ASIC1a is required for neuronal activation via low-intensity ultrasound stimulation in mouse brain
Figures
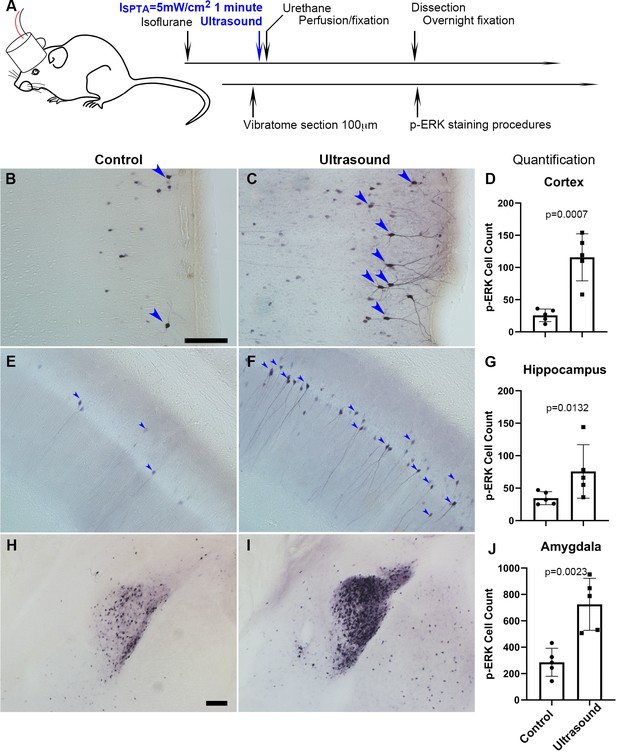
Transcranial ultrasound induces p-ERK expression in neurons of the cortex, hippocampus and amygdala of mouse brain.
(A) Illustration depicting mouse head stimulated by 1 MHz transducer which was positioned in between the mouse nasal process of maxilla and the axis of mouse ear of an anesthetized mouse. (B) Micrograph representing cortical region with basal level of p-ERK staining in sham control mice (n = 5), scale bar 100 μm. Sham control mice were handled with similar procedures of placing transducer on the head without turning on the ultrasound function generator. (C) Micrograph representing cortical region with p-ERK staining stimulated by ultrasound (ISPTA = 5 mW/cm2, 1 minute) (n = 5). (D) Quantitative bar graph of the number of p-ERK stained cells within comparable area of 1.224 mm2 (Length 1275 μm, Width 960 μm), showing significant difference (P = 0.0007) of cell count by ImageJ. (E) Micrograph representing hippocampal region with basal level of p-ERK staining in sham control mice. (F) Micrograph representing hippocampal region with p-ERK staining in mice stimulated by ultrasound. (G) Bar graph showing quantification of significantly p-ERK different cell count (1.224 mm2) (P = 0.0132) in hippocampal region. (H) Micrograph representing amygdala of sham controls. Scale bar 100 μm (I) Micrograph representing amygdala of ultrasound stimulated mice. (J) Quantification of amygdaloid significant difference (P = 0.0023) of p-ERK cell count (1.224 mm2).
-
Figure 1—source data 1
Source data for Figure 1D, G, J.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig1-data1-v2.xlsx
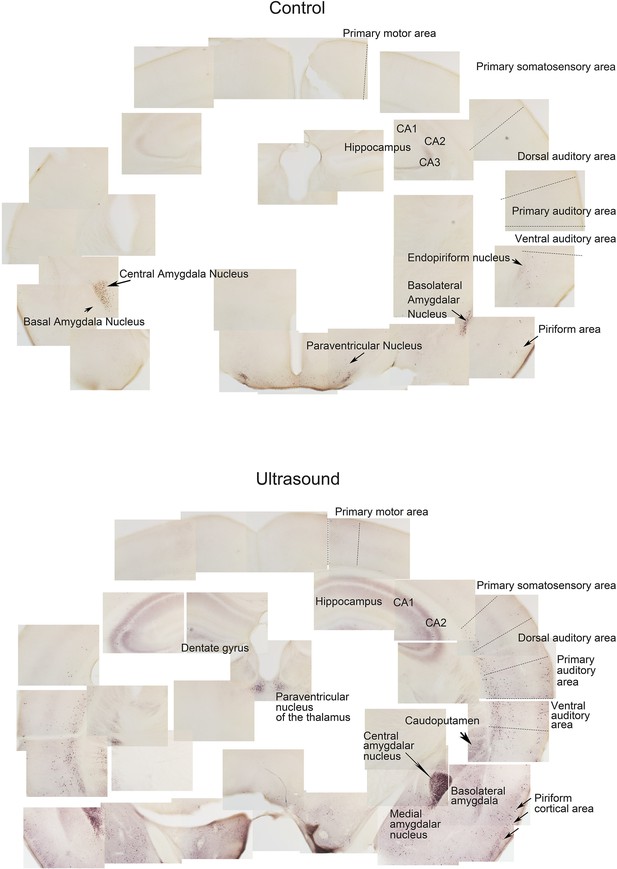
Stitched images showing p-ERK expression of brain slice containing hippocampus, cortex and amygdala of sham control v.s ultrasound stimulated mice.
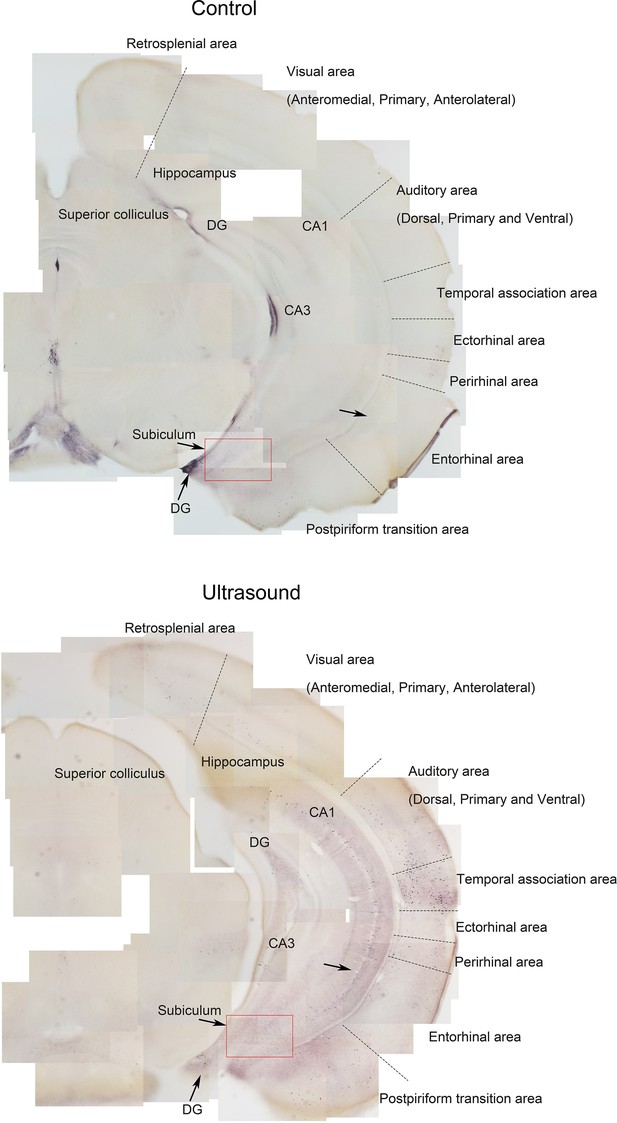
Stitched images showing p-ERK expression of hippocampus and piriform cortex of sham control v.s ultrasound stimulated mice.
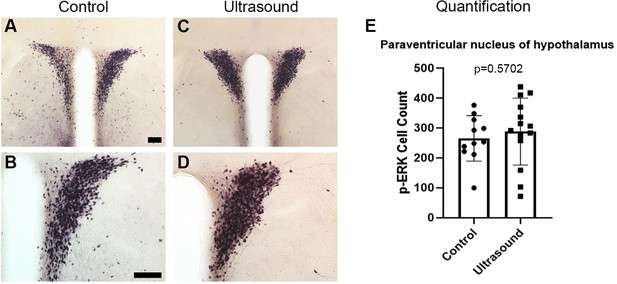
The similar p-ERK expression of paraventricular nucleus of hypothalamus (PVH) of control untreated vs ultrasound stimulated mice.
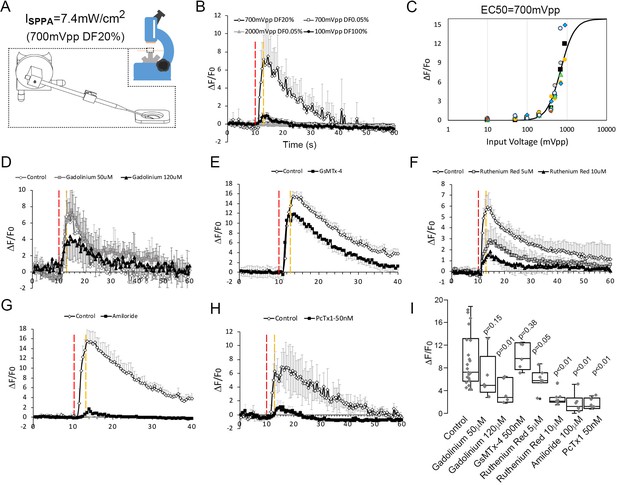
Neuronal calcium signals induced by micropipette-guided ultrasound suppressed by ASIC1a inhibitors.
(A) A micropipette positioned to the cortical neurons cultured on a 30 mm cover slips mounted to a chamber coupled to microscope platform. Calcium signals recorded from neurons stained by Invitrogen Oregon Green 488 BAPTA-1, AM cell permeant. (B) Line graphs of averaged calcium signals in four neurons stimulated by micropipette ultrasound for 3 s with an input voltage 2000mVpp, duty factor (DF) 0.05 % (n = 5); or 100mVpp, DF100% (n = 3) or 700mVpp, DF20% (n = 4). The red-dotted line denotes start of the stimulation while the yellow-dotted line denotes the end. (C) Calcium responses as a function of micropipette ultrasound in 20%DF. Dose-dependent (input voltages from 10mVpp, to 900mVpp, DF20%) responses of ultrasound with an EC50 of 700mVpp is shown (n = 5). (D) Effects of gadolinium (120 μM), a non-selective blocker of mechanically sensitive ion channels, on calcium signals in cortical neurons (n = 5). (E) Effects of GsMTx-4 (500 nM), a selective Piezo inhibitor, on calcium signals in cortical neurons (n = 4). Control n = 4. (F) Effects of Ruthenium red (10 μM), a non-selective TRP inhibitor, on calcium signals in cortical neurons (n = 4). Control n = 6. (G) Effects of amiloride (100 μm), an ASICs family inhibitor, on calcium signals in cortical neurons (n = 4). (H) Effects of PcTx1 treated (50 nM), a selective ASIC1a inhibitor, on calcium signals in cortical neurons (n = 5). (I) Statistical analyses of channel blockers on ultrasound-induced calcium signals in cortical neurons. Control n = 21, Gadolinium (50 μM) n = 5, Gadolinium (100 μM) n = 5, GsMTx-4 (500 nM) n = 5, Ruthenium red (5 μM) n = 5, Ruthenium red (10 μM) n = 5, Amiloride (100 μM) n = 5, PcTx1 (50 nM) n = 5.
-
Figure 2—source data 1
Source data for Figure 2B and C.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Source data for Figure 2D.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Source data for Figure 2E and G.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig2-data3-v2.xlsx
-
Figure 2—source data 4
Source data for Figure 2F.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig2-data4-v2.xlsx
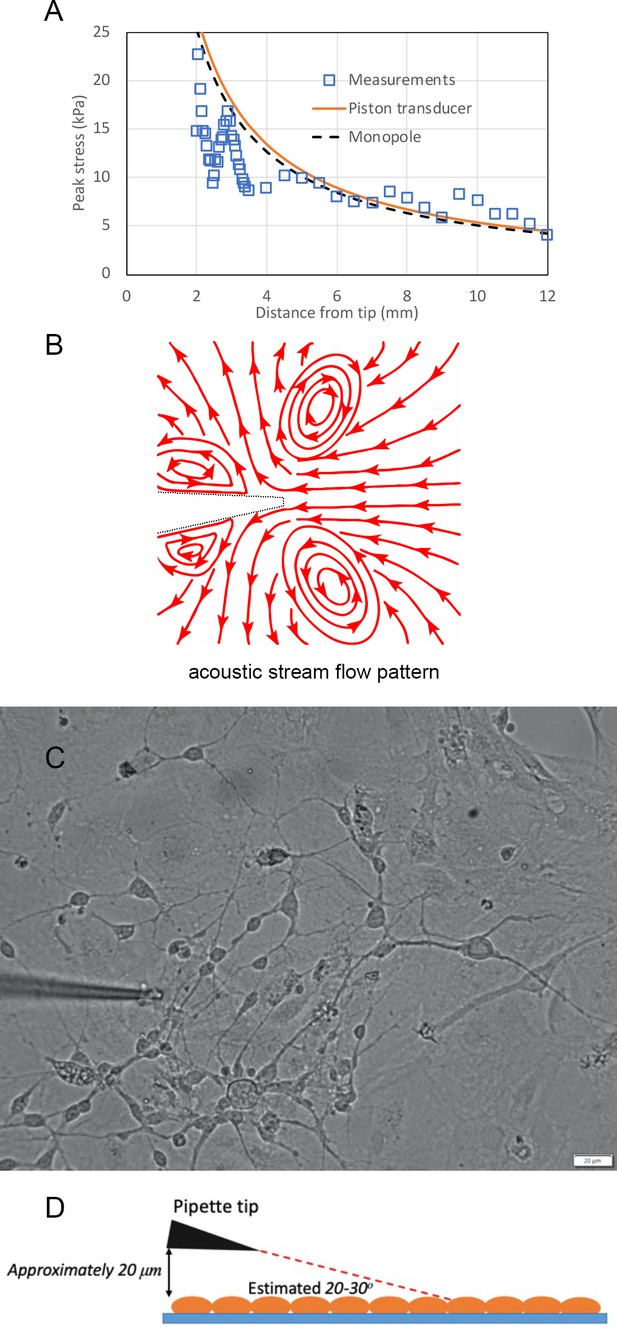
Peak pressure stress and acoustic streaming pattern of micropipette guided ultrasound.
(A) The measured and monopole modeled peak pressure stress vs distance from tip of micropipette. (B) The schematic acoustic stream flow pattern depicted from Video 1. (C) Position of micropipette for guiding ultrasound. This image is corresponding to the Video 3. The z-position of micropipette is estimated to be within 20 micrometer on top of the cultured cell layer because the micropipette tip is maintained visibly clear. (D) Cartoon illustration of the position of micropipette for guiding ultrasound. Color code: Blue, cover glass; orange, cells; black, micropipette; red dashed line representing the angle of pipette tip in reference to the horizontal cover glass.
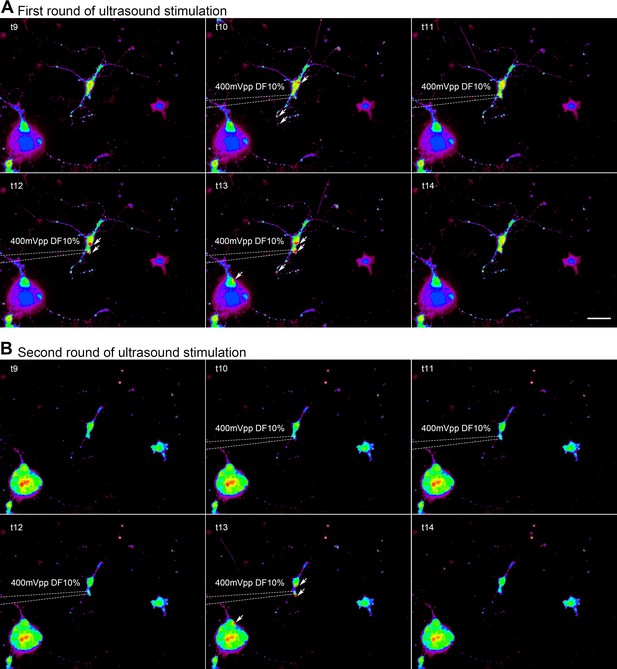
Neuronal calcium response upon micropipette guided ultrasound stimulation.
(A) Invitrogen Fura-2, AM, cell permeant (Fura-2) stained primary cultured neuron isolated from neonatal mouse cortex was stimulated using micropipette-guided ultrasound (400mVpp, duty factor 10%). Image right before stimulation, during the entire 4 seconds of stimulation and right after the stimulation were shown. Ab340nm/380 nm ratio values are spectrum color coded. Sites of obvious increased calcium are indicated by the arrows. (B) The same neuron could be activated when the cell was stimulated for the second time even though photo-bleaching effects were overwhelming. Scale bar 20 μm.
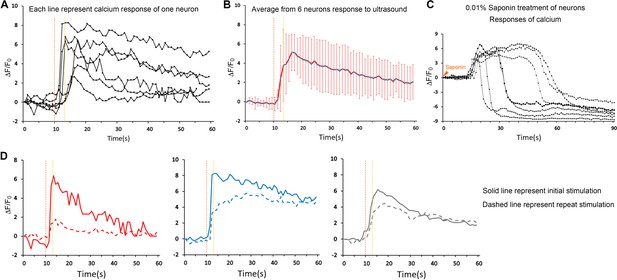
Quantification of neuronal calcium responses upon micropipette guided ultrasound stimulation.
(A) Invitrogen Oregon Green 488 BAPTA-1, AM, cell permeant (BAPTA) stained primary neurons were stimulated using micropipette guided ultrasound (700mVpp, duty factor 10%). Calcium response of each neuron induced by stimulation is plotted against time. Stimulation window is within the dotted orange and yellow lines. (B) The representative responses of the six neurons are plotted by using the averages and standard deviations. (C) The calcium responses of neurons upon cell perforation treatment were plotted against time for the internal calibration. (D) Three examples of calcium response of neurons comparing initial stimulation (solid line) and repeated stimulation (dashed line) upon micropipette guided ultrasound (700mVpp, duty factor 10%).
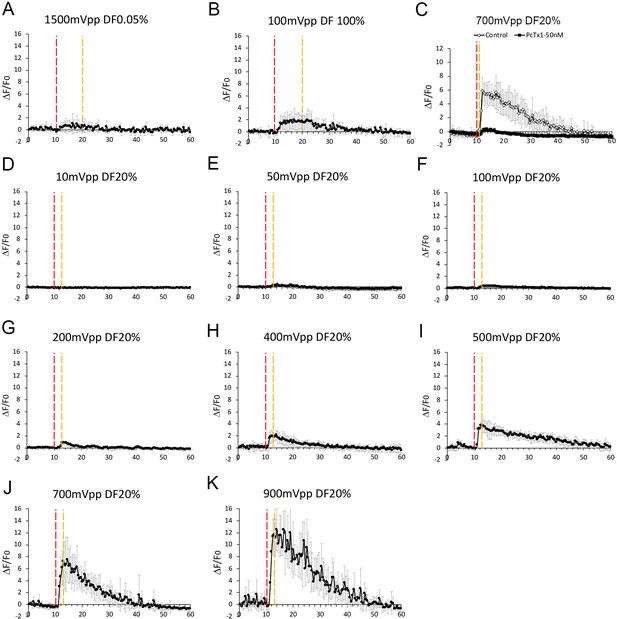
Input voltage and duty factor settings for ultrasound dose and the neuronal calcium responses.
(A) The ultrasound-induced calcium response of 1500mVpp at 0.05% duty factor for 10 s was minimal. The physical effect of pressure stress is dominant in this stimulation condition.n = 4. (B) The ultrasound induced calcium response of 100mVpp at 100% duty factor for 10 s was also minimal. The physical effect of acoustic streaming is dominant in this stimulation condition. n = 3. (C) The elevated ultrasound-induced calcium response of 700mVpp at 20% duty factor for as short as 1 s. The response was diminished by applying PcTx1 (50 nM), an ASIC1a inhibitor. (C) Control n = 5, PcTx1 n = 5. (D–K) The dose response of ultrasound induced calcium response from 10 mVpp to 900 mVpp at 20% duty factor. The ultrasound stimulation time were 3 ss. (D) n = 8. (E) n = 5. (F) n = 5. (G) n = 5. (H) n = 6. (I) n = 5. (J) n = 5. (K) n = 3.
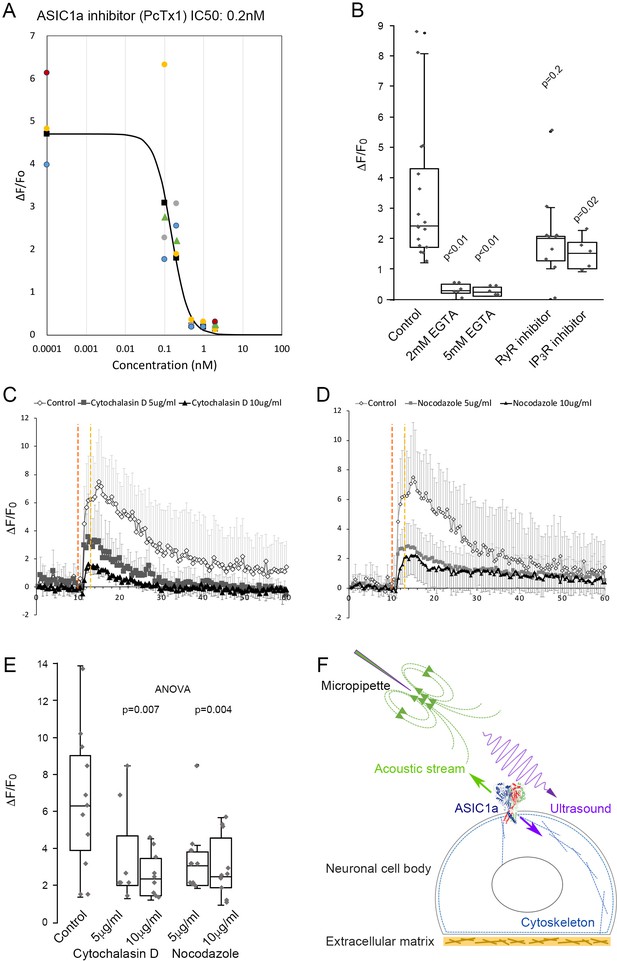
ASIC1a as a mechanoreceptor responsive to mechanical stimuli with combined ultrasound and acoustic streaming.
(A) PcTx1 dose-dependent inhibition curve of calcium responses induced by micropipette ultrasound of 700mVpp, DF20% for 3 s (n = 5). (B) Whisker plots showing comparison of peak ΔF/F0 within 3–5 s upon micropipette ultrasound (700mVpp, DF20%, 3 s) stimulation in the untreated control primary neurons (n = 16), 2- or 5 mM EGTA-treated neurons (n = 6 or 4, respectively), RyR inhibitor JTV519 fumarate (10 μM) treated neurons (n = 10), or IP3R inhibitor (-)-Xestonspongin C (1 μM)-treated neurons (n = 5). Student t-test with p value compared to control listed above the whisker plot. (C) Graph showing calcium response of actin polymerization inhibitor Cytochalasin D (5–10 μg/ml)-treated neurons (n = 7 and n = 9, respectively) compared to untreated control (n = 5). (D) Calcium signals showing the effect of the microtubule assembly inhibitor nocodazole (5–10 μg/ml) on neurons (n = 8 and n = 10, respectively). (E) Whisker plots showing comparison of peak ΔF/F0 within 3–5 s upon micropipette ultrasound (700mVpp, DF20%, 3 s) stimulation in the untreated control primary neurons (n = 11), cytochalasin D 5- or 10 μg/ml treated neurons (n = 7 or n = 9, respectively), nocodazole 5- or 10 μg/ml treated neurons (n = 8 or n = 10, respectively). Statistical p values of one-way ANOVA analysis were listed to show the significance of treatment. (F) Cartoon depicting ultrasound stimulating ASIC1a in the cell body of a neuron under the micropipette ultrasound stimulation. Green arrow represents the pulling force of acoustic stream and purple arrow represents the compression force of ultrasound that results in cytoskeletal rearrangement.
-
Figure 3—source data 1
Source data for Figure 3A.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Source data for Figure 3B.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig3-data2-v2.xlsx
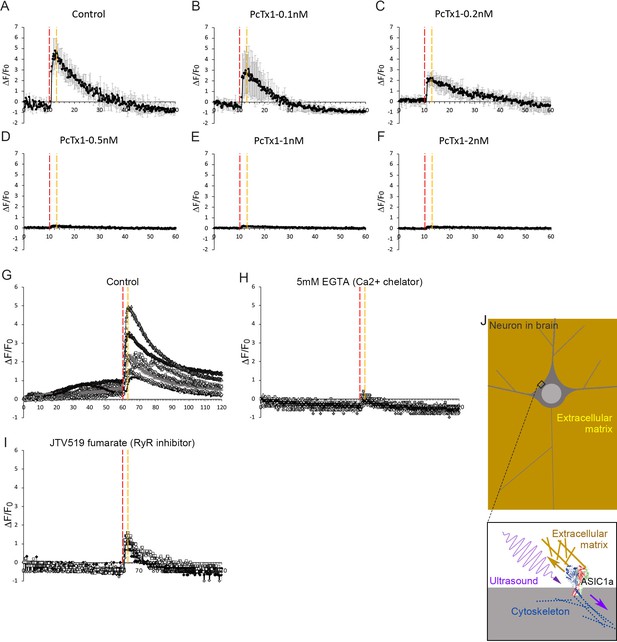
A–FThe dose-dependent calcium response of PcTx1, an ASIC1a inhabitation, under micropipette stimulation (700mVpp, 20%DF, check).
The ultrasound stimulation time was 3 seconds. (A) n = 3. (B) n = 4. (C) n = 4. (D) n = 3. (E) n = 3. (F) n = 5. (G) This figure is not used in main manuscript, n = 7. (H) The EGTA (a calcium chelator) inhibit the calcium response of ultrasound stimulation (5 mM), n = 5. (I) The JVT519 fumerate, a RyR inhibitor, also moderately inhibit calcium response of ultrasound stimulation (dose), n = 5. (J) A schematic mechanotransduction model for in vivo circumstance.
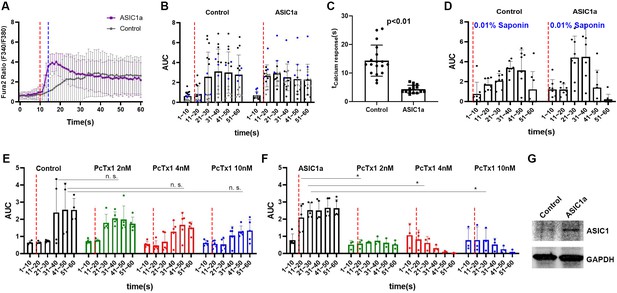
ASIC1a overexpression showed a fast calcium response upon micropipette-guided ultrasound stimulation in CHO cells.
(A) Invitrogen Fura-2, AM, cell permeant (Fura-2) stained CHO cells. Fluorescence ratios of Fura-2 emission at wavelengths 340 nm/380 nm (F340/380nm) were recorded and averaged line graphs were shown. F340/380nm ratio values plotted against time were shown in Figure 4—figure supplement 1A, B. Ultrasound stimulation is indicated by the red dashed lines at time point 10 s for a duration of 3 s. The blue dashed line indicates the time of ultrasound termination. Control n = 18, ASIC1a overexpressed n = 15. (B) Area under curve (AUC) of F340/380nm were plotted in a 10 s bin manner. Each dot represents a single cell quantified. Three batches of experiments were represented by three different colors. Refer to Supplementary file 1 for the two-way ANOVA analysis of this graph. Control n = 14, ASIC1a overexpressed n = 15. (C) Calcium response time determined by the maximum F340/380nm was significantly shortened by ultrasound stimulation compared to the sham transfected controls. Control n = 18, ASIC1a overexpressed n = 15. (D) Cell perforation treatment with 0.1% saponin in HHBS after the experiments for internal calibration of maximal response. Refer to Supplementary file 1 for the two-way ANOVA analysis of this graph. Control n = 7, ASIC1a overexpressed n = 7. (E) CHO cells calcium response either with or without PcTx1 treatments. Refer to Supplementary file 1 for the two-way ANOVA analysis of this graph. Control n = 4, PcTx1 (2 nM) n = 5, (4 nM) n = 5, (10 nM) n = 5. (F) ASIC1a overexpressing CHO cells either with or without PcTx1 treatments. Refer to Supplementary file 1 for the two-way ANOVA analysis of this graph. Control n = 5, PcTx1 (2 nM) n = 3, (4 nM) n = 3, (10 nM) n = 3. (G) Western analysis of ASIC1a comparing the sham transfected control and Asic1-transfected cells. GAPDH detection serves as an internal control.
-
Figure 4—source data 1
Source data for Figure 4A.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Source data for Figure 4B and C.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Source data for Figure 4D.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig4-data3-v2.xlsx
-
Figure 4—source data 4
Source data for Figure 4E.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig4-data4-v2.xlsx
-
Figure 4—source data 5
Source data for Figure 4F.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig4-data5-v2.xlsx
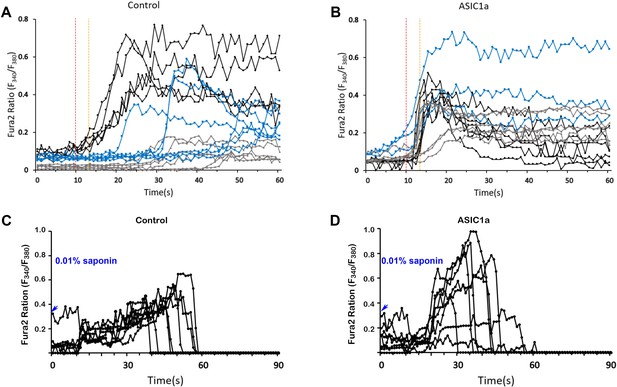
Effect of ASIC1a over expression in CHO cells on calcium response upon micropipette guided ultrasound stimulation.
(A) Invitrogen Fura-2, AM, cell permeant (Fura-2) stained CHO cells stimulated by micropipette guided ultrasound (400mVpp, duty factor 10%). Each line represents the calcium response of a single cell upon ultrasound stimulation. Three different colors represent three different batches of cells. (B) Calcium response of ASIC1a overexpressing CHO cells. The transfection efficiency was monitored by either tdTomato fluorescence or western analysis. (C) Calcium response in CHO cells caused by cell perforation treatment at the end of the experiments (n-7). Calcium response of Asic1 transfected cells upon cell perforation treatment (0.01% Saponin in HHBS).
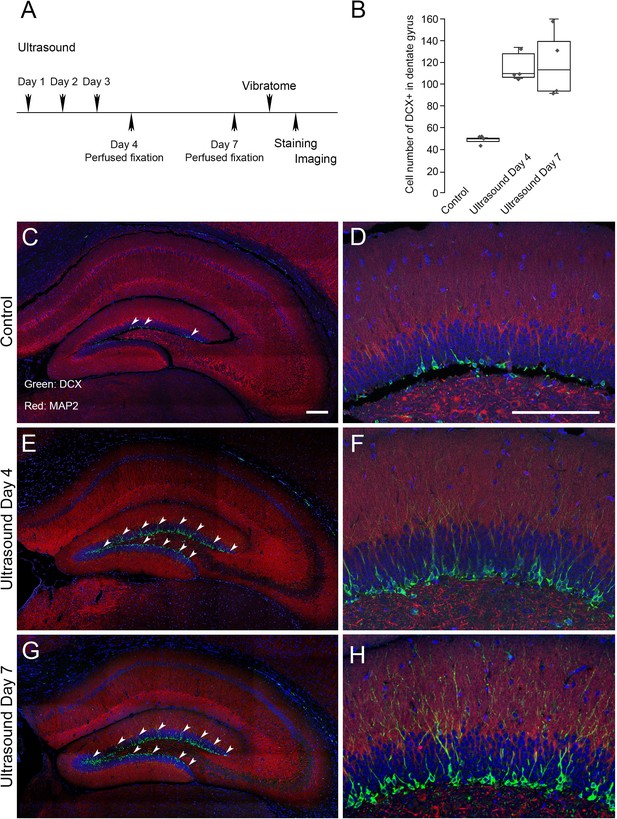
Neurogenesis in dentate gyrus induced by repeated transcranial ultrasound treatments.
(A) The mice were treated three consecutive days by ultrasound of 4 mW/cm2, 1% for 5 min, subsequently perfused fixed at day 4 or day 7 and brains were dissected from the head and sectioned for immunofluorescence procedures. The DCX staining in dentate gyrus of treated mice were compared to control untreated one. (B) Cell count with clear DAPI stained nucleus surrounded by DCX markers compared in control, day 4 and day 7 post-ultrasound treatments. Statistical analysis: p = 0.0013 and F ratio = 15.18 in one-way ANOVA (n = 4). (C, D) Representative micrograph of untreated mice. The vibratome coronal brain sections (100 μm) of dentate gyrus region immunofluorescently stained for DCX (green) and MAP2 (red). Blue color indicates DAPI stained nuclei. Representative micrograph showing. (E, F) Representative micrograph from ultrasound treated mice fixed at day 4. (G, H) Representative micrograph from ultrasound treated mice fixed at day 7. Scale bar 200 μm.
-
Figure 5—source data 1
Source data for Figure 5B.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig5-data1-v2.xlsx
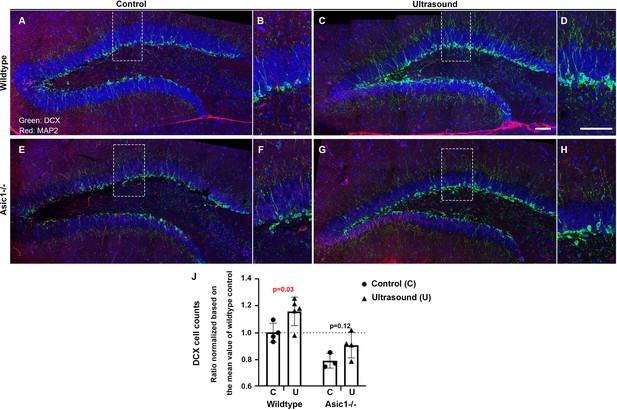
DCX staining-positive cells are increased but partially compromised by Asic1-/- after consecutive 3 days of ultrasound treatments.
(A) Micrographs stitched to show the representative DCX staining pattern (green fluorescence) of the dentate gyrus (DG) in the 5 weeks old mice of wildtype sham treated controls. (B) Magnified DCX positive cells in wildtype control DG. (C, D) DCX-positive cells increased significantly upon three continuous days of ultrasound treatments. (E, F) Representative stitched micrographs of sham treated Asic1-/- dentate gyrus. (G, H) The increase of DCX staining upon ultrasound stimulation partially compromised by Asic1-/-. (I) Quantitative analysis of DCX cell counts/mm in the 100 μm brain slices with clear DCX and DAPI staining using confocal microscopy scanning stacks of 8–10 z-planes. Scale bar 100 μm. There were two batches of mice of 5 weeks and 7 weeks old and the cells counts from all the z-stacks were normalized based on the mean value of wildtype controls to include both batches of mice. Each data point represents quantification of one animal; wildtype control n = 4, wildtype ultrasound treated n = 5, Asic1-/- control n = 3, Asic1-/- ultrasound stimulated n = 4. Refer to Supplementary file 1 for the two-way ANOVA analysis of this graph.
-
Figure 6—source data 1
Source data for Figure 6J.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig6-data1-v2.xlsx
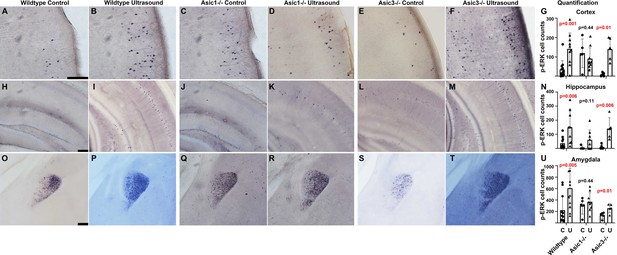
Asic1-/- suppressed the p-ERK cell count increases in cortex, hippocampus, and amygdala of mouse brain.
The IHC stained brain slices of wildtype mice, Asic1-/- mice and Asic3-/- mice. Mice of all genotypes were randomly assigned to sham treatment group and ultrasound treatment group. The quantification of p-ERK-positive cells were performed using ImageJ with setting of threshold and particle sizes that representing the actual staining pattern. (A–F) Micrographs depicting p-ERK IHC staining in the cortex of the vibratome brain slices. (G) Quantification comparing cortical p-ERK-positive cells in three different genotypes of mice either mock treated or ultrasound stimulated. (H–M) IHC micrograph depicting p-ERK staining in hippocampus. (N) Quantification comparing hippocampal p-ERK-positive cells in mice with indicated the genotypes and treatments. (O–T) IHC micrographs depicting p-ERK staining in amygdala. (U) Quantification comparing p-ERK-positive cells in amygdala. Scale bar 100 μm. Each data point represents the total cell count of one mouse brain; wildtype control n = 11, wildtype ultrasound treated n = 10, Asic1-/- control n = 5, Asic1-/- ultrasound stimulated n = 9, Asic3-/- control n = 5, Asic3-/- ultrasound stimulated n = 5. Refer to Supplementary file 1 for the two-way ANOVA analysis of this graph.
-
Figure 7—source data 1
Source data for Figure 7G, N and U.
- https://cdn.elifesciences.org/articles/61660/elife-61660-fig7-data1-v2.xlsx
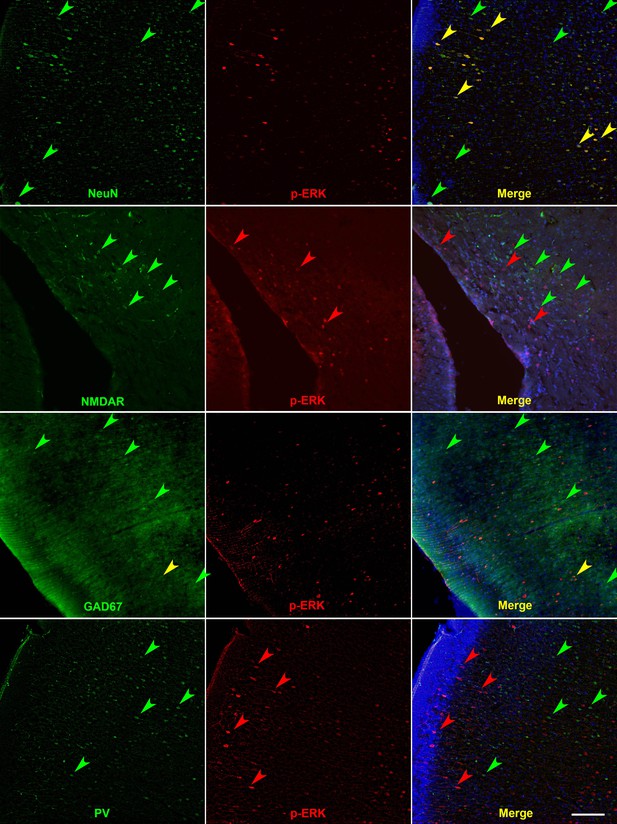
Co-staining of p-ERK with neuronal markers such as NeuN, NMDAR, GAD67, and PV (Green fluorescence).
Green fluorescent signals (e.g. green arrows) surrounding DAPI-positive nuclei that were found to co-stain (e.g. yellow arrows) with p-ERK (e.g. red arrows) were counted: NeuN+ in p-ERK population was 94% (197/209), GAD67+ in p-ERK population was 4.5% (10/223), and PV+ in p-ERK population was 0.9% (2/211). Scale bar 100 μm.
Videos
Neuronal calcium signal cannot be induced by micropipette guided ultrasound with 2000mVpp input voltage and duty cycle 0.05%.
This setting induced produce predominantly ultrasound stimulation. When the dash line depicted micropipette tip appeared in the video, ultrasound function generator was turned on.
Micropipette-guided ultrasound stimulation of neuronal calcium elevation.
The setting was 700mVpp input voltage and duty cycle 20% for 3 s. When the dash line depicted micropipette tip appeared in the video, ultrasound function generator was turned on. The setting generated both ultrasound and acoustic streaming effects.
Micropipette-guided ultrasound stimulation of neuronal calcium response.
Ultrasound with 250mVpp input voltage and continuous waves stimulation. When the dash line depicted micropipette tip appeared in the video, ultrasound function generator was turned on. The setting generated both ultrasound and acoustic streaming effects.
Micropipette-guided ultrasound with 400mVpp input voltage and duty factor 10% induced the neuronal calcium signals captured by Fura-2 imaging methods.
Calcium elevations were measured by fluorescence ratios of Fura-2 emission at wavelengths 340 nm/380 nm (F340/380nm). Spectrum color coded fluorescence ratios in which the red color represents the highest ratio while purple color represents the lowest ratio of F340/380nm. When the dash line depicted micropipette tip appeared in the video, ultrasound function generator was turned on. The ultrasound setting generated both ultrasound and acoustic streaming effects.
CHO cells calcium response induced by micropipette guided ultrasound.
Right panel was CHO cells overexpressing ASIC1a while left panel was showing transfection sham control cells. Calcium elevations were measured by fluorescence ratios of Fura-2 emission at wavelengths 340 nm/380 nm (F340/380nm). Spectrum color coded fluorescence ratios in which the red color represents the highest ratio while purple color represents the lowest ratio of F340/380nm. When the dash line depicted micropipette tip appeared in the video, ultrasound function generator was turned on. The ultrasound setting was 600mVpp input voltage and duty cycle 10% for 3 s. This setting generated both ultrasound and acoustic streaming effects.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Other | Wildtype C57BL/6 J (Mus musculus) | The Jackson Laboratory | Stock number: 000664 | Experimental Animal Facility, Institute of Biomedical Sciences, Academia Sinica IACUC 12-03-332, 20-06-1492 |
| Other | Asic1a-/-(specific targeting alternative spliced isoform Asic1a-/-) C57BL6/J (Mus musculus) | Eur J Neurosci 2015 Jun; 41(12):1553–68. doi:10.1111/ejn.12905. | Asic1-/- | Experimental Animal Facility, Institute of Biomedical Sciences, Academia Sinica IACUC 12-03-332, 20-06-1492 |
| Other | Asic3-/- C57BL6/J (Mus musculus) | Nat Commun 2016 May; 7:11,460. doi:10.1038/ncomms11460. | Asic3-/- | Experimental Animal Facility, Institute of Biomedical Sciences, Academia Sinica IACUC 12-03-332, 20-06-1492 |
| cell line (Cricetulus griseus) | Epithelial like Chinese Hamster ovary cells | ATCC | CCL-61 (RRID:CVCL_0214) | |
| Transfected construct (Discosoma sp.) | pCMV - mCherry | Gift from Dr. Huang, Yi-Shuian, IBMS, Academia Sinica, Taiwan | Vector control | 1 μg DNA: 3 μl Lipofectamin 2000 in 1 ml opti-MEM medium |
| Transfected construct (Mus musculus) | pT7-RFP-N2- T▲T - P2A - mASIC1a (alternative spliced isoform Asic1a) | The cDNA of mouse ASIC1a was subcloned into the plasmid (pT7-RFP-N2) | Asic1 | 1 μg DNA: 3 μl Lipofectamin 2000 in 1 ml opti-MEM medium |
| Antibody | pERK; Rabbit polyclonal [polyclonal Phospho-p44/42 MAPK (ERK1/2) (Thr202/Tyr204)] | Cell Signaling Technology | #9,101 (RRID:AB_331646) | IHC (1:500) |
| Antibody | Doublecortin; (Rabbit polyclonal) | Cell Signaling | #4,604 (RRID:AB_561007) | ICC (1:200) |
| Antibody | MAP2; (Mouse monoclonal) | Thermo Fisher Scientific | MA5-12823 (RRID:AB_10982160) | ICC (1:200) |
| Antibody | Secondary biotinylated goat-anti-rabbit antibodies; (Goat monoclonal) | Vector Laboratories | BA-1000–1.5 (RRID:AB_2313606) | (1:1000) |
| Antibody | Alexa Fluor 488-conjugated goat anti-rabbit IgG; (Goat monoclonal) | Thermo Fisher Scientific | 16–237 (RRID:AB_436053) | (1:100) |
| Antibody | Alexa Fluor 555-conjugated goat anti-mouse IgG; (Goat monoclonal) | Thermo Fisher Scientific | A-21422 (RRID:AB_141822) | (1:100) |
| Commercial assay or kit | IHC Reagent; Avidin-Biotin pre-mix solution | Vector Laboratories | A-2004–5 (RRID:AB_2336507) | (1:200) |
| Commercial assay or kit | IHC staining kit; DAB Peroxidase Substrate Kit with Nickel, 3,3’-diaminobenzidine | Vector Laboratories | SK-4100 (RRID:AB_2336382) | According to instruction manual |
| Commercial assay or kit | Live cell imaging staining reagent for calcium; Invitrogen Oregon Green 488 BAPTA-1, AM, cell permeant | Thermo Fisher Scientific | O6807 | 5 μM |
| Commercial assay or kit | Live cell imaging staining reagent for calcium; Invitrogen Fluo-4, AM, FluoroPure | Thermo Fisher Scientific | F23917 | 5 μM |
| Commercial assay or kit | Live cell imaging staining reagent for calcium; Fura-2, AM, cell permeant | Thermo Fisher Scientific | F1221 | 5 μM |
| Chemical compound, drug | GsMTx-4 | Abcam Inc | ab141871 | 500 nM |
| Chemical compound, drug | Amiloride | Sigma-Aldrich | A7410-1G | 100 μM |
| Chemical compound, drug | PcTx1 | Tocris | #5,042 | 0.1–50 nM |
| Chemical compound, drug | Ruthenium Red | Tocris | #1,439 | 5–10 μM |
| Chemical compound, drug | RyR inhibitor, JTV519 fumarate | Tocris | #4,564 | 10 μM |
| Chemical compound, drug | Thapsigargin | Sigma-Aldrich | T9033 | 100 nM |
| Chemical compound, drug | Cytochalasin D | Tocris | #1,233 | 5 μg- 10 μg/ml |
| Chemical compound, drug | Nocodazole | Tocris | #1,228 | 5 μg- 10 μg/ml |
| Software, algorithm | Image J | Abràmoff, M. D., Magalhães, P. J., & Ram, S. J. (2004). Image processing with ImageJ. Biophotonics international, 11(7), 36–42. | Particle analysis | Set threshold and define particle sizes |
| Software, algorithm | GraphPad Prism 8 | Swift, M. L. (1997). GraphPad prism, data analysis, and scientific graphing. Journal of chemical information and computer sciences, 37(2), 411–412. | Student t-test |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61660/elife-61660-transrepform1-v2.docx
-
Supplementary file 1
Supplementary files 1A-H.
- https://cdn.elifesciences.org/articles/61660/elife-61660-supp1-v2.docx





