Cavin3 released from caveolae interacts with BRCA1 to regulate the cellular stress response
Figures
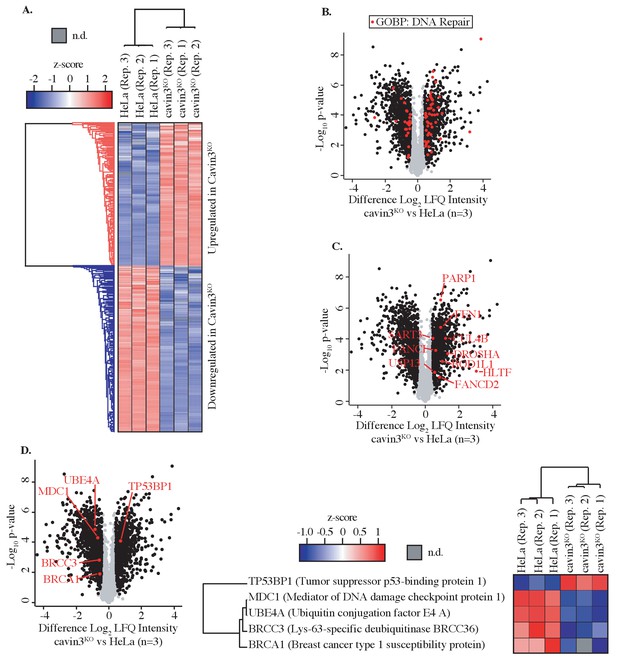
Global proteome analysis of cavin3 KO HeLa cells by label-free quantitative proteomics.
(A) Z-score for HeLa WT and cavin3 KO cells (replicates Rep. 1–3) showing upregulated proteins (red) and downregulated proteins (blue). (B) Volcano plot showing proteins (red dots) identified by Gene Ontology Biological Process (GOBP) involved in DNA repair. (C) Volcano plot showing DNA repair proteins upregulated in cavin3 KO cells. (D) Volcano plot showing proteins of the BRCA1 A-complex, BRCA1, BRCC3, MDC1, and UBE4A downregulated in cavin3 HeLa KO cells and upregulation of 53BP1 with a heatmap analysis of the expression of each of these proteins in replicate (Rep. 1–3) HeLa WT and cavin3 KO cells.
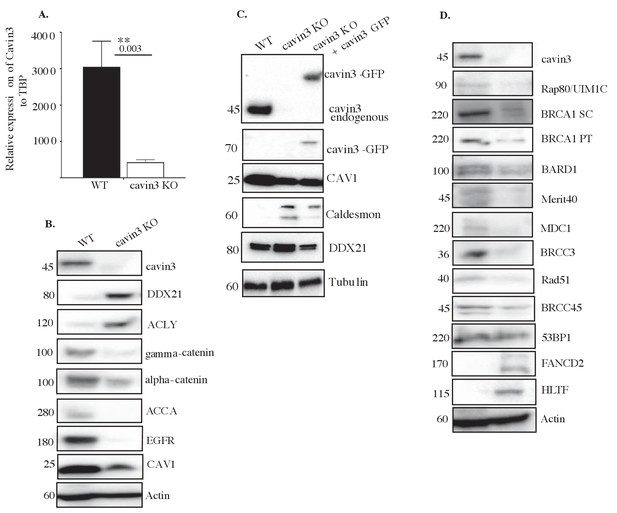
General characterization of cavin3 KO HeLa cells.
(A) mRNA analysis of cavin3 in WT and cavin3 KO cells from three independent experiments performed in triplicate samples as mean ± SD using Student's t-test, **p<0.01. (B) Representative western blot analysis of equally loaded lysates for cavin3, DDX21, ACLY, gamma-catenin, alpha-catenin, ACCA, EGFR, CAV1, and Actin in WT and cavin3 KO HeLa cell. (C) Western blot analysis of equally loaded lysates from WT, cavin3 KO, and cavin3 KO transfected with cavin3 for cavin3, GFP, CAV1, Caldesmon, DDX21, and Tubulin as the loading control. Western blots are representative of three independent experiments. (D) Representative western blot analysis of lysates from WT, and cavin3 KO cells were western blotted for cavin3, RAP80/UIM1C, BRCA1 Santa Cruz (SC), BRCA1 Proteintech (PT), BARD1, Merit40, MDC1, BRCC3, Rad51, BRCC45, 53BP1, FANCD2, HLTF, and Actin as the loading control.
-
Figure 1—figure supplement 1—source data 1
Raw western data for HeLa WT and cavin3 KO cells with molecular weight markers for Figure 1—figure supplement 1B.
(A) Western blot analysis of anti-rabbit cavin3, (B) anti-rabbit CAV1, (C) anti-rabbit ACLY, (D) anti-mouse alpha-catenin, (E) anti-rabbit ACCA, and (F) anti-rabbit EGFR antibodies in (1) WT HeLa cells and (2) cavin3 KO cells.
- https://cdn.elifesciences.org/articles/61407/elife-61407-fig1-figsupp1-data1-v2.pdf
-
Figure 1—figure supplement 1—source data 2
Raw western data for HeLa WT, cavin3 KO, and cavin3 KO with cavin3-GFP cells with molecular weight markers for Figure 1—figure supplement 1C.
(A) Western blot analysis of anti-rabbit cavin3, (B) anti-rabbit CAV1, (C) anti-mouse GFP, (D) anti-rabbit DDX21, (E) anti-rabbit Caldesmon and (F) anti-Tubulin antibodies in (1) HeLa WT, (2) cavin3 KO cells, and (3) cavin3KO + cavin3 GFP-expressing cells.
- https://cdn.elifesciences.org/articles/61407/elife-61407-fig1-figsupp1-data2-v2.pdf
-
Figure 1—figure supplement 1—source data 3
Raw western data for HeLa WT, cavin3 KO, and cavin3 KO with cavin3-GFP cells with molecular weight markers for Figure 1—figure supplement 1C.
(A) Western blot analysis of anti-rabbit cavin3, (B) anti-rabbit CAV1, (C) anti-mouse GFP, (D) anti-rabbit DDX21, (E) anti-rabbit Caldesmon, and (F) anti-Tubulin antibodies in (1) HeLa WT, (2) cavin3 KO cells, and (3) cavin3KO + cavin3 GFP-expressing cells.
- https://cdn.elifesciences.org/articles/61407/elife-61407-fig1-figsupp1-data3-v2.pdf
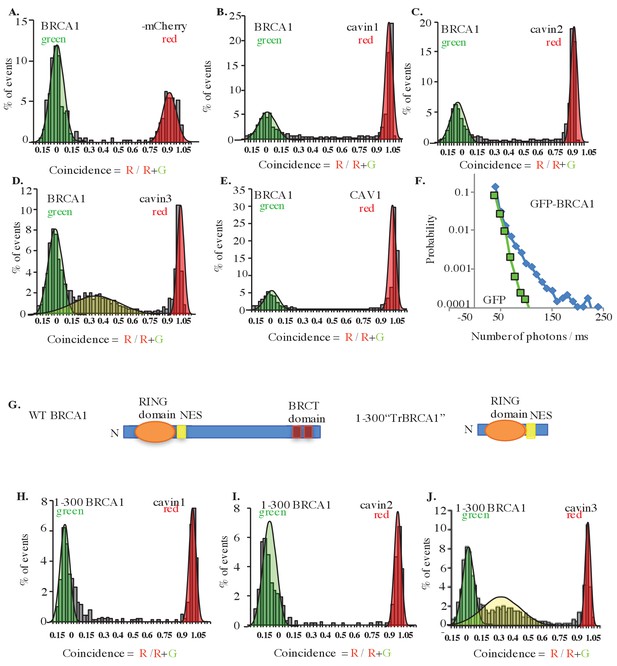
Single-molecule analysis of BRCA1 with cavin3-mCherry in MCF7 cells.
(A) Two-color single molecule fluorescence coincidence of BRCA1-GFP with (A) mCherry control, (B) mCherry-cavin1, (C) mCherry-cavin2, (D) mCherry-cavin3, and (E) mCherry-CAV1 coexpressed in MCF7 cells. The green curve represents BRCA1-GFP-only events, the red curve represents mCherry-only events, and the yellow curve represents BRCA1-GFP + Cherry events. (F) Distribution of burst brightness measured for BRCA1-GFP (blue) and GFP control (green). (G) Schematic representation of domain organization of full-length wildtype (WT) BRCA1 and the truncated (Tr) 1–300 BRCA1 constructs. NES: nuclear export signal; BRCT domain: BRCA1 C terminus domain; N: N terminus. (H–J). Two-color single-molecule fluorescence coincidence of 1–300 BRCA1 with (H) cavin1, (I) cavin2, and (J) cavin3 expressed in Leishmania cell-free lysates. More than 1000 events were collected in all cases.
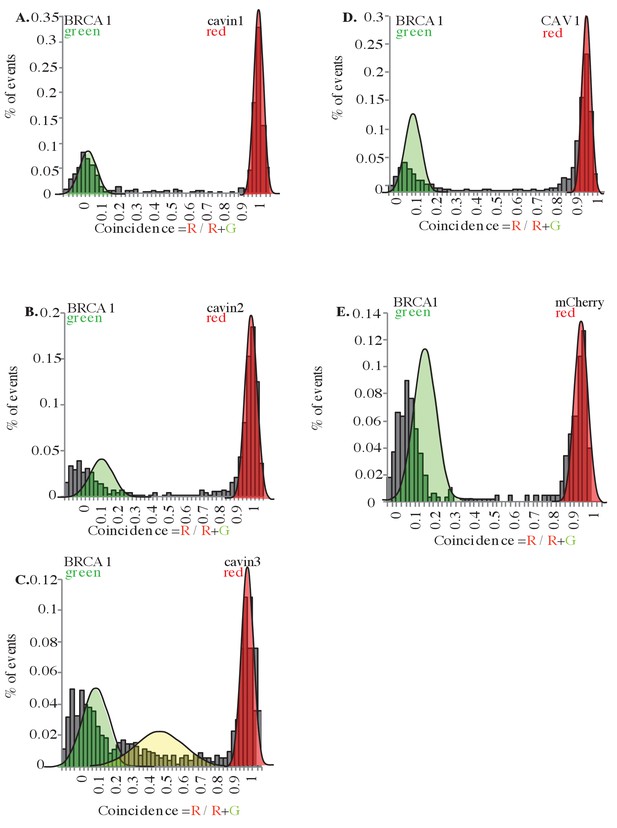
Single-molecule analysis in MDA-MB231 cells.
Two-color single-molecule fluorescence coincidence of BRCA1-GFP with mCherry tagged (A) cavin1, (B) cavin2, (C) cavin3, (D) CAV1, and (E) mCherry control expressed in MDA-MB231 cells. The green curve represents BRCA1-GFP-only events, the red curve represents mCherry-only events, and the yellow curve represents BRCA1-GFP + Cherry events. More than 1000 events were collected.
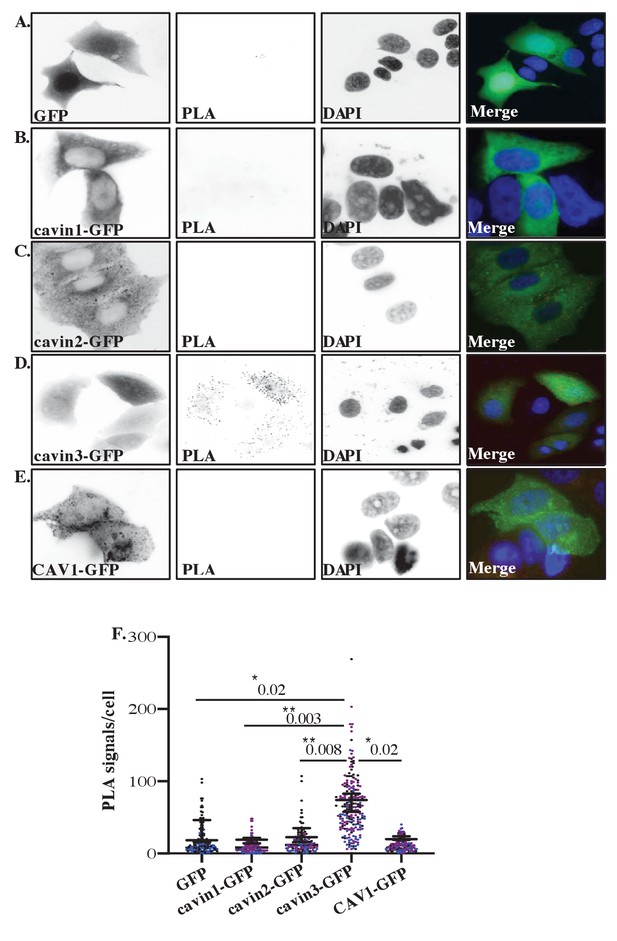
Proximity ligation assay (PLA) analysis of cavin3 and BRCA1 interaction in MCF7 cells.
(A–E) Immunofluorescence microscopy in combination with PLA for protein-protein interactions (red dots) within single cells of stably expressing (A) MCF7-GFP, (B) MCF7-cavin1-GFP, (C) MCF7-cavin2-GFP, (D) MCF7-cavin3-GFP, and (E) MCF7-CAV1-GFP using monoclonal GFP and polyclonal BRCA1 antibodies. DNA was counterstained with DAPI (blue). Scale bars represent 10 μm. (F). Number of red dots/PLA signals in 40–50 cells for each MCF7-GFP-expressing cell line was quantified from three independent experiments using a nested ANOVA. Each biological replicate is color-coded, and the mean ± SEM is presented as a black bar. **p<0.05, **p<0.01.
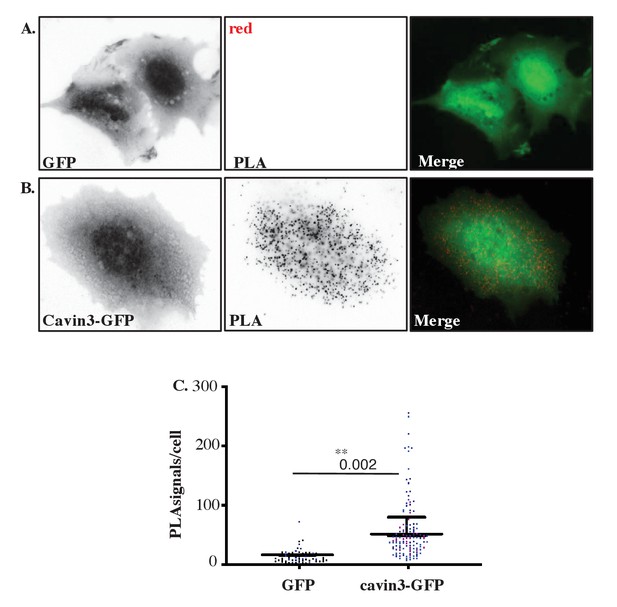
Proximity ligation assay (PLA) demonstrates cavin3 and BRCA1 interaction in MCF7 cells.
(A) Immunofluorescence microscopy in combination with PLA to detect and visualize protein-protein interactions (red dots) within single cells of stably expressing MCF7-GFP and (B) MCF7-cavin3-GFP cells using monoclonal BRCA1 (MS 110) and polyclonal GFP antibodies. DNA was counterstained with DAPI (blue). Scale bars represent 10 μm. (C) Number of red dots/PLA signals in 40–50 cells for each MCF7-GFP-expressing cell line quantified from three independent experiments using a nested ANOVA. Each biological replicate is color-coded with the mean ± SEM presented as a black bar. **p<0.01.
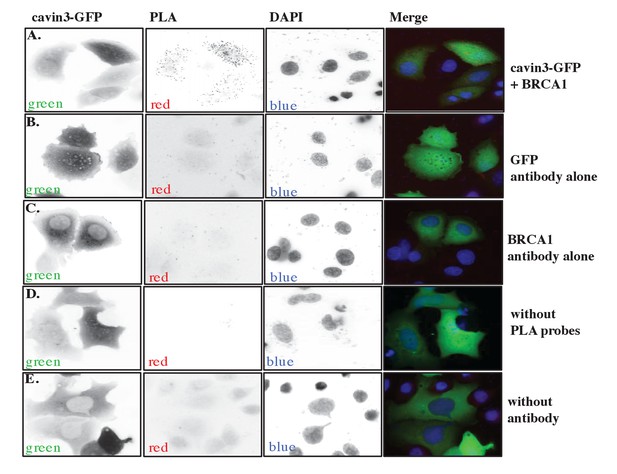
Proximity ligation assay (PLA) controls.
(A) Fluorescence microscopy analysis of PLA signals generated in MCF7 cells transfected with cavin3-GFP using mouse GFP and rabbit BRCA1 antibodies from Figure 3D, (B) GFP antibody alone, (C) BRCA1 antibody alone, (D) the absence of PLA probes, or (E) primary antibody controls. Representative images are from at least two independent experiments as shown.
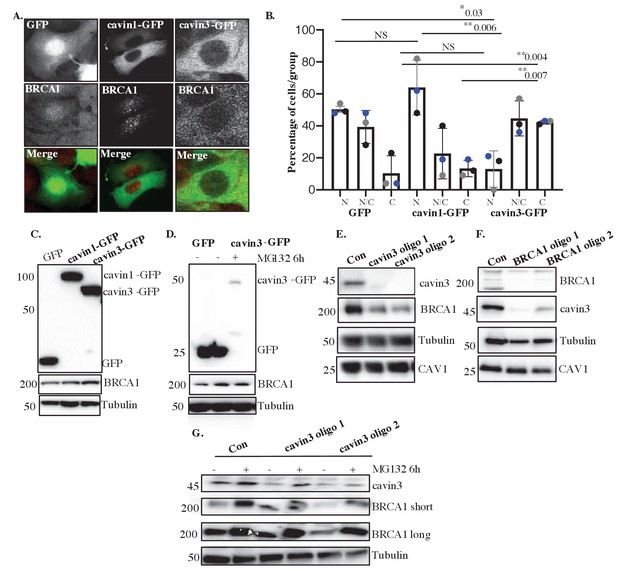
Cavin3 regulates BRCA1 protein expression and localization.
(A) Representative image of MCF7 cells stably expressing GFP alone, cavin1-GFP, and cavin3-GFP fixed and stained with a BRCA1 antibody. (B) Percentage of MCF7 cells showing strictly nuclear, nuclear-cytoplasmic, or cytoplasmic localization of BRCA1 was counted for 50 cells from 4 to 5 independent experiments as mean ± SD using a one-way ANOVA and Bonferroni’s multiple comparisons test. Each biological replicate was color-coded. NS: not significant, *p<0.05, **p<0.01. (C) Lysates from stably expressing MCF7 cells western blotted for GFP, BRCA1, and Tubulin as a load control. (D) MCF7-GFP and MCF7-cavin3-GFP cells, untreated (-) or treated with MG-132 for 6 hr. Lysates were western blotted with GFP, BRCA1, and Tubulin antibodies as a loading control. (E) A431 cells treated with control siRNAs (Con) or two siRNAs specific to cavin3. Lysates were western blotted using cavin3, BRCA1, CAV1 antibodies, and Tubulin as the loading control. (F) A431 cells treated with control siRNAs or two siRNAs specific to BRCA1. Lysates were western blotted using cavin3, BRCA1, CAV1 antibodies, and Tubulin as the loading control. (G) A431 cells treated with control (Con) or siRNAs specific to cavin3, untreated or treated with MG132 for 6 hr. Lysates were western blotted using cavin3, BRCA1, and Tubulin as a loading control. Quantitation of all blots in Figure 4 is provided in Figure 4—figure supplement 1A–E.
-
Figure 4—source data 1
Raw western data for MCF7 cells with molecular weight markers for Figure 4C.
(A) Western blot analysis of anti-rabbit BRCA1, (B) anti-mouse Tubulin, and (C) anti-mouse GFP antibodies in (1) GFP lysates, (2) cavin1-GFP lysates, and (3) cavin3-GFP lysates.
- https://cdn.elifesciences.org/articles/61407/elife-61407-fig4-data1-v2.pdf
-
Figure 4—source data 2
Raw western data for MCF7 cells with molecular weight markers for Figure 4D.
(A) Western blot analysis of anti-mouse GFP, (B) anti-rabbit BRCA1, and (C) anti-mouse Tubulin antibodies in (1) MCF7/GFP untreated, (2) MCF7/GFP + MG132-treated lysates, and (3) MCF7/cavin3-GFP untreated lysates.
- https://cdn.elifesciences.org/articles/61407/elife-61407-fig4-data2-v2.pdf
-
Figure 4—source data 3
Raw western data for A431 cells with molecular weight markers for Figure 4E.
(A) Western blot analysis of anti-rabbit cavin3, (B) anti-rabbit CAV1, (C) anti-mouse Tubulin, and (D) anti-rabbit BRCA1 antibodies in (1) A431 cells treated with control siRNA oligos, (2) A431 cells treated with cavin3-specific siRNA oligo 1, and (3) A431 cells treated with cavin3-specific siRNA oligo 2.
- https://cdn.elifesciences.org/articles/61407/elife-61407-fig4-data3-v2.pdf
-
Figure 4—source data 4
Raw western data for A431 cells with molecular weight markers for Figure 4F.
(A) Western blot analysis of anti-rabbit BRCA1, (B) anti-mouse Tubulin, (C) anti-rabbit cavin3, and (D) anti-rabbit CAV1 antibodies in A431 cells treated with (1) control siRNA oligos, (2) A431 cells treated with BRCA1-specific siRNA oligo 1, and (3) A431 cells treated with BRCA1-specific siRNA oligo 2.
- https://cdn.elifesciences.org/articles/61407/elife-61407-fig4-data4-v2.pdf
-
Figure 4—source data 5
Raw western data for A431 cells with molecular weight markers for Figure 4G.
(A) Western blot analysis of anti-rabbit BRCA1, (B) anti-rabbit cavin3, and (C) anti-mouse Tubulin antibodies in (1) A431 cells treated with control siRNA (control KD) oligos no treatment, (2) A431 cells treated with control siRNA oligos (control KD) and MG132 for 6 hr, (3) cavin3-specific siRNA (cavin3 KD) oligo 1 no treatment, (4) cavin3-specific siRNA (cavin3 KD) oligo 1 and MG132 for 6 hr, (5) cavin3-specific siRNA (cavin3 KD) oligo 2 no treatment, and (6) cavin3-specific siRNA (cavin3 KD) oligo 2 and MG132 for 6 hr.
- https://cdn.elifesciences.org/articles/61407/elife-61407-fig4-data5-v2.pdf
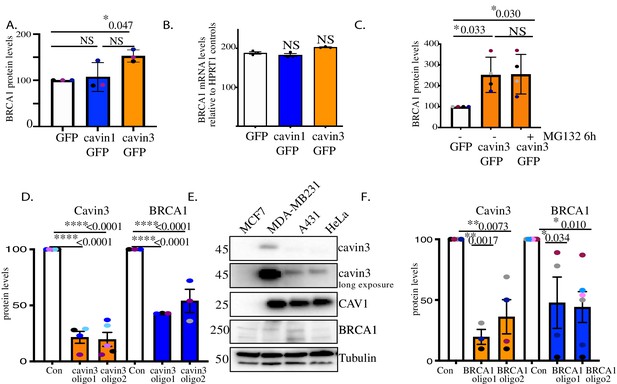
Reciprocal regulation of BRCA1 and cavin3 protein levels.
(A) Relative protein expression of BRCA1 in MCF7 cells expressing GFP (white), cavin1-GFP (blue), and cavin3-GFP (orange) using a one-way ANOVA and Bonferroni’s multiple comparisons test from three independent experiments. (B) BRCA1 gene expression was analyzed using the TaqMan Gene Expression assay as described in Materials and methods in MCF7 cells. Results are expressed as mean ± SD using a one-way ANOVA and Bonferroni’s multiple comparisons test from three independent experiments. NS: not significant. (C). Relative protein expression of BRCA1 in MCF7 cells expressing GFP or cavin3-GFP untreated (-) or treated with MG132 (+) using a one-way ANOVA and Bonferroni’s multiple comparisons test from three independent experiments. (D) Relative protein levels of cavin3 and BRCA1 in cells treated with control (Con) siRNAs or siRNAs specific to cavin3 (oligo1 and oligo2) in A431 cells using a one-way ANOVA and Bonferroni’s multiple comparisons test from 3 to 4 independent experiments. (E). Representative western blots of MCF7, MDA-MB231, A431, and HeLa cells were western blotted for protein expression of cavin3, CAV1, BRCA1, and Tubulin as the loading control. (F) Relative protein levels of cavin3 and BRCA1 in cells treated with control (Con) siRNAs or siRNAs specific to BRCA1 (oligo1 and oligo2) in A431 cells using a one-way ANOVA and Bonferroni’s multiple comparisons test from 3 to 4 independent experiments. For each experiment, each biological replicate was color-coded. NS: not significant; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 4—figure supplement 1—source data 1
Raw western data for HeLa WT and cavin3 KO cells with molecular weight markers for Figure 4—figure supplement 1E.
(A) Western blot analysis of anti-rabbit cavin3, (B) anti-mouse Tubulin, (C) anti-rabbit CAV1, and (D) anti-rabbit BRCA1 antibodies in (1) MCF7, (2) MDA-MB231, (3) A431, and (4) HeLa cells.
- https://cdn.elifesciences.org/articles/61407/elife-61407-fig4-figsupp1-data1-v2.pdf
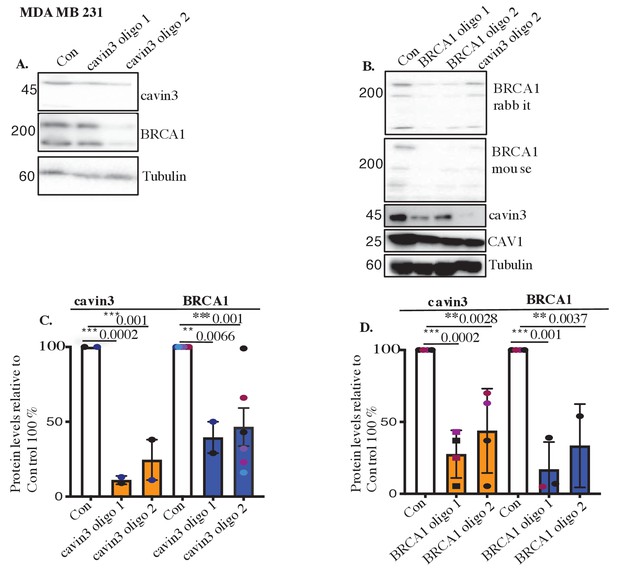
Validation of loss of cavin3 and BRCA1 in MDA-MB231 cells.
(A) Representative western blot analysis of cavin3, BRCA1, CAV1, and Tubulin as the loading control in MDA-MB231 cells following treatment with two siRNAs targeting cavin3 oligo 1 and 2 (or control). (B) Relative protein expression of cavin3 and BRCA1 in MDA-MB 231 cells transfected with cavin3 oligo 1 and cavin3 oligo 2 compared to control-treated cells from three independent experiments using a one-way ANOVA with Bonferroni’s multiple comparisons tests. Each biological replicate is color-coded. (C). Western blot analysis of BRCA1, cavin3, and Tubulin as the loading control in MDA-MB 231 cells following treatment with two siRNAs targeting BRCA1 oligo 1 and 2 (or control). (D) Relative protein expression of cavin3 and BRCA1 in MDA-MB 231 cells transfected with cavin3 oligo 1 and cavin3 oligo 2 compared to control-treated cells from three independent experiments using a one-way ANOVA with Bonferroni's multiple comparisons test. Each biological replicate is color-coded. **p<0.01, ***p<0.001.
-
Figure 4—figure supplement 2—source data 1
Raw western data for MDA-MB231 cells with molecular weight markers for Figure 4—figure supplement 2A.
(A) Western blot analysis of anti-rabbit BRCA1, (B) anti-rabbit cavin3, and (C) anti-mouse Tubulin in (1) MDA-MB231 treated with control siRNAs, (2) MDA-MB231 cells treated with cavin3-specific siRNA oligo 1, and (3) MDA-MB231 cells treated with cavin3-specific siRNA oligo 2.
- https://cdn.elifesciences.org/articles/61407/elife-61407-fig4-figsupp2-data1-v2.pdf
-
Figure 4—figure supplement 2—source data 2
Raw western data for MDA-MB231 cells with molecular weight markers for Figure 4—figure supplement 2B.
(A) Western blot analysis of anti-rabbit CAV1, (B) anti-rabbit cavin3, (C) anti-rabbit cavin1, (D) anti-mouse Tubulin, (E) anti-rabbit BRCA1, and (F) anti-mouse BRCA1 in (1) MDA-MB231 cells treated with control siRNAs, (2) MDA-MB231 cells treated with BRCA1-specific siRNA oligo 1, (3) MDA-MB231 cells treated with BRCA1-specific siRNA oligo, and (4) MDA-MB231 cells treated with cavin3-specific oligo 1.
- https://cdn.elifesciences.org/articles/61407/elife-61407-fig4-figsupp2-data2-v2.pdf
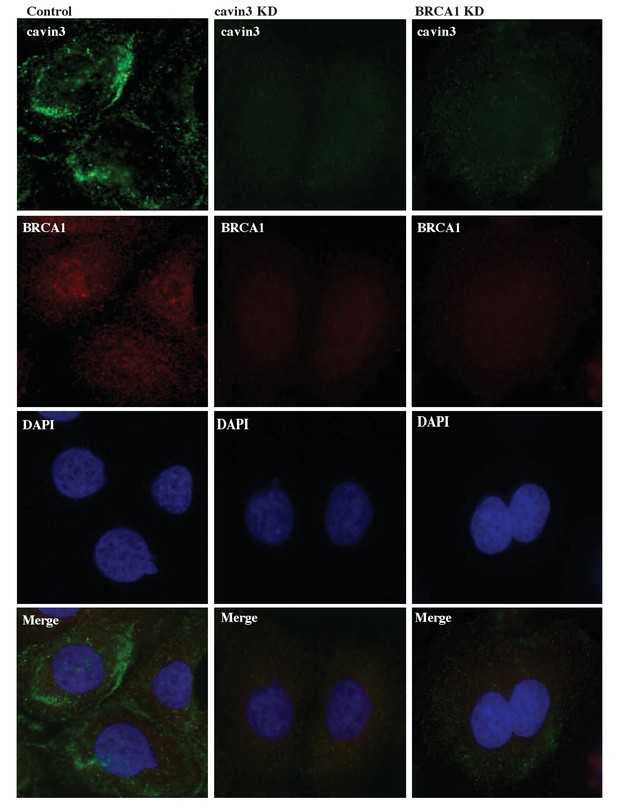
Reciprocal loss of BRCA1 and cavin3 in A431 cells.
Representative immunofluorescence images of control knockdown, cavin3 knockdown, and BRCA1 knockdown cells for cavin3 (green), BRCA1 (red), and DAPI (nuclei). Images are representative of three independent experiments.
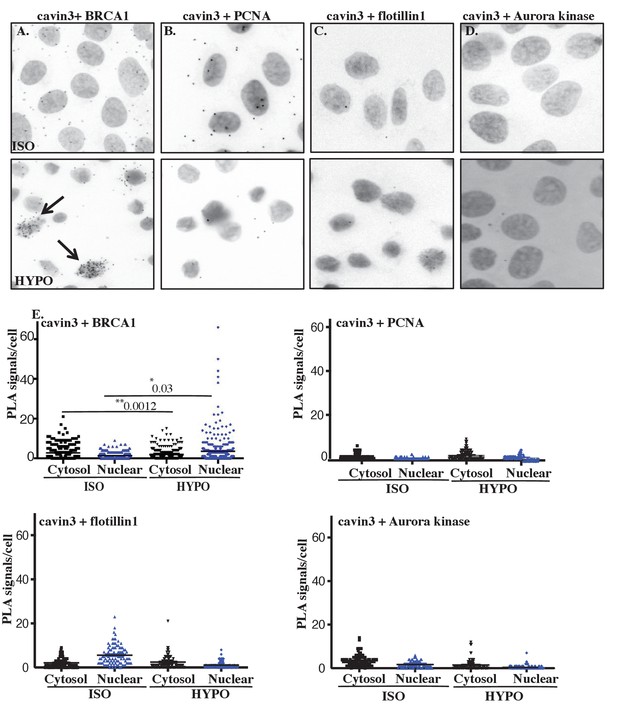
Cellular swelling of A431 cell causes an increase in the BRCA1-cavin3 interaction.
(A) A431 cells were treated with isotonic (ISO) or hypo-osmotic (HYPO) medium, and proximity ligation assay (PLA) was performed using cavin3 and BRCA1, (B) cavin3 and PCNA, (C) cavin3 and flotillin1, and (D) cavin3 and Aurora kinase antibodies as controls for PLA. DNA was counterstained with DAPI (blue). Scale bars represent 10 μm. (E) Total number of PLA signals in the cytosol and the nucleus of cells as defined by DAPI staining in 50 cells for each pair of antibodies quantified from three independent experiments using a nested ANOVA with the mean ± SEM represented by the black bar, *p<0.05, **p<0.01.
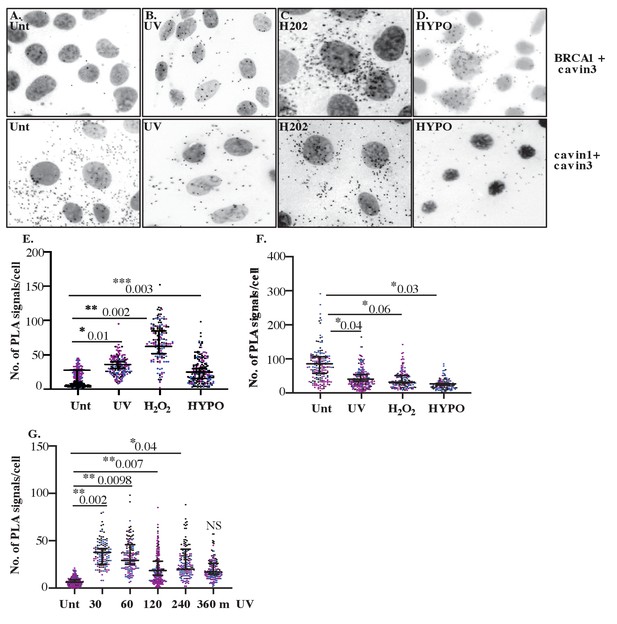
Close association between cavin3 and BRCA1 in A431 cells after stress treatment.
(A) Immunofluorescence microscopy in combination with proximity ligation assay (PLA) visualization of endogenous protein-protein interactions (red dots) within A431 cells in (A) untreated (Unt.) cells, (B) UV treated and a chase time of 30 min, (C) 200 μM H2O2 (H2O2) for 30 min, and (D) hypo-osmotic treatment (HYPO) for 10 min. Top panel: BRCA1 and cavin3; bottom panel: cavin1 and cavin3. (E) PLA signals/cell for cavin3-BRCA1 association in 50 cells/biological replicate with three independent experiments. (F) PLA time-course analysis after UV treatment and a chase time up to 360 min in 50 cells/biological replicate with three independent experiments. (G) PLA signals/cell for cavin1-cavin3 association in 50 cells/biological replicate with three independent experiments. All data was quantified from three independent experiments using a nested ANOVA. Each biological replicate is color-coded with the mean ± SEM presented as a black bar. NS: not significant; *p<0.05, **p<0.01, ***p<0.001.
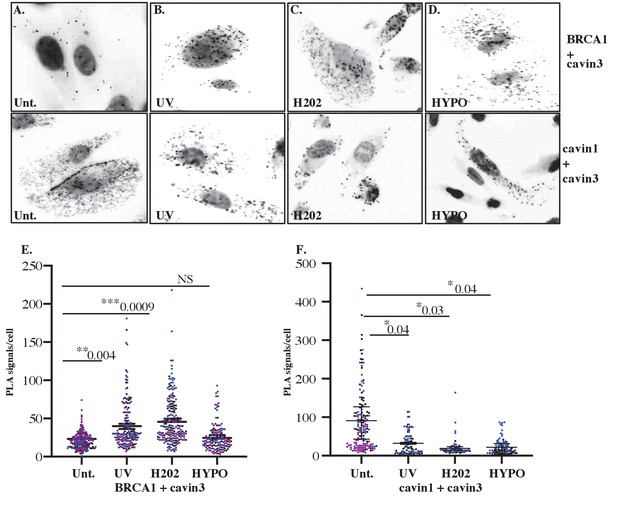
Close association of cavin3 and BRCA1 in MDA-MB231 cells after stress treatment.
(A) Immunofluorescence microscopy in combination with proximity ligation assay (PLA) visualization of endogenous protein-protein interactions (red dots) within MDA-MB231 cells in untreated (Unt.) cells, (B) UV treatment (2 min) and a 30 min chase time, (C) 200 μM H2O2 for 30 min, and (D) hypo-osmotic treatment (90% H2O in Dulbecco’s modified Eagle’s medium [DMEM]) for 10 min. (E) Number of red dots/PLA signals in 40–50 cells for cavin-BRCA1 was quantified from three independent experiments and is presented as mean ± SEM using a nested ANOVA. Each biological replicate is color-coded with the mean presented as a black bar. (F) Number of red dots/PLA signals in 40–50 cells for cavin3-cavin1 was quantified from three independent experiments and is presented as mean ± SEM using a nested ANOVA. Each biological replicate is color-coded with the mean presented as a black bar. *p<0.05, **p<0.01, ***p<0.001.
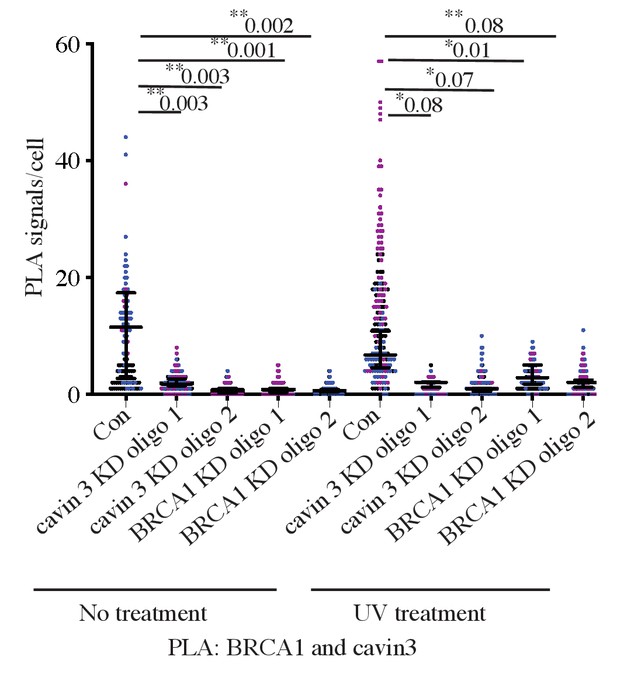
Proximity ligation assay (PLA) controls for cavin3 and BRCA1 PLA antibodies in A431 cells.
A431 cells treated with control (Con) or siRNAs specific to cavin3 or BRCA1 (oligo 1 and 2). Cells were left untreated or subjected to UV treatment. Cells were subject to immunofluorescence microscopy in combination with PLA using monoclonal mouse BRCA1 and polyclonal rabbit cavin3 antibodies. The number of PLA signals in 40–50 cells was quantified from three independent experiments using a nested ANOVA. Each biological replicate is color-coded with the mean ± SEM presented as a black bar. *p<0.05, **p<0.001.
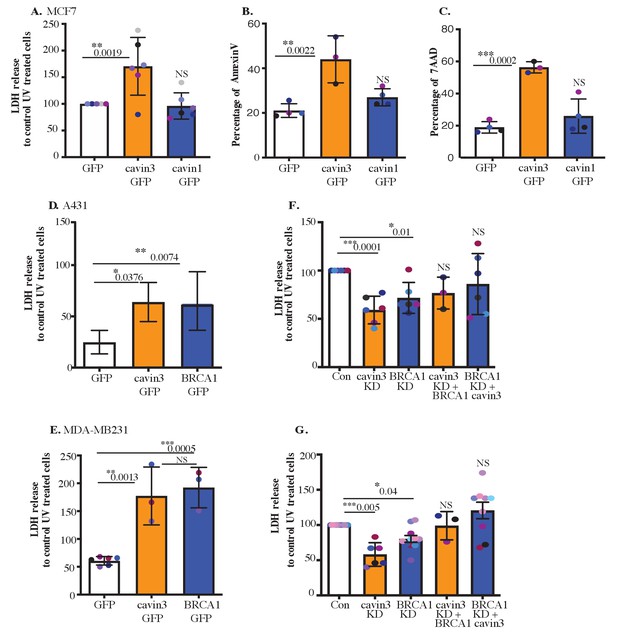
Cavin3 potentiates BRCA1 functions in apoptosis.
(A) LDH release of MCF7-GFP, cavin3-GFP, and cavin1-GFP cells subjected to UV treatment and a 6 hr chase. LDH release is expressed as a percentage to control GFP cells from six independent experiments presented as mean ± SD using a one-way ANOVA and Bonferroni’s multiple comparisons test. (B) Annexin V-positive cells after UV treatment and a 6 hr recovery time in MCF7 cells presented as mean ± SD using a one-way ANOVA and Bonferroni’s multiple comparisons test from three independent experiments. (C) 7-AAD-positive cells after UV treatment and a 24 hr recovery time in MCF7 cells presented as mean ± SD using a one-way ANOVA and Bonferroni’s multiple comparisons test from three independent experiments. (D) A431 cells and (E) MDA-MB231 cells were transfected with GFP, cavin3-GFP, or BRCA1-GFP. Results are the relative percentage of LDH release to GFP as mean ± SD using a one-way ANOVA and Bonferroni’s multiple comparisons test from at least three independent experiments. (F) A431 cells and (G) MDA-MB231 cells were treated with control, cavin3, or BRCA1 specific siRNAs. Cavin3-depleted A431 and MDA-MB231 cells were transfected with BRCA1-GFP for 24 hr. BRCA1-depleted A431 and MDA-MB231 cells were transfected with cavin3-GFP for 24 hr. All cells were UV treated, and LDH release was measured and calculated relative to control siRNA UV-treated cells. The results represent independent experiments as mean ± SD using a one-way ANOVA and Bonferroni’s multiple comparisons test from three independent experiments. Each biological replicate is color-coded. NS: not significant; *p<0.05, **p<0.01, ***p<0.001.
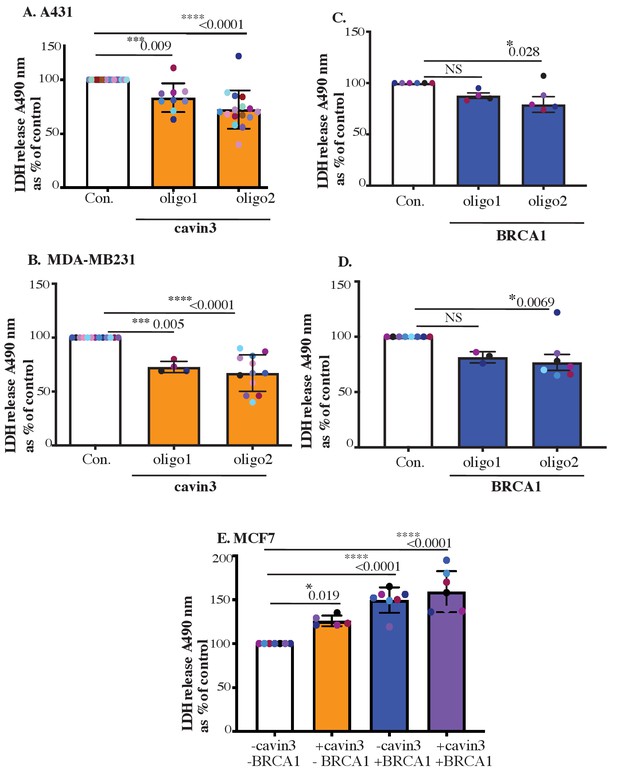
Validation of LDH release in MCF7, A431, and MDA-MB231 cells.
(A) Equal numbers of A431 cells treated with control siRNA or cavin3-specific siRNAs were subjected to UV treatment and a recovery time of 6 hr. (B) Equal numbers of A431 cells treated with control siRNAs or BRCA1-specific siRNAs were subjected to UV treatment (2 min) and a recovery time of 6 hr. (C) Equal numbers of MDA-MB231 cells treated with control siRNAs or cavin3-specific siRNAs were subjected to UV treatment and a recovery time of 6 hr. (D) Equal numbers of MDA-MB231 cells treated with control siRNAs or BRCA1-specific siRNAs were subjected to UV treatment and a recovery time of 6 hr. LDH release was measured from the cell supernatant and was calculated relative to control siRNA UV-treated cells as mean ± SD using a one-way ANOVA and Bonferroni’s multiple comparisons test. (E) MCF7 cells depleted of BRCA1 (-cavin3, -BRCA1), depleted of BRCA1 and transfected with cavin3-GFP (-BRCA1, +cavin3), left untreated (-cavin3, +BRCA1), or transfected with cavin3-GFP (+BRCA1, +cavin3). All cells were subjected to UV treatment and a 6 hr recovery time. LDH release was measured from the cell supernatant and was calculated relative to control MCF7 cells lacking both BRCA1 and cavin3 as mean ± SD using a one-way ANOVA and Bonferroni’s multiple comparisons test. Each biological replicate is color-coded. NS: not significant; *p<0.05, ***p<0.001, ****p<0.0001.
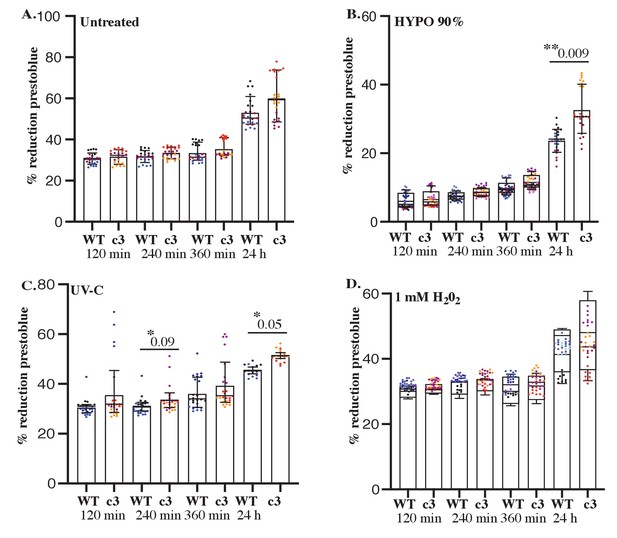
Cavin3 KO cells exhibit resistance to stressors that allow BRCA1 interaction.
Equal numbers of WT and cavin3 KO cells were either left (A) untreated, (B) treated with 90% hypo-osmotic (HYPO) medium, (C) UV-C 2 min, or (D) 1 mM H2O2 (oxidative stress). PrestoBlue reagent was added to plates immediately and were read at 570 and 600 nm at 120 min, 240 min, 360 min, and 24 hr. The % reduction PrestoBlue was calculated from eight wells/replicate experiment and is presented as the mean ± SEM using a nested ANOVA for each time point from 3 to 4 independent experiments. Each biological replicate is color-coded. *p<0.05, **p<0.01.
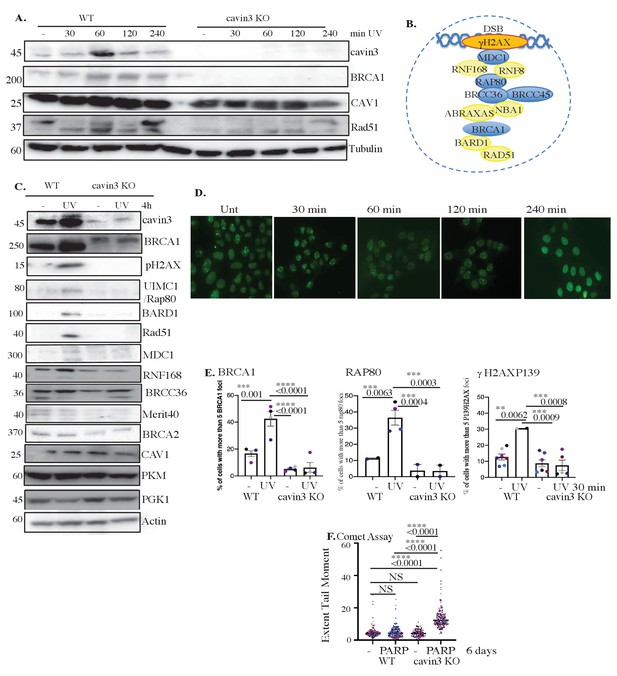
Cavin3-deficient HeLa cells exhibit abolishment of DNA repair.
(A) Representative western blot analysis of WT and cavin3 KO cells UV time course for cavin3, BRCA1, CAV1, Rad51, and Tubulin. (B) Protein components of the BRCA1 A-complex. Blue-colored circles: proteins downregulated in the label-free quantitative (LFQ) proteomics; yellow-colored circles: proteins not detected in the LFQ proteomics of cavin3 KO cells. (C) Representative western blot analysis of cavin3, BRCA1, pH2AX, UIM1C/Rap80, BARD1, Rad51, MDC1, RNF168, BRCC36, Merit40, BRCA2, CAV, PKM, PGK1, and Actin in WT and cavin3 KO HeLa cells untreated (-) or UV treated (UV) followed by a 4 hr chase. Quantitation of protein levels from three independent experiments is presented in Figure 8—figure supplement 1. (D) Representative immunofluorescence images of BRCA1 foci after UV treatment in WT HeLa cells. (E) Percentage of cells with more than five BRCA1 foci, Rap80 foci, and γH2AX foci in WT and cavin3 KO cells following UV treatment and a 30 min chase. The results are presented as mean ± SD using a one-way ANOVA and Bonferroni’s multiple comparisons test from three independent experiments. (F) WT and cavin3 KO cells untreated or treated with the PARP inhibitor (AZD2461, PARPi) 5 nM for 6 days were subjected to comet assays. The results are presented as the mean ± SEM using a one-way ANOVA and Bonferroni’s multiple comparisons test from three independent experiments. Each biological replicate is color-coded. Extent Tail Moment was calculated as described in Materials and methods. NS: not significant; **p<0.01, ***p<0.001; ****p<0.0001.
-
Figure 8—source data 1
Raw western data for HeLa WT and cavin3 KO cells time course after UV treatment with molecular weight markers for Figure 8A.
Western blot analysis of (A) anti-rabbit cavin3, (B) anti-rabbit CAV1, (C) anti-rabbit BRCA1, (D) anti-rabbit RAD51, and (E) anti-mouse Tubulin antibodies in (1) WT control, (2) WT UV 30 min chase, (3) WT UV 60 min chase, (4) WT UV 120 min chase, (5) WT UV 240 min chase, (6) cavin3 KO control, and (7) cavin3 KO UV 30 min chase, cavin3 KO UV 60 min chase, cavin3 KO UV 120 min chase, and cavin3 KO 240 min chase.
- https://cdn.elifesciences.org/articles/61407/elife-61407-fig8-data1-v2.pdf
-
Figure 8—source data 2
Raw western data for HeLa WT and cavin3 KO cells untreated or UV treatment for 4 hr with molecular weight markers for Figure 8C.
(A) Western blot analysis of anti-rabbit cavin3, (B) anti-rabbit BRCA1, (C) anti-rabbit Rad51 and γH2AX, (D) anti-rabbit BRCC36, (E) anti-sheep Merit40, (F) anti-rabbit BRCA2, (G) anti-rabbit CAV1, (H) anti-rabbit PKM, (I) anti-rabbit PGK1, and (J) anti-actin antibodies in (1) WT untreated cells, (2) WT + UV treatment and a 4 hr chase, (3) cavin3 KO cells, and (4) cavin3 KO + UV treatment and a chase 4 hr chase time.
- https://cdn.elifesciences.org/articles/61407/elife-61407-fig8-data2-v2.pdf
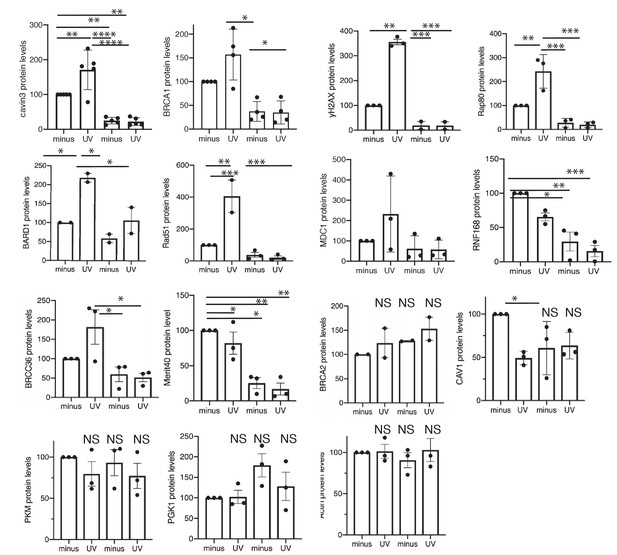
Quantitation of BRCA1-A-complex proteins in WT and cavin3 KO cells.
Densitometry analysis was performed of the protein levels of cavin3, BRCA1, P139 γH2AX, RAP80, BARD1, RAD51, MDC1, RNF168, BRCC36, Merit40, BRCA2, CAV1, PKM, PGK1, and Actin in Figure 8A, C in WT and cavin3 KO cells subjected to UV treatment and a 4 hr chase from 2 to 3 independent experiments presented as mean ± SD using a one-way ANOVA and Bonferroni’s multiple comparisons test. NS: not significant; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
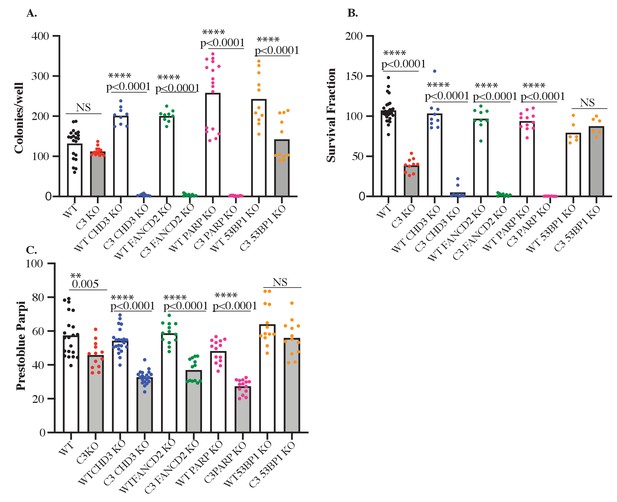
Cavin3 KO cells are sensitive to PARP inhibition, and 53BP1 loss causes PARP inhibitor reversion.
(A) WT HeLa (black dots) and cavin3 KO cells (red dots), depleted of CHD3 (blue dots), FANCD2 (green dots), PARP1 (pink dots), and 53BP1 (orange dots) seeded at low density without treatment were allowed to form colonies for 6 days. Colonies were fixed and stained with 0.5% crystal violet/20% ethanol and colonies larger than 50 cells were counted. Each dot represents the number of colonies in a six-well dish. (B) WT HeLa (black dots) and cavin3 KO cells (red dots), depleted of CHD3 (blue dots), FANCD2 (green dots), PARP1 (pink dots), and 53BP1 (orange dots) seeded at low density were treated with 5 nM AZD2461 and were allowed to form colonies for 6 days. Colonies were fixed and stained with 0.5% crystal violet/20% ethanol and colonies larger than 50 cells were counted. Each dot represents the number of colonies in a six-well dish. (C) WT HeLa (black dots) and cavin3 KO cells (red dots), depleted of CHD3 (blue dots), FANCD2 (green dots), PARP1 (pink dots), and 53BP1 (orange dots) seeded at low density were treated with 5 nM AZD2461 for 6 days followed by PrestoBlue addition and quantitation.
-
Figure 8—figure supplement 2—source data 1
Raw western data for HeLa WT and cavin3 KO cells depleted of FANCD2, PARP1, CHD3, and 53BP1 with molecular weight markers for Figure 8—figure supplement 2.
(A) Western blot analysis of anti-rabbit FANCD2 and (B) anti-mouse actin antibodies in (1) WT HeLa, (2) WT HeLa + FANCD2 KO, (3) cavin3 KO, and (4) cavin3 KO + FANCD2 KO cells. (C) Western blot analysis of anti-rabbit PARP1 and (D) anti-mouse Tubulin antibodies in (1) WT HeLa, (2) WT HeLa + PARP1 KO clone 1, (3) cavin3 KO, (4) cavin3 KO + PARP1 KO clone 1, (5) WT HeLa, (6) WT + PARP1 KO clone 3, (7) cavin3 KO, and (8) cavin3 KO + PARP1 clone 3 cells. (E) Western blot analysis of anti-rabbit CHD3 and (F) anti-mouse Tubulin antibodies in (1) WT HeLa, (2) WT + CHD3 KO clone 1, (3) WT + CHD3 KO clone 3, (4) cavin3 KO cells, (5) cavin3 KO + CHD3 clone 1, and (6) cavin3 KO + CHD3 clone 3. (G) Western blot analysis of anti-rabbit 53BP1 and (H) anti-mouse Tubulin antibodies in (1) WT HeLa, (2) WT + 53BP1 KO, (3) cavin3 KO, and (4) cavin3 KO + 53BP1 KO cells.
- https://cdn.elifesciences.org/articles/61407/elife-61407-fig8-figsupp2-data1-v2.pdf
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | MCF7 cells | ATCC | ATCC: HBT-22 RRID:CVCL_0031 | Figure 2, Figure 3, Figure 4, Figure 7A–C, Figure 3—figure supplement 1, Figure 3—figure supplement 2, Figure 4—figure supplement 1A–C, E, Figure 7—figure supplement 1E |
| Cell line (H. sapiens) | MDA-MB231 cells | ATCC | ATCC: HTB-26 RRID:CVCL_0062 | Figure 7E, G, Figure 2—figure supplement 1, Figure 4—figure supplement 1E, Figure 4—figure supplement 2, Figure 6—figure supplement 1, Figure 7, Figure 7—figure supplement 1C, D |
| Cell line (H. sapiens) | A431 cells | ATCC | ATCC:CRL-1555 RRID:CVCL_0037 | Figure 5, Figure 6, Figure 7D, Figure 4—Figure 4—figure supplement 1D-F, Figure 4—figure supplement 3, Figure 6—figure supplement 2, Figure 7—figure supplement 1A, B |
| Cell line (H. sapiens) | HeLa WT cells | ATCC | ATCC: CRM-CCL-2, RRID:CVCL_0030 | Figure 1, Figure 8, Figure 1—figure supplement 1, Figure 7—figure supplement 2, Figure 8—figure supplements 1 and 2 |
| Cell line (H. sapiens) | HeLa cavin3 KO cells | This paper | Figure 1, Figure 8, Figure 1—figure supplement 1, Figure 7—figure supplement 2, Figure 8—figure supplements 1 and 2 | |
| Antibody | 53BP1 rabbit polyclonal | GeneTex | GeneTex Cat# GTX112864 | WB 1:1000 |
| Antibody | ACCA rabbit polyclonal | Cell Signaling | Cell Signaling Cat# 3662 RRID:AB_2219400 | WB 1:5000 |
| Antibody | Actin mouse monoclonal | Millipore | Millipore Cat# MAB1501, RRID:AB_2223041 | WB 1:5000 |
| Antibody | ACLY rabbit polyclonal | Sigma-Aldrich | Sigma-Aldrich Cat# HPA028758, RRID:AB_10603575 | WB 1:2000 |
| Antibody | Aurora kinase mouse monoclonal | BD Biosciences | BD Biosciences Cat# 611082, RRID:AB_2227708 | PLA 1:100 |
| Antibody | Alexa Fluor 488 Goat anti-Rabbit IgG (H + L) | Thermo Fisher Scientific | Thermo Fisher Scientific Cat# A-11034, RRID:AB_2576217 | IF: 1:500 |
| Antibody | Alexa Fluor 546 Goat anti-Mouse IgG (H + L) | Thermo Fisher Scientific | Thermo Fisher Scientific Cat# A-11030, RRID:AB_2534089 | IF 1:500 |
| Antibody | Alexa Fluor 594 Donkey anti-Rabbit IgG (H + L) | Thermo Fisher Scientific | Thermo Fisher Scientific Cat# A-21207, RRID:AB_141637 | IF 1:500 |
| Antibody | Alexa Fluor 594 Goat anti-Mouse IgG (H + L) | Thermo Fisher Scientific | Thermo Fisher Scientific Cat# A-21203, RRID:AB_141633 | IF 1:500 |
| Antibody | BARD1 E-11 mouse monoclonal | Bio-Strategy Laboratory Products | Santa Cruz Cat# sc-74559, RRID:AB_2061237 | WB 1:500 |
| Antibody | BRCA1 C-20 rabbit polyclonal | Bio-Strategy Laboratory Products | Santa Cruz Cat# sc-642, RRID:AB_630944 | WB 1:500 IF 1:100 PLA 1:100 |
| Antibody | BRCA1 MS110 mouse monoclonal | Abcam | Abcam Cat# ab16780, RRID:AB_2259338 | WB 1:1000 IF 1:100 PLA 1:100 |
| Antibody | BRCA1 D-9 mouse monoclonal | Bio-Strategy Laboratory Products | Santa Cruz Cat# sc-6954, RRID:AB_626761 | IF1:50 |
| Antibody | BRCA1 rabbit polyclonal | Millipore | Millipore Cat# 07-434, RRID:AB_2275035 | WB 1:2000 |
| Antibody | BRCA1 rabbit polyclonal | Proteintech | Proteintech Cat# 22363-1-AP, RRID:AB_2879090 | WB 1:1000 |
| Antibody | BRCA2 rabbit polyclonal | BioVision | BioVision Cat# 3675-30T, RRID:AB_2067764 | WB 1:2000 |
| Antibody | BRCC36 rabbit polyclonal | ProScience | ProScience Cat# 4311 | WB 1:1000 |
| Antibody | BRCC45 rabbit polyclonal | GeneTex | GeneTex Cat# GTX105364, RRID:AB_1949757 | WB 1:2000 |
| Antibody | Caldesmon mouse monoclonal | BD Biosciences | BD Biosciences Cat#610660 | WB 1:3000 |
| Antibody | Catenin- alpha mouse monoclonal | Cell Signaling | Cell Signaling Cat# 2131 | WB 1:3000 |
| Antibody | Catenin-gamma mouse monoclonal | Cell Signaling | Cell Signaling Cat# 2309 | WB 1:3000 |
| Antibody | Caveolin1 (CAV1) rabbit polyclonal | BD Biosciences | BD Biosciences Cat#610060, RRID:AB_397472 | WB 1: 5000 |
| Antibody | cavin1 mouse monoclonal | Abmart, China | PLA 1:100 | |
| Antibody | cavin1 rabbit polyclonal | Sigma-Aldrich | Sigma-Aldrich Cat# AV36965, RRID:AB_1855947 | WB 1:2000 |
| Antibody | cavin3 mouse monoclonal | Novus | Novus Cat# HOO112464-MO, RRID:AB_11188730 | PLA 1:200 |
| Antibody | cavin3 rabbit polyclonal | Proteintech | Proteintech Cat# 16250-1-AP, RRID:AB_2171897 | WB 1:2000 IF 1:300 PLA 1:200 |
| Antibody | CHD3 rabbit polyclonal | GeneTex | GeneTex Cat# GTX131779, RRID:AB_2886520 | WB 1:500 |
| Antibody | DDX21 rabbit polyclonal | Novus | Novus Cat# NBP1-88310, RRID:AB_11027665 | WB 1:2000 |
| Antibody | EGFR Clone LA22 mouse monoclonal | Millipore | Millipore Cat# 05-104, RRID:AB_11210086 | WB 1:4000 |
| Antibody | FANCD2 N1 mouse monoclonal | GeneTex | GeneTex Cat# GTX116037, RRID:AB_2036898 | WB 1:500 |
| Antibody | Flotillin1 Clone 18 mouse monoclonal | BD Biosciences | BD Biosciences Cat# 610821, RRID:AB_398140 | PLA 1:100 |
| Antibody | GFP mouse monoclonal | Roche | Roche Cat#11814460001, RRID:AB_390913 | WB 1:4000 PLA 1:300 |
| Antibody | Histone H2.AX-Chip Grade | Abcam | Abcam Cat# ab20669, RRID:AB_445689 | WB 1:1000 |
| Antibody | Histone H2.AX (20E3) P139 | Cell Signaling Technology | Cell Signaling Technology Cat# 9718, RRID:AB_2118009 | IF 1:500 |
| Antibody | Histone H2.AX Chip Grade P139 rabbit polyclonal | Abcam | Abcam Cat# ab2893, RRID:AB_303388 | WB 1:3000 |
| Antibody | HLTF rabbit polyclonal | Proteintech | Proteintech Cat# 14286-1-AP, RRID:AB_2279646 | WB 1:2000 |
| Antibody | HRP-Goat anti-Mouse IgG (H + L) | Thermo Fisher Scientific | Thermo Fisher Scientific Cat# G-21040, RRID:AB_2536527 | WB 1:5000 |
| Antibody | HRP-Goat anti-Rabbit IgG (H + L) | Thermo Fisher Scientific | Thermo Fisher Scientific Cat# G-21234, RRID:AB_2536530 | WB 1:5000 |
| Antibody | HRP-rabbit anti-sheep IgG (H + L) | Abcam | Abcam Cat# ab97130, RRID:AB_10679515 | WB 1:2000 |
| Antibody | MDC1 rabbit polyclonal | Novus | Novus Cat#NB10056657, RRID:AB_838567 | WB 1:100 |
| Antibody | Merit40 sheep polyclonal | R&D Systems | R&D Systems Cat# AF6604, RRID:AB_10717577 | WB 1:500 |
| Antibody | PARP1 rabbit polyclonal | GeneTex | GeneTex Cat# GTX112864, RRID:AB_11173565 | WB 1:1000 |
| Antibody | PCNA mouse monoclonal | Millipore | Millipore Cat# NA03T, RRID:AB_2160357 | PLA: 1:100 |
| Antibody | PGK1 rabbit polyclonal | GeneTex | GeneTex Cat# GTX107614, RRID:AB_2037666 | WB 1:3000 |
| Antibody | PKM rabbit polyclonal | GeneTex | GeneTex Cat# GTX107977, RRID:AB_1951264 | WB 1:3000 |
| Antibody | Rad51 mouse monoclonal | Novus | Novus Cat# NB 100-148, RRID:AB_350083 | WB 1:1000 |
| Antibody | RAP80 D1T6Q rabbit polyclonal | Cell Signaling Technology | Cell Signaling Technology Cat# 14466, RRID:AB_2798487 | WB1:1000 IF 1:100 |
| Antibody | RNF168 rabbit polyclonal | GeneTex | GeneTex Cat# GTX118147, RRID:AB_11169617 | WB 1:1000 |
| Antibody | Tubulin (DM1A) mouse monoclonal | Abcam | Abcam Cat# ab7291, RRID:AB_2241126 | WB 1:4000 |
| Sequence-based reagent | CHD3 human | Integrated DNA Technologies | Hs.Cas9.CHD3.1.AA, strand sequence GACCGGGTCGGAAACGAAGA | |
| Sequence-based reagent | FANCD2 human | Integrated DNA Technologies | Hs.Cas9.FANCD2.1.AA, strand sequence AGTTGACTGACAATGAGTCG | |
| Sequence-based reagent | PARP1 human | Integrated DNA Technologies | Hs.Cas9.PARP1.1.AA, strand sequence GAGTCGAGTACGCCAAGAGC | |
| Sequence-based reagent | 53BP1 human | Integrated DNA Technologies | Hs.Cas9.TP53BP1.1.AA strand sequence AACGAGGAGACGGTAATAGT | |
| Sequence-based reagent | siRNAs to BRCA1 human | Life Technologies | HSS101089 HSS186096 HSS186097 | |
| Sequence-based reagent | siRNAs to cavin3 human | Life Technologies | HSS174185 HSS150811 HSS150809 | |
| Commercial assay or kit | Cytotoxicity Detection KitPLUS LDH | Sigma-Aldrich | Sigma-Aldrich: 4744934001 | |
| Commercial assay or kit | Duolink In situ PLA Probe anti-Rabbit MINUS | Sigma-Aldrich | Sigma-Aldrich Cat# DUO92005, RRID:AB_2810942 | |
| Commercial assay or kit | Duolink In situ PLA Probe anti-Mouse PLUS | Sigma-Aldrich | Sigma-Aldrich Cat# DUO92001, RRID:AB_281039 | |
| Commercial assay or kit | Duolink In situ detection reagent Orange | Sigma-Aldrich | Sigma-Aldrich: DUO92007 | |
| Commercial assay or kit | PrestoBlue Viability Reagent (x10) | Life Technologies | Life Technologies: A13261 | |
| Chemical compound, drug | AZD2461 | Sigma-Aldrich | Sigma- Aldrich: SML 1858 | |
| Chemical compound, drug | CRISPR MAX kit | Life Technologies | Life Technologies: CMAX00001 | |
| Chemical compound, drug | cOmplete, mini EDTA-free protease inhibitor cocktail | Sigma-Aldrich | Sigma-Aldrich: 11836170001 | |
| Chemical compound, drug | DMEM | Gibco/Thermo Fisher | Gibco/Thermo Fisher: 10313-021 | |
| Chemical compound, drug | FBS SERANA | Fisher Biotechnology | Fisher Biotechnology: FBS-AU-015 batch no: 18030416 | |
| Chemical compound, drug | G418 | Sigma-Aldrich | Sigma-Aldrich: 472788001 | |
| Chemical compound, drug | Hydrogen peroxide 3% (w/w) solution | Sigma-Aldrich | Sigma-Aldrich: H1009 | |
| Chemical compound, drug | L-glutamine 100X | Gibco/Thermo Fisher | Gibco/Thermo Fisher: 25030-081 | |
| Chemical compound, drug | Lipofectamine 3000 Reagent | Thermo Fisher | Thermo Fisher: L3000015 | |
| Chemical compound, drug | MG132 (Z-Leu-Leu-Leu-al) | Sigma-Aldrich | Sigma-Aldrich: C2211 | |
| Chemical compound, drug | OptiMem reduced serum medium | Thermo Fisher | Thermo Fisher: 31985070 | |
| Chemical compound, drug | PhosSTOP Phosphatase Inhibitors | Sigma-Aldrich | Sigma-Aldrich: 4906837001 | |
| Chemical compound, drug | Trypsin-EDTA (0.05%) phenol red | Gibco/Thermo Fisher | Gibco/Thermo Fisher: 25300062 | |
| Software, algorithm | GraphPad Prism | GraphPad Prism (https://graphpad.com) | RRID:SCR_015807 | Version 9 |
| Software, algorithm | ImageJ | ImageJ (http://imagej.nih.gov/ij) | RRID:SCR_003070 |
Additional files
-
Supplementary file 1
Complete label-free quantitative proteomics for cavin3 KO cells.
Complete list of proteins analyzed in cavin3 KO compared to WT HeLa cells (control). Significant (p<0.05) mean log2 transformed SILAC ratios.
- https://cdn.elifesciences.org/articles/61407/elife-61407-supp1-v2.xlsx
-
Supplementary file 2
Pathway analysis for cavin3 KO cells.
Gene Ontology Biological Process (GOBP) name of both significantly upregulated and downregulated pathways with their corresponding p-values and enrichment scores.
- https://cdn.elifesciences.org/articles/61407/elife-61407-supp2-v2.xlsx
-
Supplementary file 3
Supplementary discussion and references.
- https://cdn.elifesciences.org/articles/61407/elife-61407-supp3-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61407/elife-61407-transrepform-v2.docx





