An interactive meta-analysis of MRI biomarkers of myelin
Figures
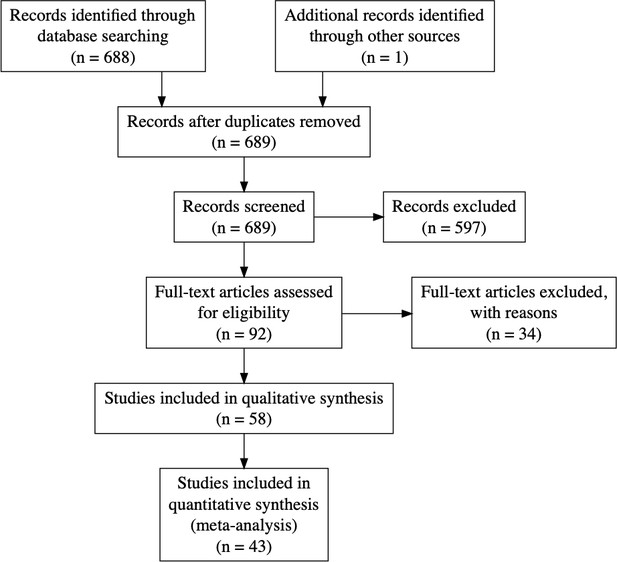
Sankey diagram representing the screening procedure (PRISMA flow chart provided in the appendix).
To see the interactive figure: https://neurolibre.github.io/myelin-meta-analysis/01/selection.html#figure-1.
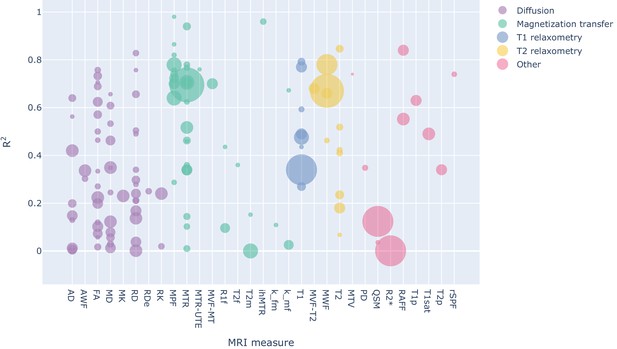
Bubble chart of R2 values between a given MRI measure and histology for each study across MRI measures, with the area proportional to the number of samples.
To see the interactive figure: https://neurolibre.github.io/myelin-meta-analysis/02/closer_look.html#figure-3.
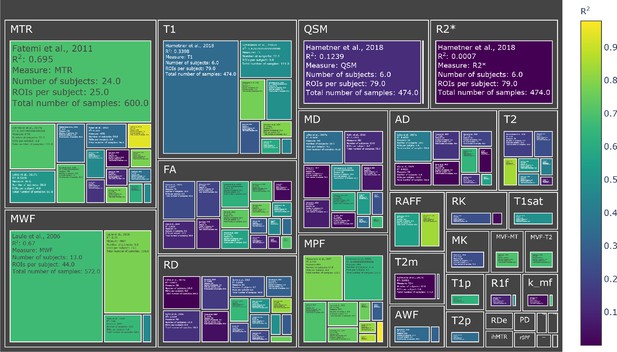
Treemap chart of the studies considered for the meta-analysis, organized by MRI measure.
The color of each box represents the reported R2 value while the size box is proportional to the sample size. To see the interactive figure: https://neurolibre.github.io/myelin-meta-analysis/02/closer_look.html#figure-4.
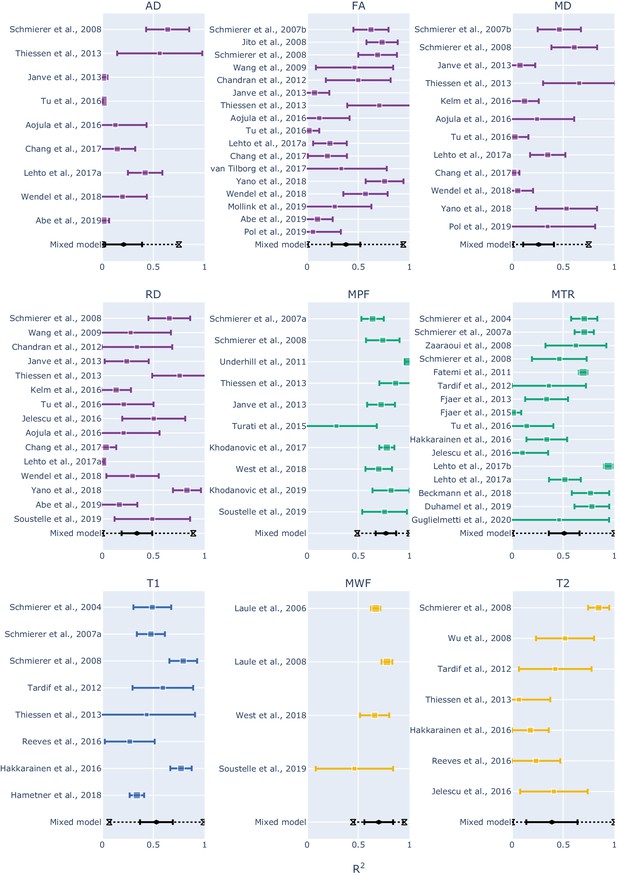
Forest plots showing the R2 values reported by the studies and estimated from the mixed-effect model for each measure.
The hourglasses and the dotted lines in the mixed-effect model outcomes represent the prediction intervals. To see the interactive figure: https://neurolibre.github.io/myelin-meta-analysis/03/meta_analysis.html#figure-5.

Results from the repeated measures meta-regression, displayed in terms of z-scores (left) and p-values (right) for each pairwise comparison across all the MRI measures.
In the z-score heatmap, each element refers to the comparison between the measure on the x axis with the one on the y axis. For example, MPF and FA (z-score = 7.14; p-value<0.0001) are statistically different, while MPF and T1 (z-score = 2.51; p-value=0.43) are not statistically different. To see the interactive figure: https://neurolibre.github.io/myelin-meta-analysis/03/meta_analysis.html#figure-6.
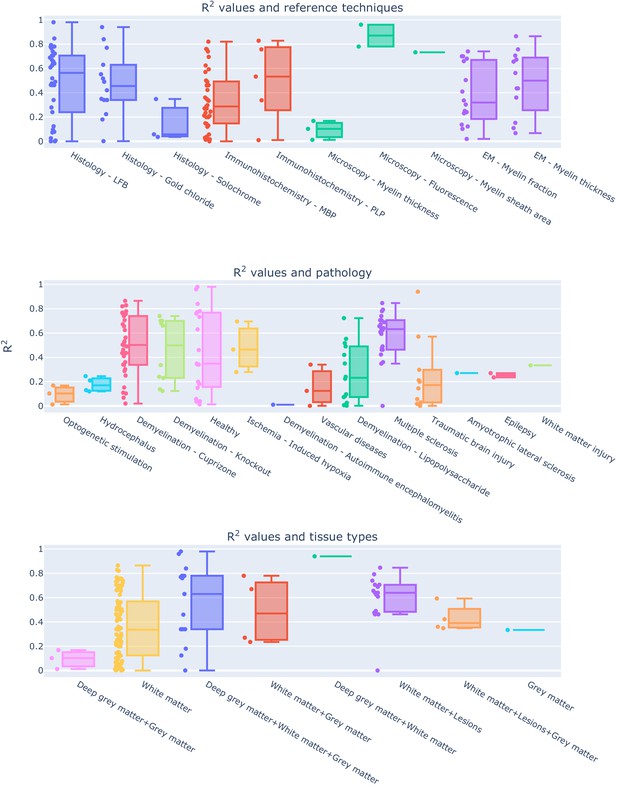
Experimental conditions and methodological choices influencing the R2 values (top: reference techniques; middle: pathology model; bottom: tissue types).
To see the interactive figure: https://neurolibre.github.io/myelin-meta-analysis/04/other_factors.html#figure-7.
Tables
Results from the mixed-effect models: for each measure we reported the number of studies, the estimate and standard error of the overall R2 distribution, the τ2 and the I2.
| Measure | Number of studies | Estimate | Standard error | Tau2 | I2 |
|---|---|---|---|---|---|
| MTR | 16 | 0.508 | 0.0691 | 0.07 | 96.03% |
| MPF | 10 | 0.7657 | 0.0455 | 0.0128 | 83.18% |
| FA | 17 | 0.3766 | 0.0663 | 0.0652 | 87.49% |
| RD | 15 | 0.3364 | 0.0679 | 0.0615 | 92.30% |
| MD | 12 | 0.2639 | 0.0679 | 0.044 | 87.35% |
| T1 | 8 | 0.5321 | 0.0692 | 0.0328 | 86.51% |
| AD | 9 | 0.2095 | 0.0802 | 0.048 | 97.69% |
| T2 | 7 | 0.3938 | 0.1023 | 0.0651 | 84.49% |
| MWF | 4 | 0.6997 | 0.0432 | 0.0041 | 73.19% |
Selected studies for qualitative analysis.
| Study | MRI measure(s) | Histology/microscopy measure | Tissue | Condition | Focus |
|---|---|---|---|---|---|
| Schmierer et al., 2004 | T1, MTR | Histology - LFB | Human | Multiple sclerosis | Brain |
| Odrobina et al., 2005 | T1, T2, T2int, MWF, M0m, MTR | Microscopy - Myelin fraction | Animal - Rat | Demyelination - Tellurium | PN |
| Pun et al., 2005 | T1, T2int, MWF | Microscopy - Myelin fraction | Animal - Rat | Demyelination - Tellurium | PN |
| Laule et al., 2006 | MWF | Histology - LFB | Human | Multiple sclerosis | Brain |
| Schmierer et al., 2007a | T1, MTR, MPF, T2m | Histology - LFB | Human | Multiple sclerosis | Brain |
| Schmierer et al., 2007b | FA, MD | Histology - LFB | Human | Multiple sclerosis | Brain |
| Jito et al., 2008 | FA | Microscopy - Myelin sheath area | Animal - Mouse | Healthy | Brain |
| Kozlowski et al., 2008 | MWF, FA, AD, RD, MD | Immunohistochemistry - MBP | Animal - Rat | Injury - Dorsal columnar transection | SC |
| Laule et al., 2008 | MWF | Histology - LFB | Human | Multiple sclerosis | Brain |
| Schmierer et al., 2008 | T1, T2, MTR, MPF, MD, FA, AD, RD | Histology - LFB | Human | Multiple sclerosis | Brain |
| Wu et al., 2008 | T2 | Histology - LFB | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Zaaraoui et al., 2008 | MTR | Immunohistochemistry - MBP | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Takagi et al., 2009 | FA, AD | EM - Myelin thickness | Animal - Rat | Degeneration - Contusive injury | PN |
| Wang et al., 2009 | FA, RD | Histology - LFB | Animal - Rat | Ischemia - Induced hypoxia | Brain |
| Zhang et al., 2009 | RD | Histology - LFB | Animal - Rat | Injury - Dorsal columnar transection | SC |
| Schmierer et al., 2010 | MTR, T2 | Histology - LFB | Human | Multiple sclerosis | Brain |
| Fatemi et al., 2011 | MTR | Immunohistochemistry - MBP | Animal - Mouse | Ischemia - Induced hypoxia | Brain |
| Laule et al., 2011 | MWF | Immunohistochemistry - MBP | Human | Multiple sclerosis | Brain |
| Underhill et al., 2011 | MPF | Histology - LFB | Animal - Mouse | Healthy | Brain |
| Chandran et al., 2012 | FA, RD | Immunohistochemistry - MBP | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Tardif et al., 2012 | T1, T2, MTR, PD | Immunohistochemistry - MBP | Human | Multiple sclerosis | Brain |
| Fjær et al., 2013 | MTR | Immunohistochemistry - PLP | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Harkins et al., 2013 | MWF, MPF | Microscopy - Myelin fraction | Animal - Rat | Edema - Hexaclorophene | SC |
| Janve et al., 2013 | MPF, R1a, k_ba, FA, RD, MD, AD | Histology - LFB | Animal - Rat | Demyelination - Lipopolysaccharide | Brain |
| Thiessen et al., 2013 | MPF, R1f, k_fm, k_mf, T2f, T2m, MD, RD, AD, FA, T1, T2 | EM - Myelin thickness | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Kozlowski et al., 2014 | MWF | Immunohistochemistry - MBP | Animal - Rat | Injury - Dorsal columnar transection | SC |
| Wang et al., 2014 | RD, RD-DBSI | Immunohistochemistry - MBP | Animal - Mouse | Demyelination - Autoimmune encephalomyelitis | SC |
| Fjær et al., 2015 | MTR | Immunohistochemistry - PLP | Animal - Mouse | Demyelination - Autoimmune encephalomyelitis | Brain |
| Seehaus et al., 2015 | FA, RD, MD | Histology - Silver | Human | Healthy | Brain |
| Turati et al., 2015 | MPF | Immunohistochemistry - MBP | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Wang et al., 2015 | RD-DBSI | Histology - LFB | Human | Multiple sclerosis | SC |
| Aojula et al., 2016 | FA, AD, RD, MD | Immunohistochemistry - MBP | Animal - Rat | Hydrocephalus | Brain |
| Hakkarainen et al., 2016 | T1, T2, MTR, T1p, T2p, RAFF | Histology - Gold chloride | Animal - Rat | Healthy | Brain |
| Jelescu et al., 2016 | RD, RK, AWF, Rde, T2, MTR | EM - Myelin fraction | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Kelm et al., 2016 | MD, RD, MK, RK, AWF | EM - Myelin fraction | Animal - Mouse | Demyelination - Knockout | Brain |
| Reeves et al., 2016 | T1, T2 | Immunohistochemistry - MBP | Human | Epilepsy | Brain |
| Tu et al., 2016 | FA, AD, RD, MD, MTR | Immunohistochemistry - MBP | Animal - Rat | Traumatic brain injury | Brain |
| Chang et al., 2017 | FA, AD, RD, MD | Immunohistochemistry - MBP | Animal - Mouse | Healthy | Brain |
| Chen et al., 2017 | MWF | EM - Myelin fraction | Animal - Rat | Injury - Dorsal columnar transection | SC |
| Khodanovich et al., 2017 | MPF | Histology - LFB | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Lehto et al., 2017a | RAFF, MTR, T1sat, FA, MD, AD, RD | Histology - Gold chloride | Animal - Rat | Demyelination - Lipopolysaccharide | Brain |
| Lehto et al., 2017b | MTR | Histology - Gold chloride | Animal - Rat | Traumatic brain injury | Brain |
| van Tilborg et al., 2018 | FA | Immunohistochemistry - MBP | Animal - Rat | White matter injury | Brain |
| Beckmann et al., 2018 | MTR | Histology - LFB | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Berman et al., 2018 | MTV | EM - Myelin fraction | Animal - Mouse | Demyelination - Knockout | Brain |
| Hametner et al., 2018 | R2*, T1, QSM | Histology - LFB | Human | Vascular diseases | Brain |
| Praet et al., 2018 | MK, RK, AK, FA, MD, RD, AD | Immunohistochemistry - MBP | Animal - Mouse | Amyloidosis | Brain |
| Wendel et al., 2018 | FA, AD, RD, MD | Immunohistochemistry - MBP | Animal - Mouse | Traumatic brain injury | Brain |
| West et al., 2018 | MPF, MWF, MVF-T2, MVF-MT | EM - Myelin fraction | Animal - Mouse | Demyelination - Knockout | Brain |
| Yano et al., 2018 | FA, RD, MD | Immunohistochemistry - PLP | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Abe et al., 2019 | FA, RD, AD | Microscopy - Myelin thickness | Animal - Mouse | Optogenetic stimulation | Brain |
| Duhamel et al., 2019 | ihMTR, MTR | Microscopy - Fluorescence | Animal - Mouse | Healthy | Brain |
| Khodanovich et al., 2019 | MPF | Immunohistochemistry - MBP | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Mollink et al., 2019 | FA | Immunohistochemistry - MBP | Human | Amyotrophic lateral sclerosis | Brain |
| Peters et al., 2019 | FA, MD | Histology - LFB | Human | Tuberous sclerosis complex | Brain |
| Pol et al., 2019 | QSM, FA, MD | Histology - Solochrome | Animal - Mouse | Healthy | Brain |
| Soustelle et al., 2019 | MPF, RD, MWF, rSPF | Immunohistochemistry - MBP | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Guglielmetti et al., 2020 | MTR, MTR-UTE | Immunohistochemistry - MBP | Animal - Mouse | Healthy | Brain |
Additional files
-
Source code 1
A Jupyter notebook in ipynb format containing the Python code used to process the data, run the analyses and generate all the figures.
In order to execute the notebook, the Python (3.7) and R (3.6) interpreters are required, as well as the R packages metafor (2.4) and multcomp (1.4), and the following Python packages: numpy (1.18.4); pandas (0.25.3); plotly (4.8.1); rpy2 (3.3.4); xlrd (1.2.0). The notebook assumes that the spreadsheet is in the same path as the notebook itself. More details are provided here: https://github.com/matteomancini/myelin-meta-analysis (Mancini, 2020; copy archived at swh:1:rev:17ca8673c9e15c54ad0b814248b69232b63c3a38).
- https://cdn.elifesciences.org/articles/61523/elife-61523-code1-v2.zip
-
Source data 1
A spreadsheet in xlsx format containing all the data and details collected for the studies considered in this systematic review.
- https://cdn.elifesciences.org/articles/61523/elife-61523-data1-v2.xlsx
-
Supplementary file 1
A multimedia file in HTML format containing an interactive version of the figures in this manuscript plus additional ones.
- https://cdn.elifesciences.org/articles/61523/elife-61523-supp1-v2.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61523/elife-61523-transrepform-v2.pdf