Phenotypic plasticity underlies local invasion and distant metastasis in colon cancer
Figures
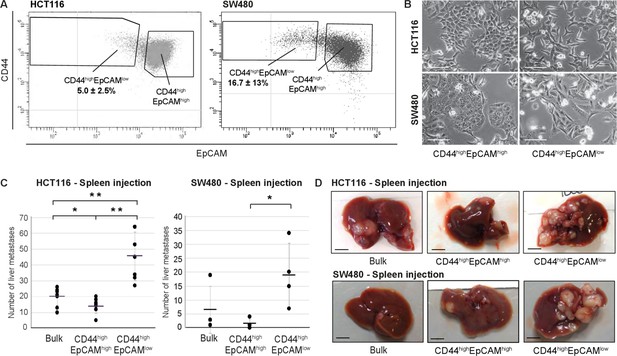
Identification and characterization of EpCAMlo cells in colon cancer cell lines.
(A) Flow cytometric analysis of the colon cancer cell lines HCT116 (left panel) and SW480 (right panel) with antibodies directed against CD44 and EpCAM. EpCAM/CD44-positive and -negative regions (gray quadrants) were defined as in Figure supplement 1 using multiple isotype controls and are shown by the quadrants in the plots. Notably, both HCT116 and SW480 revealed a continuum of different EpCAM and CD44 expression levels with a large CD44highEpCAMhigh (EpCAMhi) cluster followed by a tail of gradually decreasing EpCAM and increasing CD44 levels. By applying specific gates, cells were divided in a large EpCAMhi cluster, together with a considerably smaller CD44highEpCAMlow (EpCAMlo) subpopulation. To ensure good separation from the large EpCAMhi cluster and maximal sorting purity, EpCAMlo cells were gated as CD44hi events ≤ 60% of the EpCAM fluorescence intensity of the left border of the EpCAMhi gate and sorted from ≤50% of that value. Variable percentages of EpCAMlo cells were found to feature the HCT116 (5.0% ± 2.5%) and SW480 (16.7% ± 13%) cell lines, respectively. For the sake of simplicity, gates are shown in the figure only if they encompass sizeable percentages of cells. Graphs show representative analysis of one experiment. (B) Phase-contrast microscopy images of sorted EpCAMhi and EpCAMlo cells from HCT116 (upper images) and SW480 (lower images) cells. While EpCAMhi cells formed compact colonies with characteristic epithelial morphology, EpCAMlo cells showed a more spindle- and mesenchymal-like appearance. Scale bar: 100 µm. (C) Intrasplenic injection of bulk, EpCAMhi, and EpCAMlo cells from HCT116 (left panel) and SW480 (right panel). For each transplantation experiment, 2 × 104 cells were injected in the spleen of a recipient NSG mouse. 4 (HCT116) and 8 (SW480) weeks after injection, mice were sacrificed and individual tumors counted. Single and double asterisks indicate significant differences (p<0.05 and p<0.01, respectively). HCT116: bulk (n = 8), EpCAMhi (n = 9), and EpCAMlo (n = 7). SW480: bulk (n = 4), EpCAMhi (n = 4), and EpCAMlo (n = 4). (D) Images of mouse livers 4 (HCT116) and 8 (SW480) weeks after orthotopic injection with 104 cells. Scale bar: 5 mm.
-
Figure 1—source data 1
EpCAMlo cells among colon cancer cell lines.
The percentage of CD44hi/EpCAMlo subpopulation was determined in a panel of commonly employed colon cancer cell lines by flow cytometric analysis. Please note that, within the same cell line, these percentages can vary depending on the passage number and culture conditions. If indicated by Lindeman et al., consensus molecular subtype classification of the cell lines is shown.
- https://cdn.elifesciences.org/articles/61461/elife-61461-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Cell cycle analysis of EpCAMhi and EpCAMlo cells in HCT116 and SW480.
Cells fractions were sorted and plated in culture. After 72 hr, cells were fixed and stained with propidium iodide. Cell cycle distribution was assayed by flow cytometry. Tables demonstrate average and standard deviation of three independent experiments.
- https://cdn.elifesciences.org/articles/61461/elife-61461-fig1-data2-v2.xlsx
-
Figure 1—source data 3
Quantification of EpCAMhi/lo percentages of all liver metastases as determined by FACS.
- https://cdn.elifesciences.org/articles/61461/elife-61461-fig1-data3-v2.xlsx
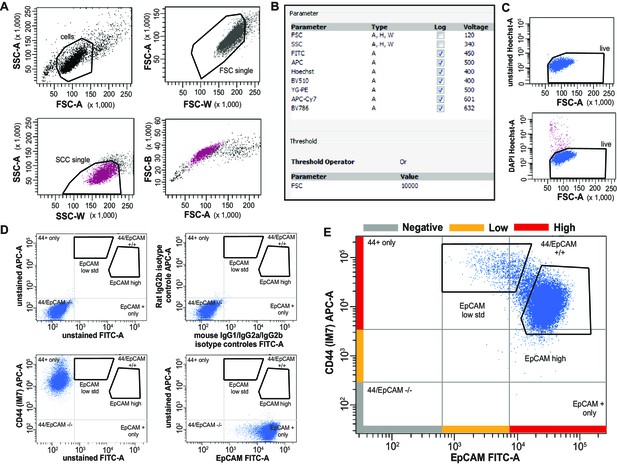
Further characterization of EpCAMlo cells in colon cancer cell lines: FACS analysis.
(A) FSC-A/SSC-A, FSC-W/FSC-A, and SSC-W/SSC-A single-cell gates (confirmed by gating on FSC-A/FSC-H). Purity of sorted single cells was confirmed by microscopy. (B) Acquisition parameters used for FACS analysis. (C) Gating strategy employed to select for live cells by DAPI staining (top: unstained cells; bottom: after DAPI staining). (D) Top: unstained and isotype controls, showing absence of relevant unspecific antibody binding. Bottom: fluorescence minus one (FMO) samples, showing the absence of uncompensated spillover between fluorescence channels. For the sake of clarity, a quadrant is placed to show the relative position of unstained and single stains in both cells lines. For the EpCAM-FITC antibody, a mouse IgG1-FITC isotype control S. Cruz sc-2855 was used; for the CD44-APC antibody, a Rat IgG2a-APC isotype control S. Cruz sc-2895 was used. Similar results were obtained with SW480 cells (not shown). (E) Full EpCAM/CD44 staining, also showing the rationale behind the definition of high and low referred to EpCAM and CD44 levels. Despite the differences in CD44 levels between EpCAMhi and EpCAMlo cells, for the sake of simplicity the populations were defined ‘low’ for a defined marker if they were mainly covering a region within the 1st log above the negative gate, high if they were above the 1st log. FSC-H: forward-scatterheight; FSC-W: forward-scatter width.
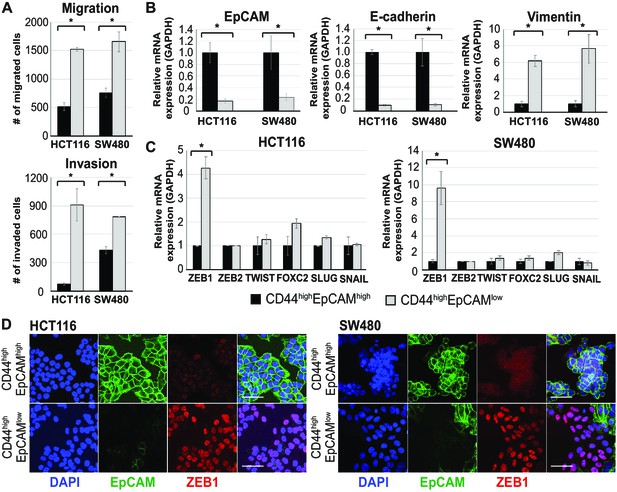
Further characterization of EpCAMlo cells in colon cancer cell lines: migration/invasion and EMT analysis.
(A) Top panel: transwell migration assay of EpCAMhi (black bar) and EpCAMlo (gray bar) cells from the HCT116 and SW480 lines. 105 cells were plated on TC-coated membrane in triplicate and left overnight before counting the number of migrated cells on the bottom side of the membrane. Each bar represents the mean ± SD of two independent experiments. Asterisks indicate significant differences (p<0.05). Bottom panel: invasion assay of EpCAMhi (black bar) and EpCAMlo (gray bar) cells from the HCT116 and SW480 lines. 105 cells were plated in triplicate on top of an extracellular matrix-coated membrane and left overnight before counting the number of cells migrated to other side of the membrane. Each bar represents the mean ± SD of two independent experiments. Asterisks indicate significant differences (p<0.05). (B) RT-qPCR expression analysis of epithelial (EPCAM and CDH1) and mesenchymal (VIM) markers in sorted EpCAMhi (black bars) and EpCAMlo (gray bars) from the HCT116 and SW480 lines. GAPDH was employed for normalization purposes. Each bar represents the mean ± SD of three independent experiments. Asterisks indicate significant differences (p<0.05). (C) RT-qPCR expression analysis of epithelial to mesenchymal transition transcription factors (ZEB1, ZEB2, TWIST, FOXC2, SLUG, and SNAIL) in EpCAMhi (black bars) and EpCAMlo (gray bars) cells. Left panel: HCT116. Right panel: SW480. GAPDH was employed for normalization. Each bar represents the mean ± SD of three independent experiments. Asterisks indicate significant differences (p<0.05). (D) Immunofluorescence (IF) analysis of EpCAMhi and EpCAMlo cells. Cells were sorted and directed plated on cover slips. After 4 days, cells were fixed with 4% paraformaldehyde and stained with antibodies against EpCAM (green) and ZEB1 (red). Nuclei were visualized by DAPI staining of DNA (blue). Scale bar: 50 µm.
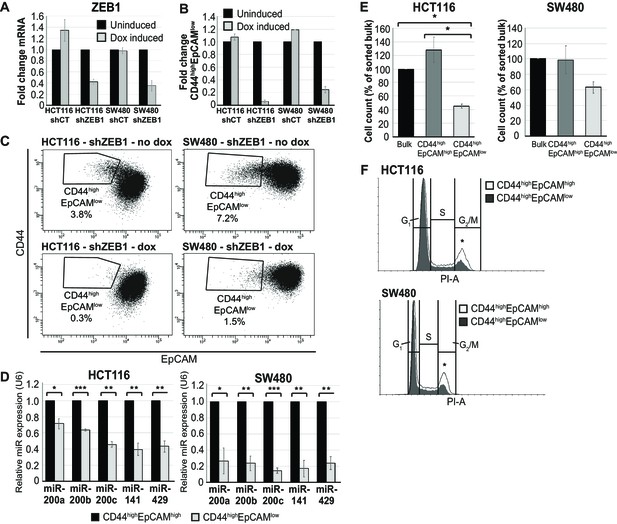
Further characterization of EpCAMlo cells in colon cancer cell lines: EMT and cell cycle analysis.
(A). qRT-PCR expression analysis of ZEB1 in HCT116 and SW480 transduced with an inducible control (shCT) or ZEB1-shRNA (shZEB1) construct. shRNA expression was induced with 1 µg/mL of doxycycline. Each bar represents the mean ± SD of three independent experiments. (B) Bar graph of flow cytometric analysis (see B). Each bar represents the relative mean ± SD of three independent experiments. (C) Representative analysis of the flow cytometric analysis of the shCT- and shZEB1-transfected HCT116 and SW480 cell lines using antibodies against CD44 and EpCAM. Cells were induced with 1 µg/mL doxycycline for 72 hr before analysis. (D) RT-qPCR expression analysis of the members of the miRNA 200 family (miR-200a, miR-200b, miR-200c, miR-141, and miR-429) in EpCAMhi (black bars) and EpCAMlo (gray bars) cells. Upper panel: HCT116. Bottom panel: SW480. U6 was employed for normalization. Each bar represents the mean ± SD of three independent experiments. Single asterisks indicate significant differences of p<0.05, double asterisks of p<0.01, and triple asterisks of p<0.001. (E) Cell proliferation assay. Sorted bulk, EpCAMhi and EpCAMlo cells were seeded in triplicate in plates and cultured in conventional medium. HCT116 and SW480 cells were harvested and number of cells was counted at 4 and 11 days, respectively. Each bar represents the mean ± SD three independent experiments. (F) Cell cycle analysis of EpCAMhi and EpCAMlo cells in HCT116 (upper panel) and SW480 (lower panel). Cell fractions were sorted and plated in culture. After 72 hr, cells were fixed and stained with propidium iodide. Cell cycle distribution was assayed by flow cytometry. Graphs show representative analysis of one experiment. Tables demonstrate average and standard deviation of three independent experiments. White graph: EpCAMhi; gray graph: EpCAMlo. Asterisks show the significant (p<0.05) differences between EpCAMhi and EpCAMlo cells in G1 and G2M-phases.
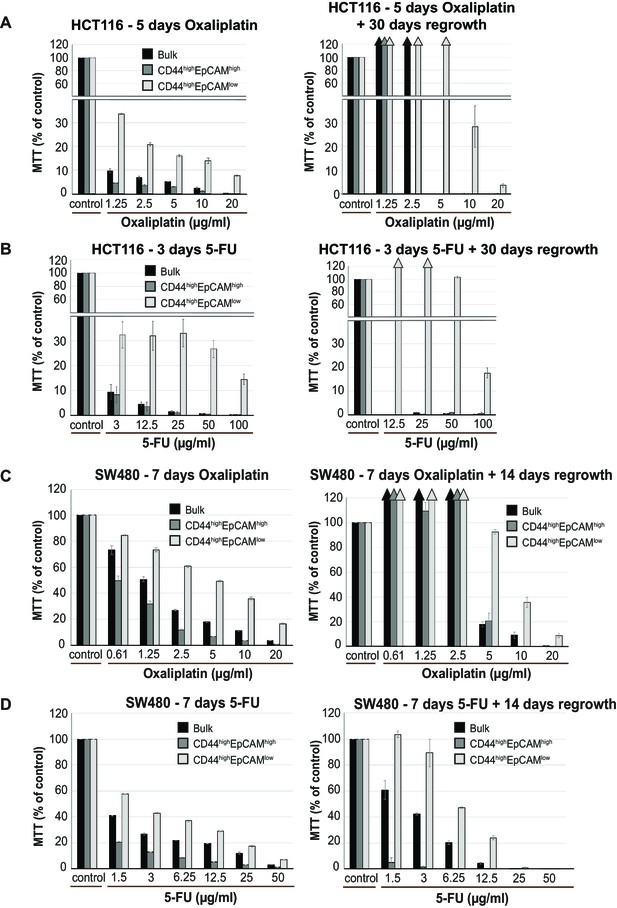
Further characterization of EpCAMlo cells in colon cancer cell lines: chemoresistance.
Bulk, EpCAMhi, and EpCAMlo HCT116 and SW480 cells were sorted and plated to recover and expand for 4 days.105 cells per well were plated in triplicate in a 96-well plate and left to attach. After 24 hr, standard cell culture medium was changed for medium containing chemotherapeutic drug oxaliplatin or 5-fluorouracil (5-FU). HCT116 cells were left with 5-FU and oxaliplatin for respectively 3 and 5 days. For each drug, SW480 cells were treated for 7 days. After removal of the chemotherapeutic drug, cells were washed with PBS and left to regrow in standard culture medium. Cell viability was assessed using the MTT upon removal of the drug, and after regrowth (ranging 13–30 days). O.D. reading was performed at 595 nm with a Microplate Reader. Background measurements were subtracted from each data point. All experiments were performed at least in duplicate for each individual cell line and drug. (A, B) Chemo-sensitivity assays using oxaliplatin (A) or 5-FU (B) in bulk (black bars), EpCAMhi (dark gray bars) and EpCAMlo (light gray bars) HCT116 cells. Bars represent the average ± SD of the O.D. corrected for the background measurements. (C, D) Chemo-sensitivity assays using oxaliplatin (C) or 5-FU (D) in bulk (black bars), EpCAMhi (dark gray bars) and EpCAMlo (light gray bars) SW480 cells. Bars represent the average ± SD of the O.D. corrected for the background measurements.
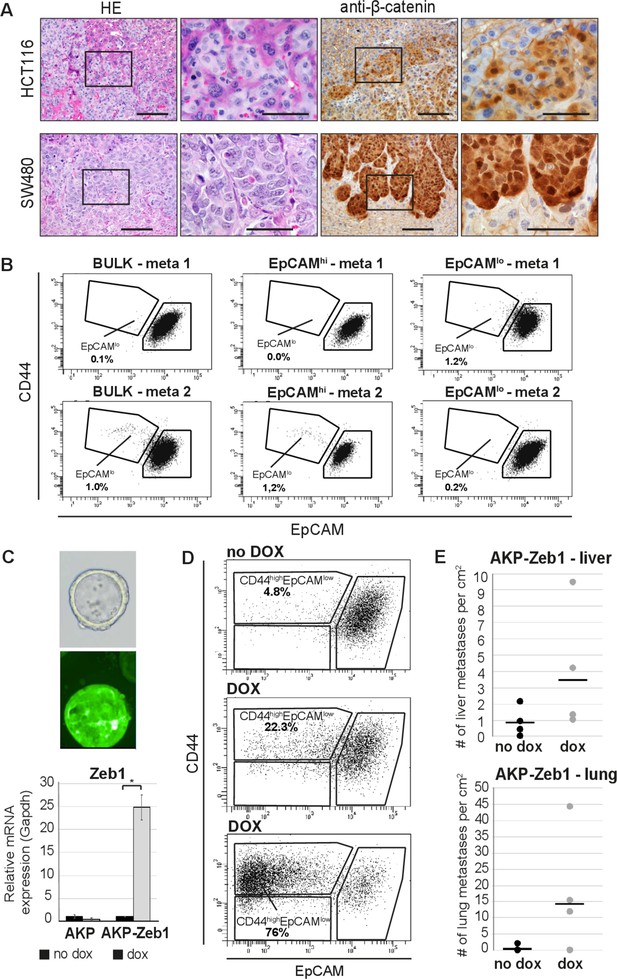
Further characterization of EpCAMlo cells in colon cancer cell lines: invasive and metastatic behaviour.
(A) Hematoxylin and eosin (H&E; first two panels) and immunohistochemistry (IHC) with antibody directed against β-catenin (third and fourth panels) in liver metastasis obtained 4 or 8 weeks after intrasplenic injection with HCT116 (upper panels) and SW480 (lower panels) cells, respectively. Second and fourth panels show zoom of the marked area in respectively first and third panels. Scale bar first and third panels: 100 µm. Scale bar second and fourth panels: 50 µm. (B) FACS analysis of liver metastases obtained by spleen injection of HCT116 bulk, EpCAMhi, and EpCAMlo cells. Upper panel: representative FACS plots. (C) Microscopic (top, left panel) analysis of AKP-Z organoids tagged with GFP and click beetle luciferase. The graph (bottom, left panel) shows the RT-qPCR expression analysis of Zeb1 in AKP (left) and AKP-Z (right) organoids upon in vitro doxycycline treatment for 48 hr. Black bars: no doxycycline treatment; gray bars: 1 μg/mL doxycycline. Each bar represents the mean ± SD of three independent experiments. (D) Upon establishment of a primary tumor in the caecum, transplanted mice were administered doxycycline in the drinking water to induce Zeb1 expression. FACS analysis of the primary tumor was performed 1 week after the start of the doxycycline. The panels show representative FACS plots of a control and two dox-treated primary tumors. (E) Quantification of the number of lung and liver metastases in uninduced (black; n = 4) and dox-induced (gray; n = 5) AKP-Z transplanted mice. Liver tissue was cut into 500 µm slices, processed for IHC, stained for β-catenin to visualize tumor cells, scanned using a NanoZoomer, and counted using NDP view software. The area of tissue analyzed was used to normalize the data.
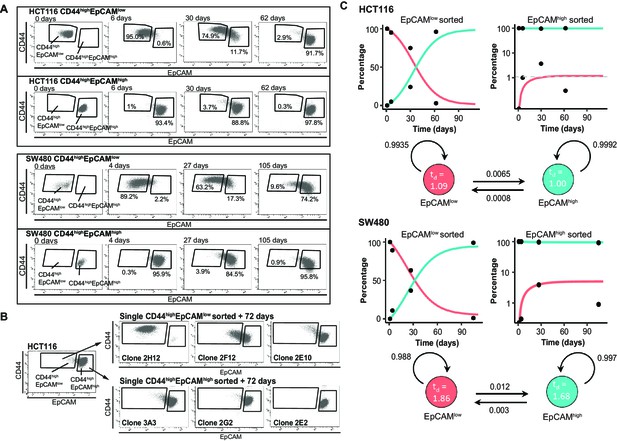
Phenotypic plasticity maintains EpCAMlo and EpCAMhi cells in a stochastic equilibrium.
(A) Analysis of plasticity of EpCAMhi and EpCAMlo cells from HCT116 (upper panel) and SW480 (lower panel). EpCAMhi and EpCAMlo cell fractions were sorted and plated in culture. At different time points, as indicated, cells were reanalyzed by flow cytometry for their levels of CD44 (y-axis) and EpCAM (x-axis) expression. (B) Flow cytometric analysis of single CD44hiEpCAMhi and CD44hiEpCAMlo HCT116 cells sorted by FACS and cultured for 72 days. Three representative individual single-cell clones per cell fraction are shown. (C) Dynamics of the EpCAMhi and EpCAMlo subpopulations from the HCT and SW480 cell lines as measured by FACS (% of total) over time. Under each graph, a schematic shows the estimated transition probabilities from the fitted two-state Markov model.
-
Figure 2—source data 1
Phenotypic plasticity maintains EpCAMlo and EpCAMhi cells in a stochastic equilibrium: clonal analysis.
The table encompasses a selection of individual single-cell sorted EpCAMhi and EpCAMlo HCT116 and SW480 clones. Single cells were sorted, grown up, and analyzed at two indicated times points by flow cytometry for their CD44 and EpCAM expression. Percentage of cells present in the set EpCAMhi and EpCAMlo gates is presented in the bar graph. Total success rate of clones grown out of single cells: HCT116 EpCAMhi 15 clones out of 24 sorted cells, HCT116 EpCAMlo: 31/48; SW480 EpCAMhi: 5/36; SW480 EpCAMlo: 11/36.
- https://cdn.elifesciences.org/articles/61461/elife-61461-fig2-data1-v2.xlsx
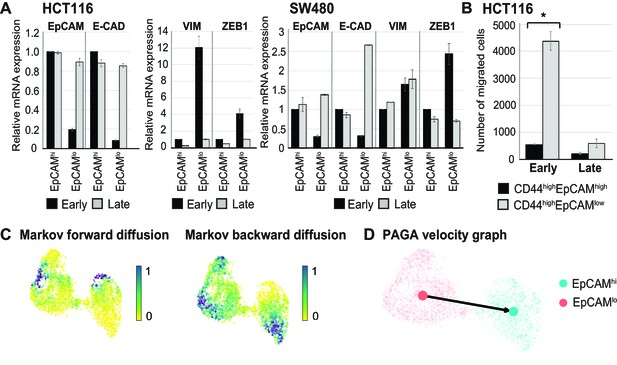
Further characterization of phenotypic plasticity in EpCAMlo and EpCAMhi cells.
(A) RT-qPCR expression analysis of EPCAM, CDH1, VIM, and ZEB1 in ‘early’ (e.g., cells that were used shortly after FACS sorting) and ‘late’ (e.g., cells that were cultured for an extended period of time before performing the experiment) sorted EpCAMhi and EpCAMlo cell from HCT116 and SW480 cell lines. GAPDH was employed for normalization purposes. Each bar represents the mean ± SD of two independent experiments. (B) Transwell migration assay of ‘early’ and ‘late’ EpCAMhi (black bar) and EpCAMlo (gray bar) cultures in HCT116. 105 cells were plated in triplicate on TC-coated membrane and left overnight before counting the number of migrated cells on the bottom side of the membrane. Each bar represents the mean ± SD of two independent experiments. Asterisks indicate significant differences (p<0.05). (C) Analysis of the HCT116 scRNAseq data as a Markov diffusion process. Markov forward (left) and backward (right) diffusion indicating the presence of sink and source points in both EpCAMhi and EpCAMlo populations. (D) Partition-based graph abstraction velocity graph mapping out the direction of velocity on a subpopulation level in HCT116.
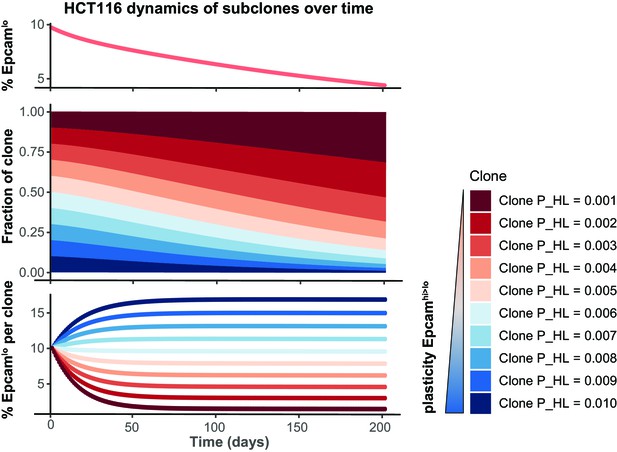
Simulation of the HCT116 two-state Markov model with a non-homogenous starting population.
The starting population was defined as 10 subclones with varying transition probability P_HL at equal proportion. The Markov model was run till day 200 and shows that the total EpCAMlo population decreases over time (top panel) because clones with lower plasticity will gain dominance in the long run (lower panels).
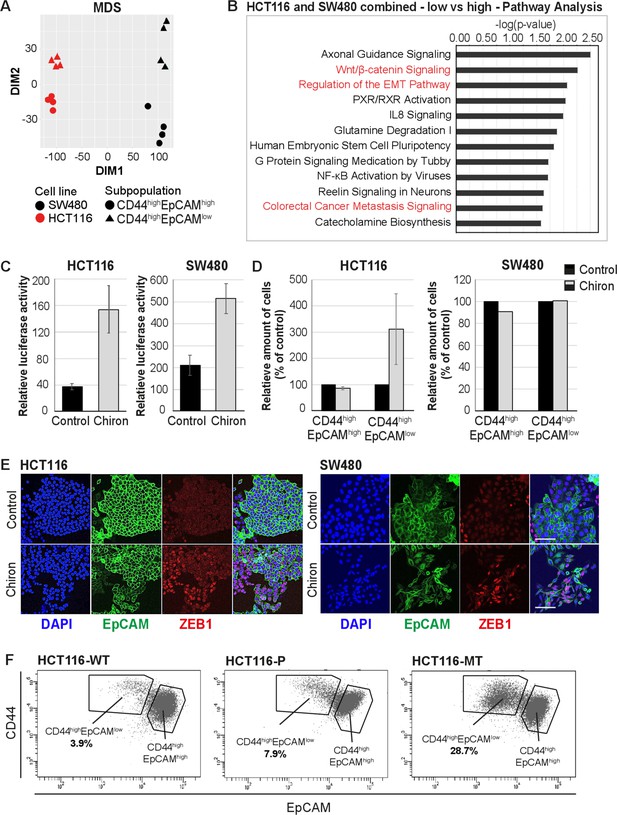
RNAseq analysis reveals enhanced Wnt signaling in EpCAMlo cells.
(A) Multidimensional scaling analysis of RNAseq profiles of EpCAMhi and EpCAMlo cells from the HCT116 and SW480 lines. Red: HCT116, black: SW480, circle: EpCAMhi, triangle: EpCAMlo. (B) Ingenuity Pathway Analysis (IPA) of the HCT116 and SW480 expression profiles from the multicell line analysis (p adjusted value <0.01; log2 fold change <–1.5 and >1.5). Red marked pathways highlight the enhanced involvement of pathways involved in epithelial to mesenchymal transition, Wnt signaling, and the formation of colon cancer metastasis in the EpCAMlo subpopulation compared to EpCAMhi cells. (C) TOP-Flash luciferase reporter analysis of Wnt signaling activity in colon cancer cell lines HCT116 and SW480 upon treatment with 4 µM Chiron for 3 days. Each bar represents the mean ± SD of two independent experiments. (D) Flow cytometric analysis using antibodies directed against CD44 and EpCAM of control and 4 µM Chiron—figure supplemented HCT116 (A) and SW480 (B) cultures. Graphs show percentage of cells within the CD44hiEpCAMhi and CD44hiEpCAMlo gates relative to the control. Each bar represents the mean ± SD of two independent experiments. (E) Immunofluorescence analysis of control and Chiron-treated HCT116 (left panel) and SW480 (right panel) cells. After 3 days of treatment, cells were fixed with 4% paraformaldehyde and stained with antibodies against EpCAM (green) and ZEB1 (red). Nuclei were visualized by DAPI staining of DNA (blue). Scale bar: 100 µm. (F) Flow cytometric analysis of three HCT116 cell lines with differential β-catenin mutation status, a parental HCT116 (HCT116-P) cell line harboring one WT and one mutant allele (Ser45 del), and two HCT116-WT and HCT116-MT cell lines harboring one WT or one mutant allele, respectively, generated by disruption of the other allele in the parental cell line (Kim et al., 2019).
-
Figure 3—source data 1
List of differentially expressed genes in EpCAMlo vs. EpCAMhi cells in HCT116 and SW480.
A total of 152 and 353 differentially regulated genes were identified between the CD44hiEpCAMhi and CD44hiEpCAMlo cells in HCT116 and SW480, respectively (p adjusted<0.01, when applying a log2 fold change of <–1.5 and > 1.5).
- https://cdn.elifesciences.org/articles/61461/elife-61461-fig3-data1-v2.xlsx
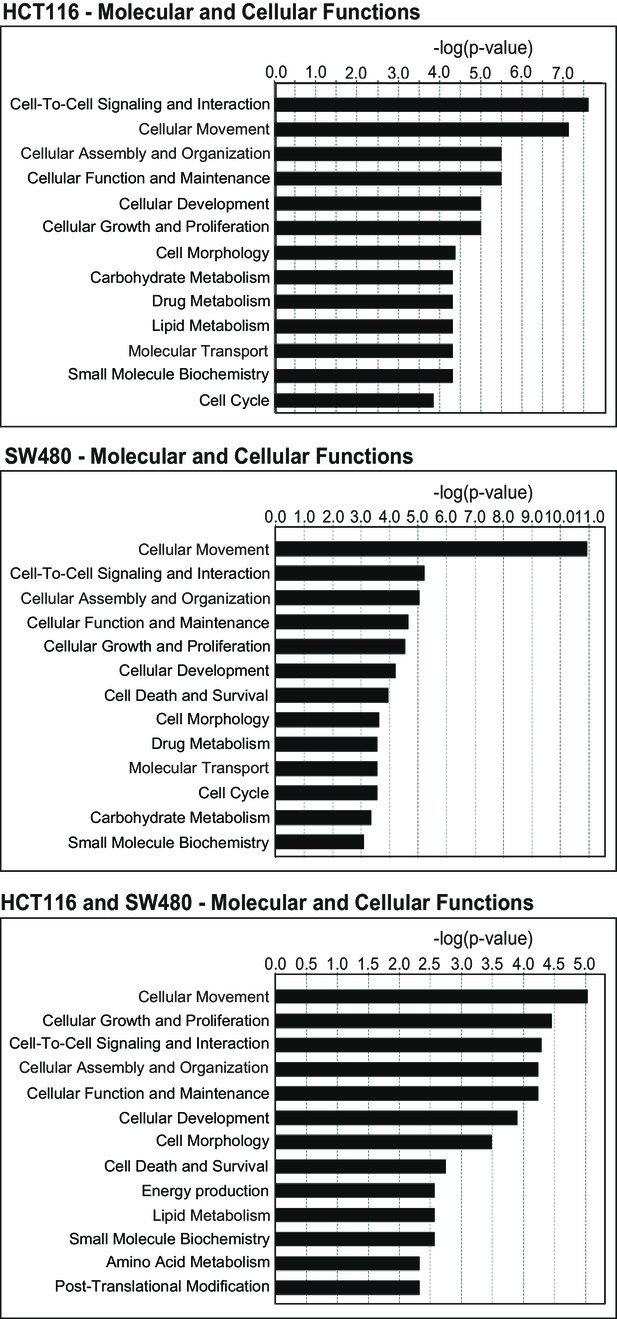
Gene ontology (GO) of molecular and cellular functions in HCT116 (upper panel), SW480 (middle), and the combined (bottom panel) gene lists.
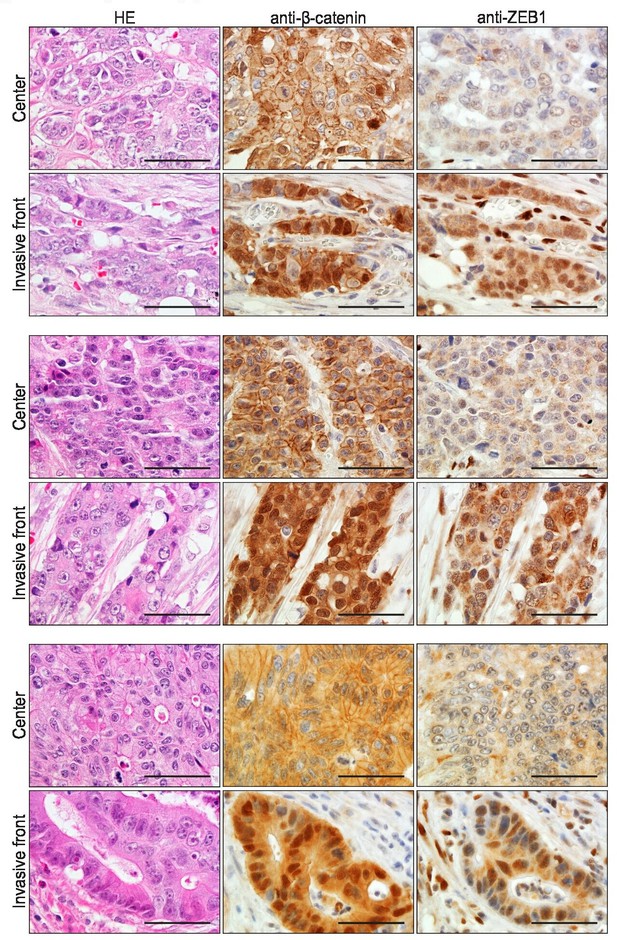
Hematoxylin and eosin (H&E) and immunohistochemistry (IHC) analyses with antibodies directed against beta-catenin and ZEB1 in consecutive sections of colon cancers from three unrelated patients with sporadic colon cancer.
For each case, areas from the tumor center and the invasive front are shown. Co-localization of nuclear β-catenin and ZEB1 expression was found in 5 out of 25 cases investigated. Scale bar: 50 µm.
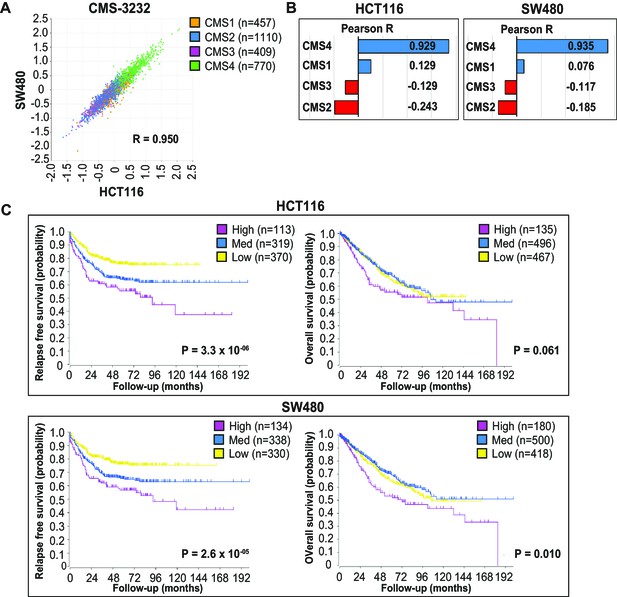
EpCAMlo gene expression profiles correlate with CMS4 colon cancer patients with shorter disease-free and overall survival.
(A) Correlation of meta-gene expression values of the signatures derived from EpCAMlo HCT116 and SW480 cells in the consensus molecular subtype (CMS)3232 human colon cancer cohort (Guinney et al., 2015). (B) Correlation of meta-gene expression values of the signatures derived from EpCAMlo HCT116 and SW480 cells with expression of CMS classifier genes positively identifying each of the four molecular subtypes. (C) Kaplan– Meier analysis. The gene sets identifying the EpCAMlo cells from both HCT116 and SW480 cell lines were used to cluster the tumors in the CMS3232 cohort into high (purple), intermediate (blue), and low (yellow) expression groups by k-means clustering. The Kaplan–Meier method was subsequently used to assess significant differences in relapse-free (left panels) and overall (right panels) survival between the generated subgroups.
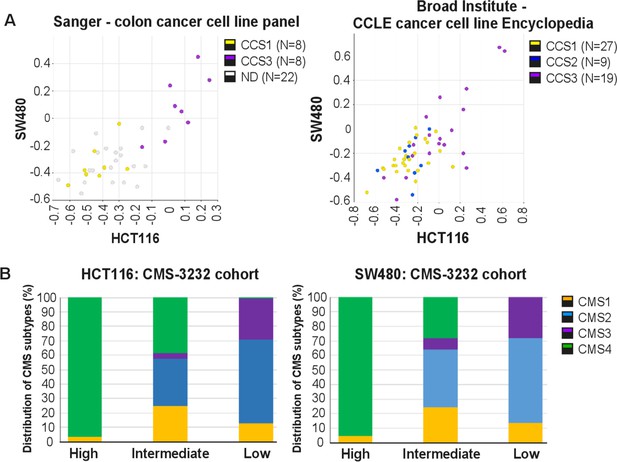
EpCAMlo expression signatures correlate with CCS3 cell lines and CMS4 colon cancer patients.
(A) Correlation of meta-gene expression values of the signatures derived from EpCAMlo HCT116 and SW480 cells in two cohorts of colon cancer cell lines in the absence of a stromal component. CCS1: epithelial-like tumor lines; CCS2: tumor lines with microsatellite instability. CCS3: mesenchymal-like tumor cell lines (De Sousa et al., 2013). (B) The gene sets identifying the EpCAMlo cells from both HCT116 and SW480 were used to cluster the tumors in the consensus molecularsubtype (CMS)3232 cohort into high, intermediate, and low expression groups by k-means clustering. All tumors in this cohort had previously been assigned to specific CMS subgroups, and then analyzed the contribution of each CMS subtype to each of the generated high, intermediate, and low expression subgroups.
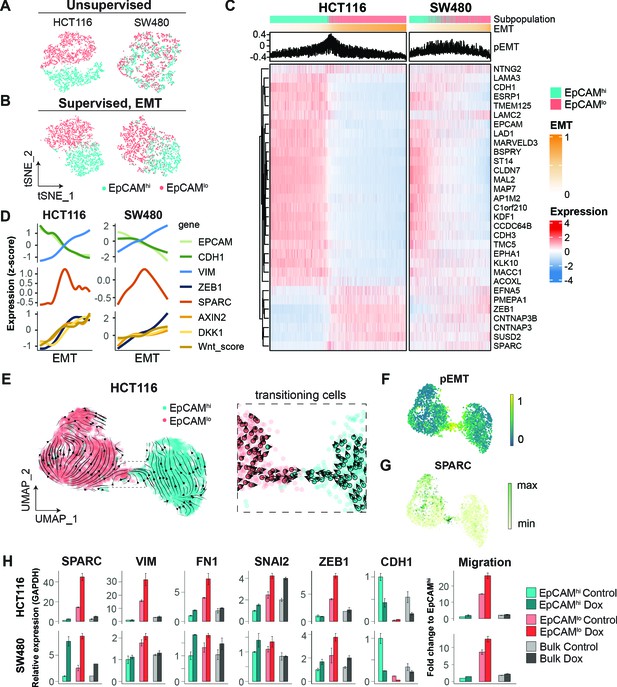
scRNAseq analysis of EpCAMlo cells reveals specific markers of partial EMT cells.
(A) tSNE of HCT116 and SW480 cells based on the variable expressed genes across EpCAMlo and EpCAMhi populations. (B) tSNE of HCT116 and SW480 cells based on genes from the epithelial to mesenchymal transition (EMT) signature (N = 107). (C) Heatmap of differentially expressed genes between EpCAMlo and EpCAMhi populations. Cells were ranked according to their EMT score (EMT = M – E). (D) Gene expression trends projected over the EMT axis. Expression values were imputed with MAGIC, scaled by their z-score, and smoothened by general additive models to visualize the gene expression trend. (E) RNA velocity analysis of the HCT116 scRNAseq data. Cells from both populations were moving in their respective state (left panel) with a minority population of transitioning cells (right panel). (F) Projection of the pEMT score on the UMAP embedding of the HCT116 cell line. (G) The expression of SPARC on the UMAP embedding of the HCT116 cell line. (H) Left panels: qPCR analysis of SPARC overexpression in the subpopulations of HCT116 and SW480. Gene expression values are relative to GAPDH and normalized to the EpCAMhi subpopulation. Right panel: quantification of the transwell migration assay upon overexpression of SPARC.
-
Figure 5—source data 1
EMT and CMS4 classifiers.
- https://cdn.elifesciences.org/articles/61461/elife-61461-fig5-data1-v2.xlsx
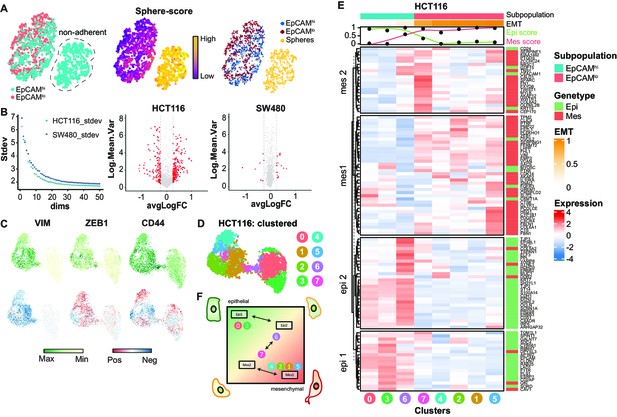
Further scRNAseq analysis of EpCAMlo cells: EMT/MET transcriptional trajectory.
(A) tSNE of the SW480 cell line indicating an additional subpopulation in the EpCAMhi population (left panel). Using a signature derived from the bulk RNAseq, this population was identified as the ‘sphere’ population (middle panel) and annotated to be excluded for further analysis (right panel). (B) Left panel: the SW480 cell line, after exclusion of the ‘sphere’ population, contains slightly higher variability compared to the HCT116 cell line, as evidenced by the variance of the top 50 principal components. Right panel: while in HCT116 most of the variable expressed genes are differentially expressed between the EpCAMhi and EpCAMlo population, this is not the case in SW480, where most of the highly variable genes do not differ between the two populations. (C) Top panels: expression values of VIM, ZEB1, and CD44 on the UMAP embedding of the HCT116 cell line. Lower panels: projection of the RNA velocity direction of the same genes. (D) HCT116 UMAP embedding annotated with the eight unsupervised clusters. (E) Heatmap of HCT116 with expression values of the epithelial to mesenchymal transition (EMT) signature averaged by the eight clusters. Clusters were ranked according to their EMT score, and genes were clustered in four distinct gene sets using k-means clustering. (F) Schematic diagram showing a transcriptional trajectory with distinct gene arrays through which pEMT cells arise.
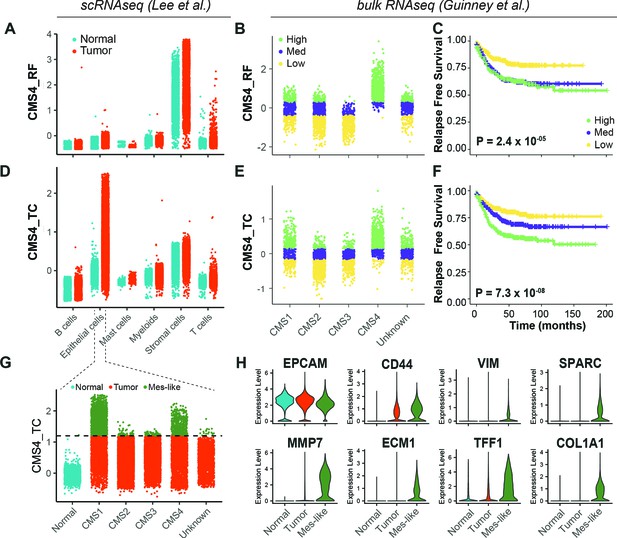
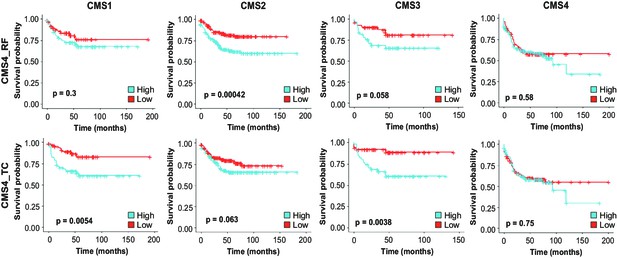
Identification of EpCAMlo cells in primary colorectal tumors.
(A) Expression of the CMS4_RF signature in the scRNAseq data from Lee et al. (N = 91,103 cells) indicates high association to the stromal cells. (B) Expression of the CMS4_RF signature in the bulk RNAseq data from Guinney et al. (N = 3232 tumors) shows association to CMS4 tumors. Tumors were grouped in three equal groups according to their association with the CMS4_RF signature. (C) Kaplan–Meier plot of the three CMS4_RF groups shows significant differences in relapse-free survival. (D) Expression of the CMS4_TC signature in the scRNAseq data reveals high association to the tumor epithelial cells. (E) Expression of the CMS4_TC signature in the bulk RNAseq data shows association to CMS1 and CMS4 tumors. Tumors were grouped in three equal groups according to their association with the CMS4_TC signature. (F) Kaplan–Meier plot of the three CMS4_TC groups shows significant differences in relapse-free survival. (G) Expression of the CMS4_TC signature in the fraction of epithelial cells (N = 24,707 cells). Cells from CMS1 and CMS4 tumors show the highest association to the CMS4_TC signature and were annotated as mes-like. (H) Violin plots of normal, tumor, and mes-like tumor cells showing expression patterns across different genes.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line(Homo sapiens) | HCT116(adult colorectal carcinoma) | ECACC | Cat# 91091005, RRID:CVCL_0291 | For sorting of subpopulations; see Materials and methods, section Flow cytometry and sorting |
| Cell line(Homo sapiens) | HCT116-WT, -P, and -MT(wild type, hetero- and homozygous for the b-catenin Ser45del mutant allele) | Author:A.K.(Kim et al., 2019) | ||
| Cell line(Homo sapiens) | SW480(adult colorectal carcinoma) | ECACC | Cat# 87092801, RRID:CVCL_0546 | For sorting of subpopulations, see Materials and methods, section Flow cytometry and sorting |
| Biological sample (Mus musculus) | APC-Kras-P53 (AKP) organoids (Apcfl/fl::KrasG12D/+::Trp53fl/R172H) | Author: J.vR (Fumagalli et al., 2017; Fumagalli et al., 2018) | ||
| Biological sample(Mus musculus) | AKP-Zeb1 (AKP-Z)(dox-inducible Zeb1 expression) | This study | See Materials and methods, section Construction of Zeb1 inducible vector | |
| Antibody | Anti-mouse CD44-APC (rat monoclonal) | BD Pharmingen | Cat# 559250, RRID:AB_398661 | FACS (1 µg/106 cells) |
| Antibody | Anti-human EpCAM-FITC (mouse monoclonal) | GeneTex | Cat# GTX30708, RRID:AB_1240769 | FACS (1 µg/106 cells) |
| Antibody | Anti-human EpCAM (mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-66020, RRID:AB_2098654 | IF (1:250) |
| Antibody | Anti-human ZEB1 (rabbit polyclonal) | Santa Cruz Biotechnology | Cat# sc-25388, RRID:AB_2217979 | IF (1:200) |
| Antibody | Anti-human β-catenin (mouse monoclonal) | BD Biosciences | Cat# 610154, RRID:AB_397555 | IHC (1:500) |
| Antibody | Anti-human ZEB1 (rabbit polyclonal) | Sigma-Aldrich | Cat# HPA027524, RRID:AB_1844977 | IHC (1:200) |
| Chemical compound, drug | 5-fluorouacil | Sigma-Aldrich | Cat# F6627 | 1.5–100 µg/mL |
| Chemical compound, drug | Oxaliplatin | Sigma-Aldrich | Cat# O9512 | 1.25–20 µg/mL |
| Commercial assay or kit | TruSeq Sample Preparation | Illumina | Cat# 15026495F | v.2 |
| Commercial assay or kit | Single Cell 3´ Reagent Kit Protocol | 10XGenomics | Cat# CG00052 | v2 chemistry |
| Software, algorithm | CellRanger | 10XGenomics | RRID:SCR_017344 | Version 2.1.1 |
| Software, algorithm | R | Seurat, GSVA, MAGIC (Stuart et al., 2019; van Dijk et al., 2018; Hanzelmann et al., 2013) | RRID:SCR_007322,RRID:SCR_021058 | Version 4.0.4 |
| Software, algorithm | Python | Velocyto, scVelo (La Manno et al., 2018; Bergen et al., 2020) | RRID:SCR_018167, RRID:SCR_018168 | Version 3.8.3 |