Molecular evidence of hybridization between pig and human Ascaris indicates an interbred species complex infecting humans
Figures
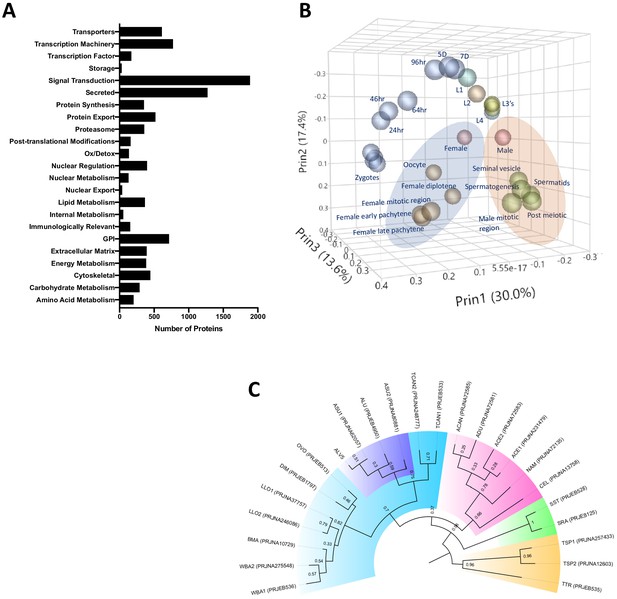
Ascaris proteome.
(A) Functional classification of the predicted proteome of A. lumbricoides (an improved proteome of Ascaris spp.), excluding proteins with unknown or uncharacterized function. (B) PCA plot based on multivariate analyses of RNA-seq data from various stages/tissues. Samples from tissues related to sperm (blue ellipse) and oocyte production (orange ellipse, see also Figure 1—figure supplement 2) cluster together. (C) Estimated tree based on orthology analyses between the predicted proteomes of publicly available nematodes. The Ascaris clade has been shaded in purple within Clade III (teal). Samples are labeled by BioProject Accession number, as well as by the first letter of the genus and the first two letters of the species name (ASU = Ascaris suum, ALU = Ascaris lumbricoides, WBA = Wuchereria bancrofti, BMA = Brugia malayi, LLO = Loa loa, DIM = Dirofilaria immitis, OVO = Onchocerca volvulus, TCAN = Toxocara canis, ACAN = Ancylostoma caninum, ADU = Ancylostoma duodenale, ACE = Ancylostoma ceylanicum, NAM = Necator americanus, CEL = Caenorhabditis elegans, SST = Strongyloides stercoralis, SRA = Strongyloides ratti, TSP = Trichinella spiralis, TTR = Trichuris trichiura). Multiple genomes for the same organism are suffixed with numerals.
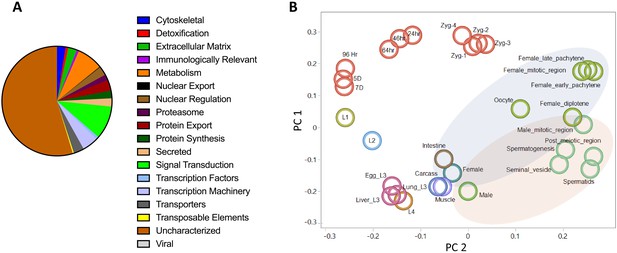
Predicted proteome and stage-specific transcriptomes of Ascaris.
(A) Functional classification of the predicted proteome of A. lumbricoides (an improved proteome of Ascaris spp.) with the majority of proteins being unknown/uncharacterized. (B) Two-dimensional principal component analysis plot illustrating the similarities in transcription profiles between the major stages (Figure 1B) and the developmental stages.
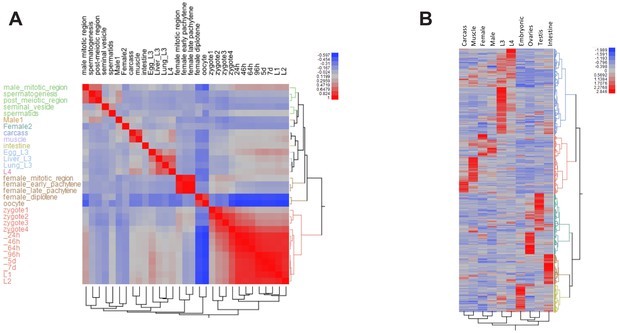
Ascaris stage-specific RNA expression heatmaps.
(A) Correlation heatmap comparing parasite transcriptomes at different life stages. (B) 1870 genes differentially expressed across the stages.
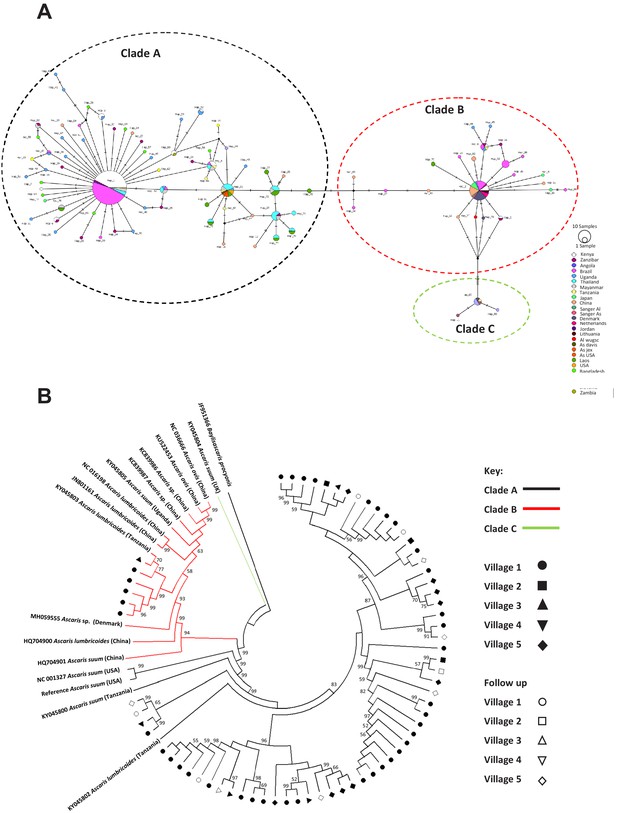
Phylogenetics of Ascaris spp based on mitochondrial sequences.
(A) Haplotype network based on the COl mitochondrial gene. Notches on the lines separating samples represent the number of nucleotide changes between the worms represented, details on the origins of haplotypes can be found in Supplementary file 4; (B) Maximum likelihood phylogenetic (ML) reconstruction of Ascaris complete mitochondrial genomes, constructed under the conditions of the GTR model and 1000 bootstrap replicates were used to provide nodal supports. The tree was constructed using all mitochondrial genomes assembled from the Kenyan worm specimens and all other published reference Ascaris mitochondrial genomes and Baylisascaris procyonis was used as the outgroup. The three major clades A, B, and C were identified by color hue, and the majority of the Kenyan worms clustered in clade A. Each village was represented by a distinct shape and unfilled shapes represented worms sequenced from specific villages post-anthelminthic treatment.
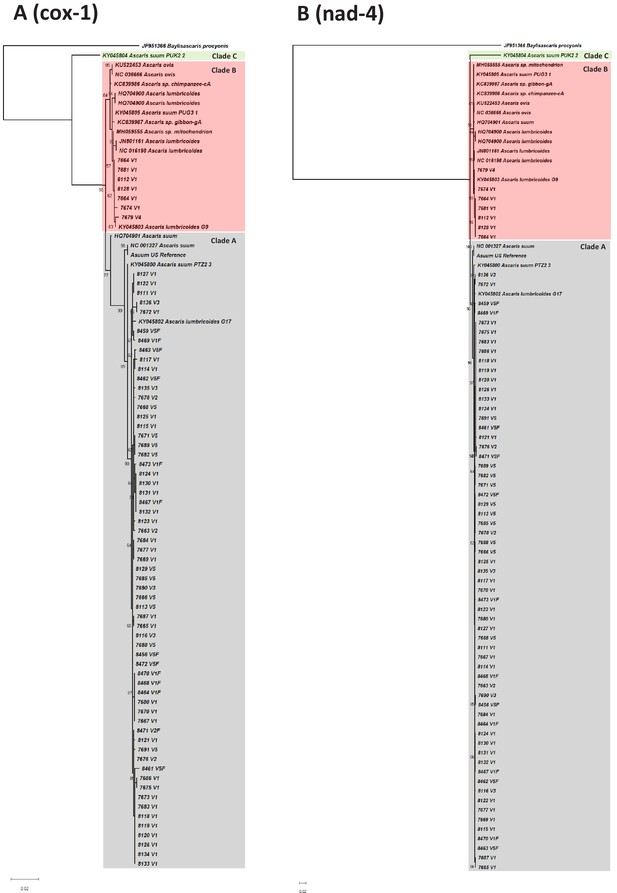
Phylogenetic trees based on cox-1 and nad-4.
Maximum likelihood phylogenetic analyses of the (A) cox-1 and (B) nad-4 genes using RaxML under the conditions of the GTR model with nodal support values generated through 1000 bootstrap replicates. The trees were generated using complete sequences of the genes extracted from the 68 Kenyan Ascaris mitochondrial genomes generated in this study and other published reference genomes. The most diverse region was the non-coding control region, both between worms from different villages in this study and between Kenyan and other worms. The nad-4 gene had a nucleotide diversity of π = 0.008827; that of the cox-1 barcoding gene was 0.006243.
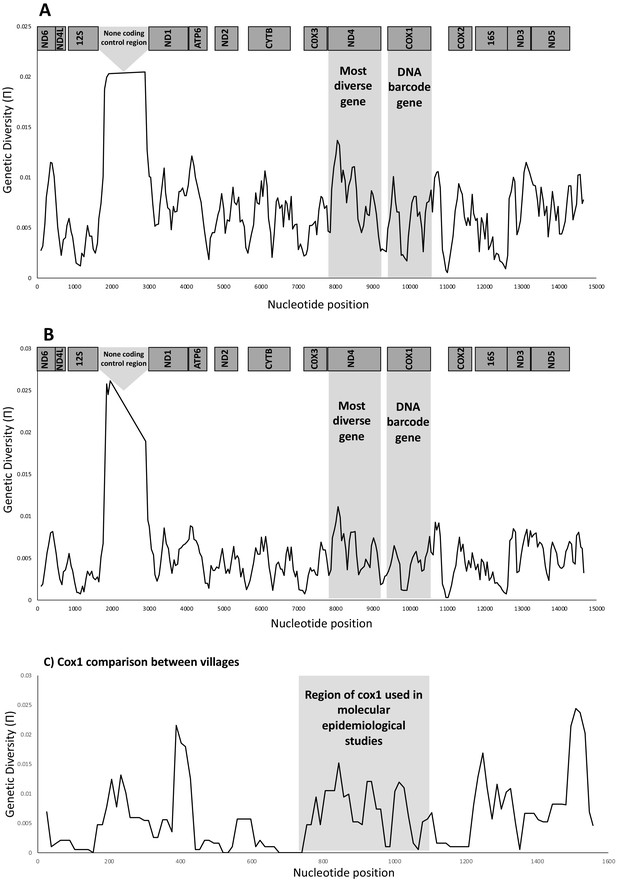
Sliding window analyses.
(A) Comparison between Kenyan samples and reference mitochondrial genomes of Ascaris lumbricoides and Ascaris suum, (B) Comparison between villages, (C) cox-1 comparison between villages. Despite differences in their molecular diversity, phylogenetic analyses based on the nad-4 and cox-1 genes revealed the same overall topology: distinct A. lumbricoides- and A. suum-type clades.
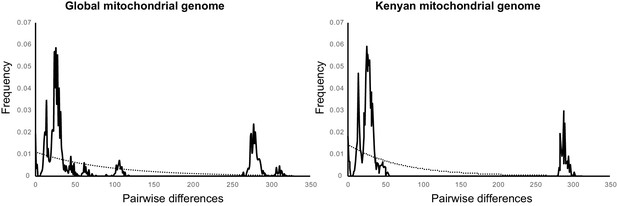
Evidence of Ascaris population expansion.
The pairwise nucleotide differences between worm samples (solid line) are compared to the binomial function that would most closely represent a theoretical stable population (dotted line). Additional information is available in Supplementary file 8.
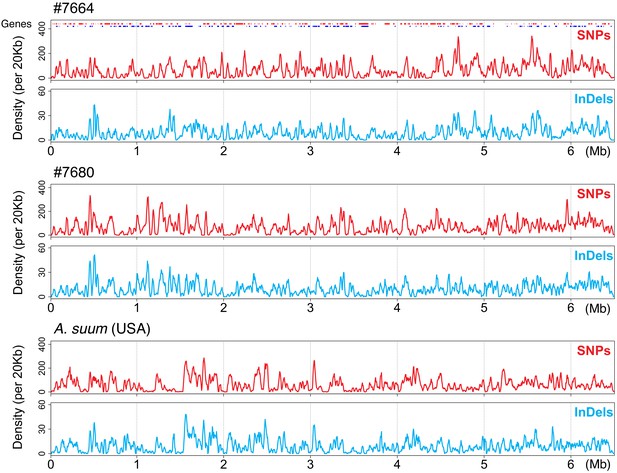
Ascaris SNPs and insertion/deletions (indels) maps of representative chromosomal fragments.
An assembled 6.5 Mb Ascaris lumbricoides chromosome fragment (ALgV5R006), with the frequency of identified SNPs and indels plotted for one representative A. lumbricoides-like worm from this study (#7664) and one A. suum-like worm (#7680). Genes are shown on the top of the plot, with red and blue indicating genes transcribed from forward and reverse strands, respectively. The y-axis shows the frequency of SNPs and indels for a 20 kb window size (with a 4 kb sliding window in x-axis). Note the profiles and the frequency between SNPs and indels are highly consistent within individual worms.
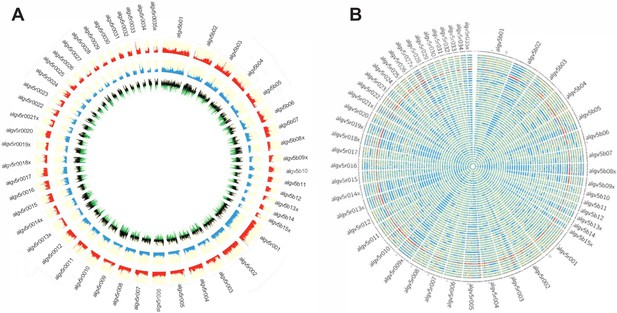
Genetic diversity of the Ascaris specimens.
(A) Circos plot depicting the genetic diversity of the Ascaris specimens. Outside track (red histograms) shows the total SNP diversity across the genome (first 50 largest scaffolds) in 10 kb sliding windows. Blue bar plot indicates the measured degree of polymorphism (π) (Nei and Li, 1979) within the Ascaris population in 10 kb sliding windows. The innermost track with black-green histogram plots the Tajima, 1989 values which reflect the difference between the mean number of pairwise differences (π) and the number of segregating sites using a sliding window of 10 kb. (B) The Circos-plot of the genome-wide distribution of heterozygous and homozygous SNPs in 10 kb blocks identified long stretches of homozygosity among the different Ascaris specimens, except 119_3, which is predominantly heterozygous throughout and was isolated from village 3. Red color = >90% of heterozygous SNPs, blue = >90% of homozygous SNPs, yellow = 50% heterozygous, 50% homozygous SNPs. Each track represents a single specimen.
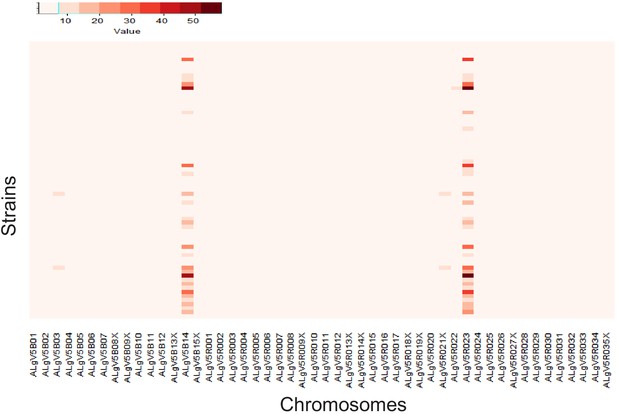
Somy analysis of the Ascaris worm specimens.
The ploidy of the Ascaris specimens are represented in a heatmap. Ploidy was calculated by averaging the count of aligned reads in 10 kb sliding windows across the genome after reference mapping against ALV5. The ploidy data suggest that Ascaris is completely diploid (close to 2 n), except at two scaffolds ALgB5B14 and ALgv5RO23, where the majority of specimens show elevated ploidy. X-axis shows the first 50 largest scaffolds involved in this study and the y-axis shows the specimens (ordered by code number 1–68).
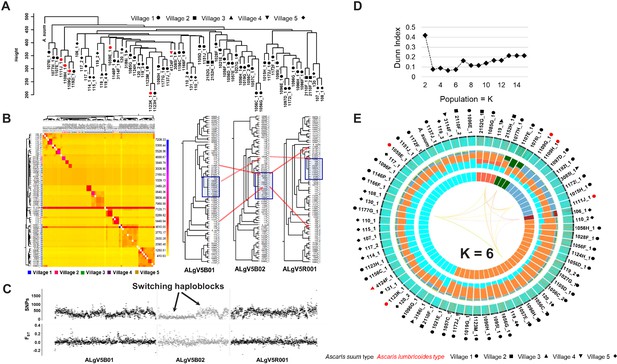
Comparative genomics and population genetic structure of Ascaris.
(A) Hierarchy phylogenetic tree of Ascaris specimens. Phylogenetic tree was constructed with genome wide SNPs (at 10x coverage) from 68 Ascaris specimens, including the A. suum reference (outgroup). Height = number of SNPs per site. Red symbol = A. lumbricoides mitochondrion genome. Black symbol = A. suum mitochondrion genome. Samples were collected from five different villages: Circle = village 1, square = village 2, upside triangle = village 3, downside triangle = village 4, diamond = village 5. (B) Heatmap clustering the co-inheritance of ancestral blocks by Bayesian method using genome wide shared haplotype segments among the Ascaris genomes. scale = posterior coincidence probability. Hierarchical clustering and phylogenetic relationships are based on percent shared haplotype segments in scaffolds ALgV5B01, ALgV5B02, and ALgV5R001. Red arrows show examples of genetic recombination demonstrated by phylogenetic incongruence in the tree topology based on shared ancestry among blue highlighted specimens (n = 13). (C) Pairwise SNPs and FST estimates in scaffolds ALgV5B01, ALgV5B02, and ALgV5R001 indicate a switching of haplotypes (black arrows), and genetic hybridization among the blue highlighted specimens (n = 13) in the phylogenetic tree depicted in Figure 2B. X-axis = total SNPs/10 kb in SNPs plot or FST/10 kb in FST plot. (D) Estimation of the number of ancestral populations (K) based on Dunn Index (Dunn, 1973). (E) Population genetic structure and admixture clustering analysis of the Ascaris genomes obtained by POPSICLE using K = 6 different color hues in the innermost concentric circle of the Circos plot. The middle concentric circle shows the relative percentage of each genetic ancestry within each genome (represented by the color hues for K = 6). The outermost concentric circle shows the genome wide local admixture profile of each worm in 10 kb sliding windows. The following geometric shapes represent villages, and the color for each shape identifies the mitochondrion genome each sample possesses: Black = A. suum; red = A. lumbricoides; Circle = village 1; square = village 2; upside triangle = village 3; downside triangle = village 4; diamond = village 5.
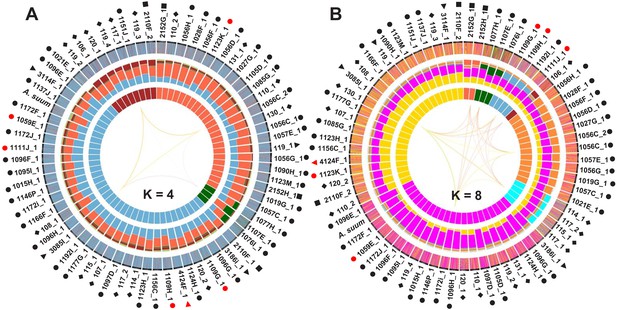
Admixture clustering and current population genetic structure of Ascaris were determined.
Data analyzed with POPSICLE with an ancestral population size = 4 (A) and 8 (B) in 10 kb sliding windows as described in Figure 4E.
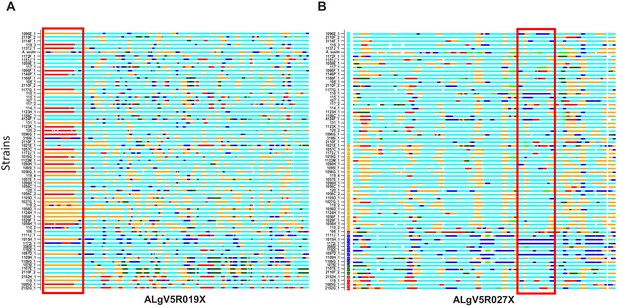
Local admixture clustering and genome wide analysis of inheritance of haploblocks of Ascaris obtained by POPSICLE (Shaik et al., 2018).
Based on ancestral population K = 6. X-axis = specimens. Red highlighted box indicates the introgression of large haplotype blocks of defined parentage among the different specimens of Ascaris in scaffolds ALgV5R019X (A) and ALgV5R027X (B). Many examples exist whereby specimens that are in linkage disequilibrium at ALgV5R019X possess different haplotypes in ALgV5R027X (for example 1107E_1 vs. 2110F_2) indicating both segregation as well as recombination in the evolution of the samples. The local admixture patterns reveal extensive genetic hybridization among different strains of Ascaris. Color assignment is depicted based on Figure 4E.
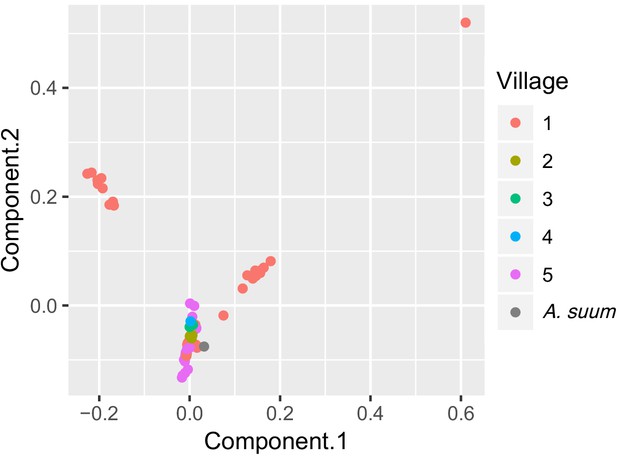
PCA plot of worms sequenced for five Kenyan villages.
Each point is color-coded by village-of-origin and plotted according to the first and second principal components, based on genome sequences. Worms from village #1 are found in each of three clusters, and two clusters contain only worms from village #1.
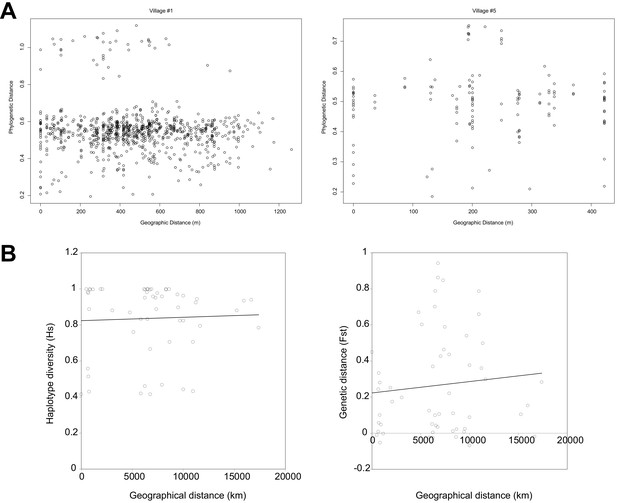
Plot of phylogenetic distances compared to geographic distances.
(A) For village #1 and village #5. (B) Plot of diversity versus geographical distance (Hs on left, Fst on right). Genetic distances based on cox-1 genes are plotted against the geographic distances between the places from which these worms were collected.
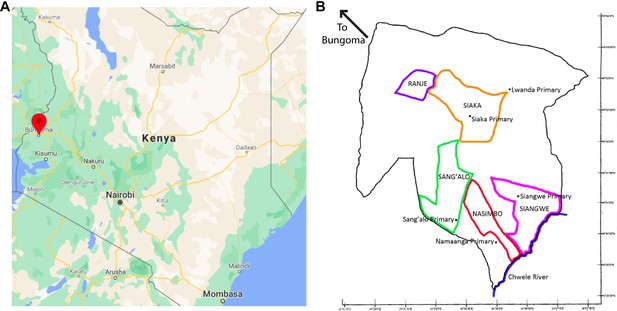
Map of Bungoma and West Sang’alo Sub-District.
(A) Bungoma town is shown by a red marker in a map of Kenya (Google Maps). (B) This map highlights the area covered by the four study villages and the pilot study village (Ranje). The locations of five primary schools where data collection occurred are marked. These primary schools were chosen because of the number of students attending these schools from the study villages. However, some of these schools are not located within the study villages.
Tables
Ascaris germline genome assemblies.
| Features | A. lumbricoides de novo | A. lumbricoides semi-de novo* | A. lumbricoides reference-based | A. suum† (Wang et al., 2017) | A. suum† (Jex et al., 2011)§ |
|---|---|---|---|---|---|
| Assembled bases (Mb) | 269.2 | 307.9 | 296.0 | 298.0 | 272.8 |
| N50 (Mb) | 0.29 | 4.77 | 4.63 | 4.65 | 0.41 |
| N50 number | 269 | 21 | 21 | 21 | 179 |
| N90 (Mb) | 0.04 | 0.95 | 0.91 | 0.92 | 0.08 |
| N90 number | 1112 | 74 | 75 | 75 | 748 |
| Total scaffold number | 8111 | 412 | 415 | 415 | 29,831 |
| Largest scaffold length (Mb) | 1.9 | 13.9 | 13.2 | 13.4 | 3.8 |
| Protein-coding genes | 17,011‡ | 17,105‡ | 17902 | 18,025 | 18,542‡ |
-
* Exhibits ~23 Mb of sequence gaps and 15.4 Mb of unplaced sequence in 4072 short contigs.
† The three A. lumbricoides assemblies constructed here are compared to the A. suum assemblies from Australia (Jex et al., 2011) and the United States (Wang et al., 2017).
-
‡21–23% are only partial genes based on the annotation from A. suum (Wang et al., 2017).
§ The sample for sequencing is derived from a mixture of the germline and somatic genomes (after DNA elimination).
Effects of host, household, village, and time point on the genetic variation of Ascaris.
| Nuclear genome phylogeny* | Mitochondrial genome phylogeny | |||||
|---|---|---|---|---|---|---|
| R† | p-value | p-adjusted (Bonferroni) | R† | p-value | Samples† | |
| Individual | 0.933 | 0.001 | 0.004 | 0.996 | 0.095 | 68 worms from 60 people |
| Household | 0.020 | 0.110 | 0.440 | 0.011 | 0.340 | 68 worms from 43 houses |
| Village | 0.052 | 0.001 | 0.004 | 0.013 | 0.335 | Five villages with 43, 17, 4, 3, and one individual each |
| Time point | 0.018 | 0.162 | 0.648 | 0.024 | 0.100 | 55 at baseline and 13 post-deworming |
-
* Results based on PERMANOVA using phylogenetic distances among worms. Results were largely similar using a distance matrix generated from the PCA plot (Figure 6) and using the Multi-Response Permutation Procedure (MRPP) method (Supplementary file 9).
† Since some worms did not have metadata associated with each variable examined, and some variables were over-represented in the sample (for example, 43 of 68 worms came from a single village) the samples are specified in this column.
Additional files
-
Supplementary file 1
Characteristics of genome assemblies.
Reference A. lumbricoides genomes generated as part of this study (1 and 3) are compared with reference genomes for A. suum generated previously (2 and 4).
- https://cdn.elifesciences.org/articles/61562/elife-61562-supp1-v1.xlsx
-
Supplementary file 2
Proteome annotation.
While ~94.6% of the genes can be transferred to both genomes, over 20% of the transferred genes are only partial matches and are fragmented supporting the view that the de novo and semi de novo A. lumbricoides assemblies are highly fragmented.
- https://cdn.elifesciences.org/articles/61562/elife-61562-supp2-v1.xlsx
-
Supplementary file 3
Description of worm from which each sample was sequenced.
The sex of the worm (based on morphological identification) and the part of the worm (germline vs somatic) is listed. Some hosts donated multiple worms.
- https://cdn.elifesciences.org/articles/61562/elife-61562-supp3-v1.xlsx
-
Supplementary file 4
cox-1 haplotype list.
- https://cdn.elifesciences.org/articles/61562/elife-61562-supp4-v1.xlsx
-
Supplementary file 5
X4 ratio analyses of Clades A and B using complete mitochondrial genomes used to construct the phylogeny in Figure 2b.
- https://cdn.elifesciences.org/articles/61562/elife-61562-supp5-v1.docx
-
Supplementary file 6
Demographic analyses using Tajima’s D and Fu’s F statistic across complete mitochondrial genomes as a detection for the signature of population expansion events.
Whether all sequences collected globally, or just sequences collected in Kenya as part of this study were examine, the Tajima’s D value was negative and significant (indicating an excess of low frequency polymorphisms) and the Fu’s Fs was positive but not significant (potentially indicating a deficiency in diversity as would be expected in populations that has recently undergone a bottle neck event).
- https://cdn.elifesciences.org/articles/61562/elife-61562-supp6-v1.docx
-
Supplementary file 7
Number of heterozygous and homozygous SNPs in each of the 68 worms from Kenya sequenced.
- https://cdn.elifesciences.org/articles/61562/elife-61562-supp7-v1.xlsx
-
Supplementary file 8
Reference mitochondrion genomes.
- https://cdn.elifesciences.org/articles/61562/elife-61562-supp8-v1.xlsx
-
Supplementary file 9
Supplement to Table 2 using alternative measures of phylogenetic distance.
- https://cdn.elifesciences.org/articles/61562/elife-61562-supp9-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61562/elife-61562-transrepform-v1.docx





