D) are shown. n = 5038, 4056 and 2366 tracks from 20 cells in two independent experiments. Mann-Whitney U test: ***p=0.0002, ns, no significant difference.
-
Figure 7—source data 1
An Excel sheet with numerical data on the quantification of the effect of MAP7 family proteins silencing on the number of Rab6 vesicle runs and on the distribution of Rab6 vesicle speeds represented as plots in Figure 7C,D.
- https://cdn.elifesciences.org/articles/61302/elife-61302-fig7-data1-v2.xlsx
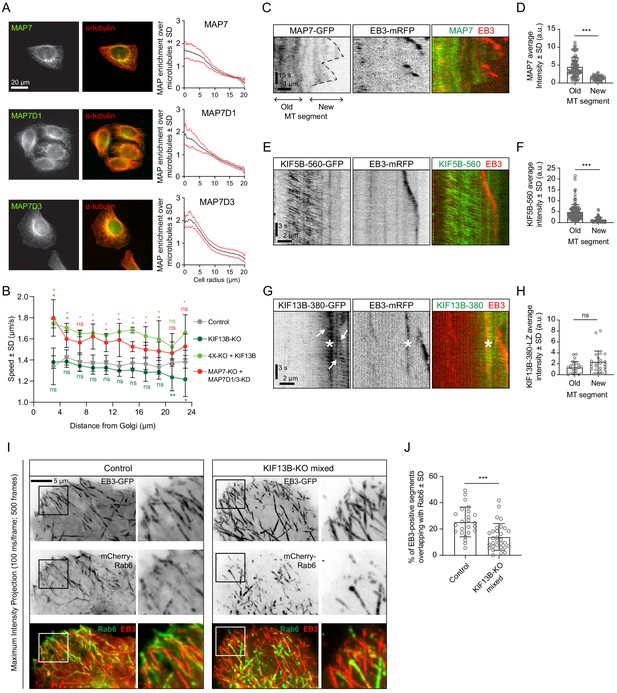
KIF13B promotes Rab6 vesicle transport to freshly polymerized MT ends.
(A) Staining of HeLa cells for endogenous MAP7, MAP7D1 or MAP7D3 together with α-tubulin and quantification of the relative enrichment of MAP7 protein signal intensity over the tubulin signal calculated and plotted against the distance from the cell center. n = 50, 48 and 41 cells, respectively, from 4, 4 and 3 independent experiments. (B) Automatic tracking of GFP-Rab6A or mCherry-Rab6A labeled vesicles was performed on data obtained by TIRFM imaging in control, KIF13B-KO, KIF13B-expressing 4X-KO or MAP7-KO+MAP7D1/3-KD HeLa cells. Tracking results were analyzed using the SAID plugin of MTrackJ as described in the Methods to extract the velocity of the Rab6 runs in relation to their distance from the Golgi. n = 49, 47, 30 and 40 cells, respectively, in 5, 5, 3 and 4 independent experiments. Mann-Whitney U test: **p=0.0079, *p<0.032, ns, no significant difference. (C, E, G) Kymographs illustrating the dynamics of a MT labeled with EB3-mRFP together with MAP7-GFP (C), KIF5B-560-GFP (E) or KIF13B-380-LZ-GFP (G) imaged using TIRFM. In (G), arrows indicate movement of KIF13B-380-LZ-GFP along MTs and the asterisk (*) marks the spot where another EB3-mRFP/KIF13-380-LZ-GFP-positive MT crosses the analyzed MT. (D, F, H) Quantification of the average intensity of MAP7-GFP (D), KIF5B-560-GFP (F) and KIF13B-380-LZ-GFP (H) on old (further than 3 µm from plus end) and new (within 2 µm from plus end) MT segments visualized with EB3-mRFP (mean ± SD). D: n = 109 (14 cells in two independent experiments) and n = 47 (14 cells in two experiments). F: n = 155 (19 cells in two independent experiments) and n = 71 (19 cells in two experiments). H: n = 24 (15 cells in three experiment) and n = 24 (15 cells in three experiment). Mann-Whitney U test: ***p<0.0001, ns, no significant difference. (I) Maximum intensity projections (500 consecutive frames, 100 ms interval) of control or KIF13B-KO mixed HeLa cells expressing mCherry-Rab6A and EB3-GFP. Magnified views of the boxed areas are shown on the right. Colors in the merged images were inverted for display purposes. (J) Quantification of the percentage of EB3-GFP-positive MT segments colocalizing with mCherry-Rab6A vesicles. Measuring has been performed in MetaMorph software using Measure Colocalization option on Threshold images. n = 24 and 30 cells, respectively, in three independent experiments. Mann-Whitney U test: ***p<0.0003.
-
Figure 8—source data 1
An Excel sheet with numerical data on the quantification of MAP7 family proteins enrichment on MTs along the cell radius, the effect of kinesin and MAP7 family proteins silencing on vesicle speed relative to distance from the Golgi, MAP7, KIF5B-560 and KIF13B-380 intensity on old and new MT segments, and the effect of KIF13B silencing on the co-localization of EB3-GFP-positive MT segments with Rab6 vesicles represented as plots in Figure 8A,B,D,F,H,J.
- https://cdn.elifesciences.org/articles/61302/elife-61302-fig8-data1-v2.xlsx
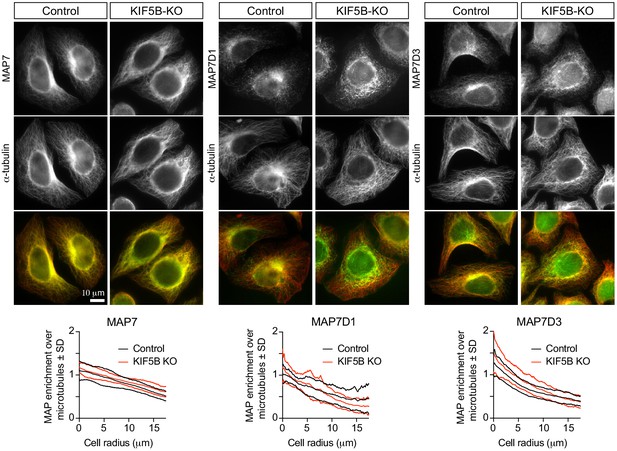
KIF5B knockout does not affect the localization of MAP7 family proteins.
Staining of control and KIF5B-KO HeLa cells for endogenous MAP7, MAP7D1 or MAP7D3 together with α-tubulin, and quantification of the relative enrichment of MAP7 proteins signal intensity over the tubulin signal calculated and plotted against the distance from the cell center. n = 26 (control) and 24 (KIF5B-KO), n = 24 (control) and 21 (KIF5B-KO), and n = 23 (control) and 19 (KIF5B-KO), respectively, from two independent experiments.
-
Figure 8—figure supplement 1—source data 1
An Excel sheet with numerical data on the quantification of the effect of KIF5B silencing on MAP7 family proteins enrichment on MTs along the cell radius represented as plots in Figure 8—figure supplement 1.
- https://cdn.elifesciences.org/articles/61302/elife-61302-fig8-figsupp1-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HeLa S3 (Kyoto) | ATCC | CCL-2.2 | |
| Cell line (Homo sapiens) | MRC5 | ATCC | CCL-171 | |
| Cell line (Homo sapiens) | KIF5B-KO HeLa | This paper | CRISPR/Cas9 generated monoclonal HeLa line | |
| Cell line (Homo sapiens) | KIF5B/KIF13B-KO HeLa | This paper | CRISPR/Cas9 generated monoclonal HeLa line | |
| Cell line (Homo sapiens) | 4X (KIF5B/KIF13B/KIF1B/KIF1C)-KO HeLa | This paper | CRISPR/Cas9 generated monoclonal HeLa line | |
| Cell line (Homo sapiens) | MAP7-KO HeLa | Hooikaas et al., 2019; PMID:30770434 | ||
| Transfected construct (Homo sapiens) | GFP-Rab6A | Matanis et al., 2002; PMID:12447383 | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | GFP-Eg5 | Jiang et al., 2012; PMID:22885064 | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | pßactin-PEX3-mRFP | Kapitein et al., 2010b; PMID:20923648 | Expression construct transfected inMRC5/peroxisome trafficking assay | |
| Transfected construct (Homo sapiens) | KIF5B(1-807)-GFP-FRB | Kapitein et al., 2010b; PMID:20923648 | Expression construct transfected inMRC5/peroxisome trafficking assay | |
| Transfected construct (Homo sapiens) | GFP-Rab11 | Hoogenraad et al., 2010; PMID:21057633 | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | NPY-GFP | Schlager et al., 2010; PMID:20360680 | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | KIF13B(1-444)-GFP-FRB | Lipka et al., 2016; PMID:26758546 | Expression construct transfected in MRC5/peroxisome trafficking assay | |
| Transfected construct (Homo sapiens) | KIF13A-GFP | Schou et al., 2017; PMID:28134340 | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | FKBP-mCherry-Rab6A | Schlager et al., 2014; PMID:25176647 | Expression construct transfected in HeLa, termed mCherry-Rab6A in this paper | |
| Transfected construct (Homo sapiens) | TagBFP-Rab6A | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | GFP-KIF13B | gift from gift from Dr. Athar Chishti (University of Illinois College of Medicine, Chicago, USA) Venkateswarlu et al., 2005; PMID:15923660 | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | TagRFP-T-Rab6A | gift from Dr. Yuko Mimori-Kiyosue (RIKEN Center for Developmental Biology, Japan) | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | GFP-KIF13B | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | GFP-KIF13B Δ motor (393–1826) | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | GFP-KIF13B-FHA/MBS (364–1013) | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | GFP-KIF13B C1 (440–1826) | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | GFP-KIF13B C2 (607–1826) | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | GFP-KIF13B C3 (752–1826) | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | GFP-KIF13B C4 (1014–1826) | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | GFP-KIF13B C5 (607–1623) | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | GFP-KIF13B C6 (993–1292) | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | GFP-KIF13B-CAP-Gly (1623–1826) | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | mCherry-KIF13B | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | KIF5B-GFP | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | KIF1B | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | KIF1C | This paper | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | PAUF-mRFP | Wakana et al., 2012; PMID:22909819 | Expression construct transfected in HeLa | |
| Transfected construct (Homo sapiens) | streptavidin-KDEL-SBP-GFP-E-Cadherin | Boncompain et al., 2012; PMID:22406856 | Expression constructtransfected in HeLa/RUSH assay | |
| Transfected construct (Homo sapiens) | streptavidin-KDEL-solubleGFP-SBP | Boncompain et al., 2012; PMID:22406856 | Expression construct transfected in HeLa/RUSH assay | |
| Antibody | anti-Rab6 (Mouse monoclonal) | Schiedel et al., 1995; PMID:8521955 | IF (1:300) | |
| Antibody | anti-Ku80 (Mouse monoclonal) | BD Biosciences | Cat#:611360, RRID:AB_398882 | WB (1:2000) |
| Antibody | anti-EB1 (Mouse monoclonal) | BD Biosciences | Cat#:610252; RRID:AB_2276073 | IF (1:200) |
| Antibody | anti-EEA1 (Mouse monoclonal) | BD Biosciences | Cat#:610456, RRID:AB_397829 | IF (1:100) |
| Antibody | anti-MAP7 (Mouse polyclonal) | Abnova | Cat#:H00009053-B01P, RRID:AB_10714227 | IF (1:300) WB (1:1000) |
| Antibody | anti-KIF1B (Rabbit polyclonal) | Bethyl | Cat#:A301-055A, RRID:AB_2131416 | WB (1:500) |
| Antibody | anti-KIF1C (Rabbit polyclonal) | Cytoskeleton | Cat#:AKIN11-A, RRID:AB_10708792 | WB (1:300) |
| Antibody | Anti-KIF5B/UKHC (Rabbit polyclonal) | Santa Cruz Biotechnology | Cat#:SC28538, clone H50, RRID:AB_2280915 | WB (1:1000) |
| Antibody | anti-Eg5 (Rabbit polyclonal) | Abcam | Cat#:ab6119, RRID:AB_941397 | WB (1:500) |
| Antibody | anti-KLC1 (Rabbit polyclonal) | Santa Cruz Biotechnology | Cat#:sc25735, clone H75, RRID:AB_2280879 | IF (1:200) WB (1:1000) |
| Antibody | Anti-MAP7D1 (Rabbit polyclonal) | Atlas antibodies | Cat#:HPA028075,RRID:AB_10603778 | IF (1:300) WB (1:1000) |
| Antibody | anti-Map7D3 (Rabbit polyclonal) | Atlas antibodies | Cat#:HPA035598,RRID:AB_10671108 | IF (1:300) WB (1:1000) |
| Antibody | Anti-GFP (Rabbit polyclonal) | Abcam | Cat#: ab290, RRID:AB_303395 | FACS (1:500) |
| Antibody | Anti-FAK (Phospho-Tyr397) (Rabbit polyclonal) | Biosource | Cat#:MBS003561 | IF (1:200) |
| Antibody | Anti-α-tubulin YL1/2 (Rat monoclonal) | Abcam | Cat#: ab6160, RRID:AB_305328 | IF (1:300) |
| Antibody | Anti-KIF13B (Rabbit polyclonal) | This paper | WB (1:500) | |
| Antibody | Alexa Fluor 488-, 594- and 647- secondaries | Molecular Probes | IF (1:300) | |
| Antibody | IRDye 680LT and 800CW secondaries | Li-Cor Biosciences | WB (1:15000) | |
| Sequence-based reagent | siRNA against luciferase (control) | CGTACGCGGAATACTTCGA | ||
| Sequence-based reagent | siRNA against Eg5 | This paper | GAGCCCAGATCAACCTTTA | |
| Sequence-based reagent | siRNA against MAP7D1 | Hooikaas et al., 2019, PMID:30770434 | TCATGAAGAGGACTCGGAA | |
| Sequence-based reagent | siRNA against MAP7D3 | Hooikaas et al., 2019, PMID:30770434 | AACCTACATTCGTCTACTGAT | |
| Sequence-based reagent | gRNA targeting sequence against KIF13B | This paper | TGCGGATACGACCCATGAAC | |
| Sequence-based reagent | gRNA targeting sequence against KIF5B | This paper | CCGATCAAATGCATAAGGCT | |
| Sequence-based reagent | gRNA targeting sequence against KIF1B | This paper | GCTGGTCTCTCGAGAATTGA | |
| Sequence-based reagent | gRNA targeting sequence against KIF1C | This paper | GCTGGTCTCACGGGCGTTAA | |
| Sequence-based reagent | gRNA targeting sequence against MAP7 | Hooikaas et al., 2019; PMID:30770434 | CGCCCTGCCTCTGCAATTTC | |
| Commercial assay or kit | HiPerfect | Qiagen | #301104 | |
| Commercial assay or kit | FuGENE6 | Promega | #E2691 | |
| Chemical compound, drug | Thymidin | Sigma-Aldrich | #T1895 | |
| Chemical compound, drug | Puromycin | InvivoGen | #ant-pr5b | |
| Chemical compound, drug | Biotin | Sigma-Aldrich | #B4639 | |
| Chemical compound, drug | Rapalog | Clontech | AP21967 | |
| Software, algorithm | ImageJ | ImageJ (http://imagej.nih.gov/ij/) | RRID:SCR_003070 | |
| Software, algorithm | FlowJo | FlowJo (https://flowjo.com) | (RRID:SCR_008520) | |
| Software, algorithm | GraphPad Prism | GraphPad Prism (https://graphpad.com) | RRID:SCR_015807 | |
| Software, algorithm | ImageJ detection of molecules plugin | Chazeau et al., 2016; PMID:26794511 | ||
| Software, algorithm | ImageJ SOS and SAID plugins | https://imagescience.org/meijering/software/beta/ Yao et al., 2017; PMID:28324611 | ||
| Software, algorithm | MATLAB code for track analysis | https://doi.org/10.6084/m9.figshare.c.5177636.v1 | ||
| Software, algorithm | MATLAB simpletracker code | https://www.github.com/tinevez/simpletracker | ||
| Other | Mitotracker Red | Invitrogen | #M7512 |