Proteomics reveals synergy between biomass degrading enzymes and inorganic Fenton chemistry in leaf-cutting ant colonies
Figures
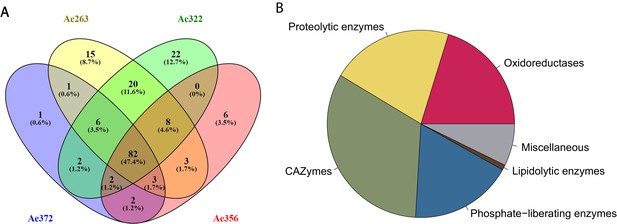
Statistics of the fecal fluid proteome of Acromyrmex echinatior.
(A) Venn diagrams, constructed using the web application Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/index.html) showing the overlap of protein profiles identified in the four fecal fluid samples obtained from colonies Ae263, Ae322, Ae356, and Ae372. (B) Pie chart showing the abundances of proteins across colonies assigned to six functional categories based on the label-free quantification (LFQ) values provided by MaxQuant.
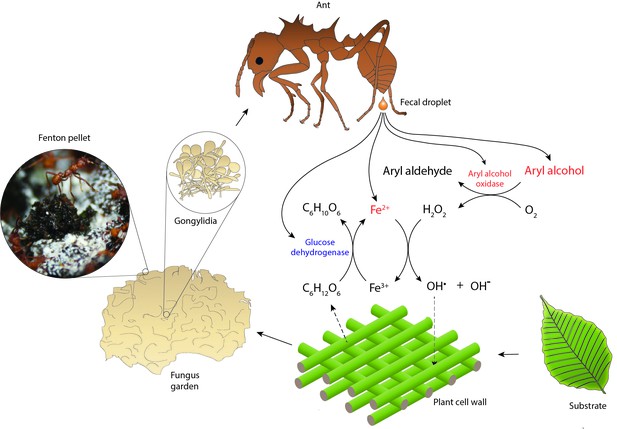
Hypothesized reaction scheme for the generation of hydroxyl radicals when ant fecal fluid interacts with chewed leaf fragments in temporary substrate pellets, based on the presence of enzymes in the total fecal fluid proteome (Supplementary file 1).
Fungal enzymes produced in gongylidia of the symbiotic garden-cultivar that the ants ingest pass unharmed through the gut to end up in the fecal fluid (Boyd and Martin, 1975b; Kooij et al., 2014b; Schiøtt et al., 2010; De Fine Licht et al., 2013; Kooij et al., 2016). After droplets of fecal fluid are deposited and become exposed to oxygen, the fungal oxidoreductases produce hydrogen peroxide while aryl alcohols are converted to aryl aldehydes. The hydrogen peroxide then reacts with reduced iron (Fe2+) to produce hydroxyl radicals (OH•) in a Fenton reaction, which aggressively breaks down cell walls of the plant substrate. Oxidized iron (Fe3+) can then be reduced again by ant-encoded glucose dehydrogenase, using glucose released via plant cell wall decomposition as electron donor. The leaf substrate is initially concentrated in green pellets of ca. 3 mm diameter distributed across the top of fungus gardens, which turn black in a few hours when subjected to Fenton-mediated degradation (inset image). Compounds ultimately derived from the fungal symbiont are in red text and compounds directly produced by the ants in blue.
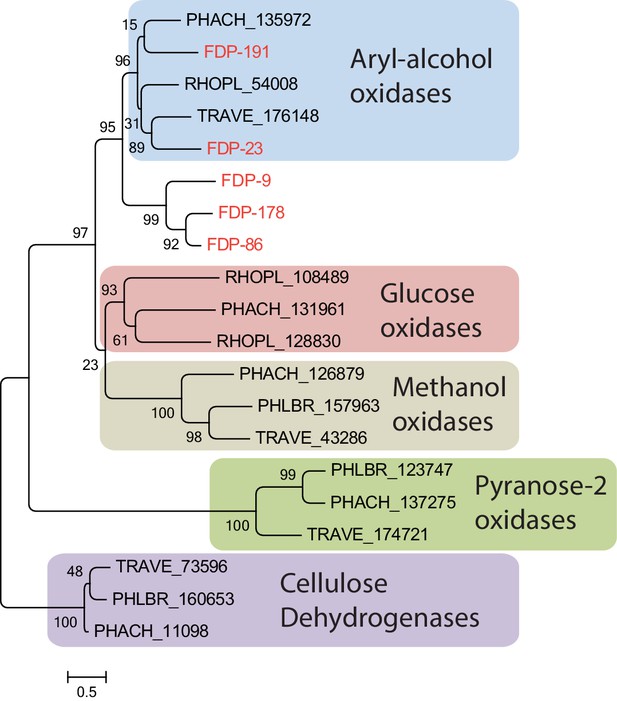
Gene tree of the five fungal glucose-methanol-choline (GMC) oxidoreductases identified in the ant fecal fluid (present study) and in representative other basidomycete fungi, all assigned to functional groups based on a previous study (Ferreira et al., 2015).
All fungal GMC oxidoreductases from the fecal fluid (Protein IDs 9, 23, 86, 178, and 191 in Table 1; red text) clustered among the known aryl-alcohol oxidases. Other closely related functional groups are glucose oxidases, methanol oxidases, pyranose-2 oxidases, and cellulose dehydrogenases, which were retrieved in Phanerochaete chrysosporium (PHACH), Rhodonia placenta (RHOPL), Trametes versicolor (TRAVE), and Phlebia brevispora (PHLBR), but not in fecal fluid of the leaf-cutting ant A. echinatior. Note that the ant-encoded glucose dehydrogenase is also a GMC oxidoreductase, but would sequence-wise not fit into this phylogeny of fungal proteins. Numbers are aLRT SH-like support values for nodes. The scale bar represents 0.5 substitutions per site.
-
Figure 3—source data 1
Amino acid sequences used for the gene tree shown in Figure 3.
- https://cdn.elifesciences.org/articles/61816/elife-61816-fig3-data1-v2.xlsx
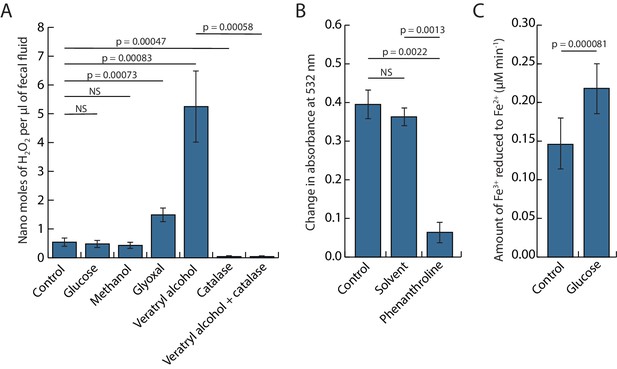
Bioassays to demonstrate that inorganic.
Fenton chemistry must be taking place when ant fecal fluid is exposed to oxygen while being deposited on leaf pulp pellets chewed by the ants, testing the key interactions hypothesized in Figure 2. (A) Bar plot showing the concentrations (means ± SE across six colonies) of hydrogen peroxide in fecal fluid after adding potential substrates (glucose, methanol, glyoxal, or veratryl alcohol) of glucose-methanol-choline (GMC) oxidoreductases with or without the hydrogen peroxide degrading enzyme catalase. One-way ANOVA showed a highly significant overall effect of treatments: F6,36 = 14.597, p=2.863e-08, and pairwise post hoc t-tests on matching samples from the same ant colonies (corrected for multiple testing with the Holm–Bonferroni method) confirmed both the enhancing effects of veratryl alcohol and glyoxal and the inhibiting effect of catalase (p-values in plot unless non-significant) (NS). (B) Deoxyribose assays showing that ant fecal fluid has the capacity to produce hydroxyl radicals (means ± SE across six colonies). Phenanthroline is known to work as an iron chelator and significantly reduced degradation of 2-deoxy-D-ribose while the solvent (methanol) of phenanthroline did not. Paired t-tests followed the same protocol as in the A-panel except that the overall ANOVA was omitted because there were only three means to compare. (C) A Ferrozine assay (means ± SE across six colonies) showing the capacity of ant fecal fluid to reduce Fe3+ to Fe2+, confirming that addition of glucose increases the rate of iron reduction. Statistics as in the B-panel.
-
Figure 4—source data 1
Data used for Figure 4A, B, and C.
- https://cdn.elifesciences.org/articles/61816/elife-61816-fig4-data1-v2.xlsx
Tables
Fecal fluid proteins that were found in three or four of the examined colony samples (Figure 1), and thus likely to belong to the core fecal fluid proteome.
For each protein the predicted function, its fungal or ant origin, the number of samples containing the protein, the relative abundance of the protein, and an identifier number are listed. Proteins were assigned to one of six functional categories (Figure 1). Although a number of oxidoreductases are listed in the CAZy database in the subcategory of auxiliary activities we have for the present study kept them separate. This is because it remains ambiguous whether they are true CAZymes if they are defined as redox enzymes that ‘act in conjunction with CAZymes’. This separation also resolved the problem that some of the oxidoreductases listed in Table 1 are not covered by the CAZy database. Question marks indicate that annotations were inconclusive.
| Category | Protein ID | Predicted function | Source | Samples with protein | Relative abundance |
|---|---|---|---|---|---|
| Oxidoreductases (including CAZymes with auxilliary activity) | 15 | Laccase | Fungus | 4 | 106.0 |
| 97 | Laccase | Fungus | 4 | 431.8 | |
| 180 | Laccase | Fungus | 4 | 14.7 | |
| 184 | Laccase | Fungus | 4 | 21.4 | |
| 35 | 4-carboxymuconolactone decarboxylase, alpha-beta hydrolase | Fungus | 4 | 4.3 | |
| 81 | Copper radical oxidase, glyoxal oxidase, galactose oxidase | Fungus | 4 | 8.9 | |
| 186 | Copper radical oxidase, glyoxal oxidase, galactose oxidase | Fungus | 4 | 8.3 | |
| 98 | FAD/FMN-containing isoamyl alcohol oxidase | Fungus | 4 | 41.1 | |
| 9 | GMC oxidoreductase, aryl-alcohol oxidase | Fungus | 4 | 5.9 | |
| 86 | GMC oxidoreductase, aryl-alcohol oxidase | Fungus | 4 | 10.3 | |
| 178 | GMC oxidoreductase, aryl-alcohol oxidase | Fungus | 4 | 6.9 | |
| 191 | GMC oxidoreductase, aryl-alcohol oxidase | Fungus | 4 | 18.3 | |
| 23 | GMC oxidoreductase, aryl-alcohol oxidase | Fungus | 3 | 1.9 | |
| 145 | GMC oxidoreductase, glucose dehydrogenase | Ant | 4 | 82.2 | |
| 149 | Succinate dehydrogenase (ubiquinone) flavoprotein subunit | Ant | 3 | 4.8 | |
| Proteolytic enzymes | 6 | Aspartic peptidase A1A, polyporopepsin | Fungus | 4 | 12.0 |
| 111 | Aspartic peptidase A1A, saccharopepsin | Fungus | 4 | 74.3 | |
| 125 | Metallopeptidase M1, ERAP2 aminopeptidase | Ant | 4 | 11.3 | |
| 112 | Metallopeptidase M14A, zinc carboxypeptidase | Ant | 4 | 27.4 | |
| 144 | Metallopeptidase M14A, zinc carboxypeptidase | Ant | 4 | 7.3 | |
| 141 | Metallopeptidase M28D, carboxypeptidase Q | Ant | 4 | 15.9 | |
| 188 | Metallopeptidase M35, deuterolysin | Fungus | 4 | 23.9 | |
| 206 | Metallopeptidase M35, peptidyl-lys metallopeptidase | Fungus | 4 | 218.6 | |
| 207 | Metallopeptidase M35, peptidyl-lys metallopeptidase | Fungus | 4 | 11.4 | |
| 56 | Metallopeptidase M36 | Fungus | 4 | 4.3 | |
| 133 | Serine protease S1A, chymotrypsin | Ant | 4 | 56.0 | |
| 134 | Serine protease S1A, chymotrypsin | Ant | 4 | 3.8 | |
| 135 | Serine peptidase S1A, chymotrypsin-2 | Ant | 4 | 8.8 | |
| 36 | Serine peptidase S8A, cuticle degrading peptidase | Fungus | 4 | 7.2 | |
| 103 | Serine peptidase S8A, cerevisin | Fungus | 4 | 51.6 | |
| 16 | Serine peptidase S10, carboxypeptidase | Fungus | 3 | 2.0 | |
| 14 | Serine peptidase S10, carboxypeptidase | Fungus | 4 | 1.3 | |
| 24 | Serine peptidase S10, carboxypeptidase OcpA | Fungus | 4 | 31.5 | |
| 95 | Serine peptidase S10, serine-type carboxypeptidase F | Fungus | 4 | 11.7 | |
| 199 | Serine peptidase S28, carboxypeptidase | Fungus | 4 | 6.3 | |
| 33 | Serine peptidase S53, grifolisin | Fungus | 4 | 20.5 | |
| 64 | Serine protease S53, grifolisin | Fungus | 4 | 192.4 | |
| Phosphate-liberating enzymes | 74 | Acid phosphatase, 3-phytase A | Fungus | 3 | 7.2 |
| 0 | Acid phosphatase, nucleotidase | Fungus | 4 | 16.9 | |
| 61 | Acid phosphatase, nucleotidase | Fungus | 4 | 24.8 | |
| 75 | Alkaline phosphatase | Fungus | 4 | 432.8 | |
| 100 | Alkaline phosphatase | Fungus | 4 | 14.8 | |
| 109 | Phytase esterase-like | Fungus | 4 | 105.1 | |
| 59 | Phytase esterase-like | Fungus | 3 | 19.2 | |
| 193 | Phytase, histidine acid phosphatase domain | Fungus | 4 | 6.5 | |
| 187 | Phytase, phosphoglycerate mutase, histidine acid phosphatase | Fungus | 4 | 25.2 | |
| 72 | PLC-like phosphodiesterase | Fungus | 3 | 3.5 | |
| 37 | Ribonuclease T1 | Fungus | 4 | 5.6 | |
| 34 | Ribonuclease T2 | Fungus | 4 | 11.5 | |
| 73 | GH3, beta-glucosidase | Fungus | 4 | 52.0 | |
| 99 | GH3, beta-xylosidase | Fungus | 4 | 76.0 | |
| 55 | GH5, endocellulase | Fungus | 3 | 3.7 | |
| 179 | GH5, exo-1,3-beta-glucanase | Fungus | 4 | 9.8 | |
| 183 | GH5, mannan endo-1,4-beta-mannosidase F | Fungus | 4 | 7.6 | |
| 13 | GH5, mannan endo-1,4-beta-mannosidase | Fungus | 3 | 11.7 | |
| 38 | GH10, endo-1,4-beta-xylanase | Fungus | 3 | 3.9 | |
| Carbohydrate active enzymes | 121 | GH12, xyloglucanase | Fungus | 3 | 4.8 |
| 132 | GH13, alpha-glucosidase, maltase | Ant | 3 | 2.9 | |
| 123 | GH15, glucoamylase | Fungus | 4 | 66.5 | |
| 88 | GH17, 1,3-beta-glucanase | Fungus | 4 | 290.8 | |
| 8 | GH18, chitinase | Fungus | 3 | 16.8 | |
| 66 | GH20, betahexosaminidase | Fungus | 3 | 1.3 | |
| 214 | GH25?, lysozyme | Fungus | 4 | 102.4 | |
| 25 | GH27, alpha-galactosidase | Fungus | 4 | 27.9 | |
| 83 | GH28, endo-polygalacturonase | Fungus | 4 | 83.3 | |
| 80 | GH29, alpha-L-fucosidase | Fungus | 3 | 1.8 | |
| 104 | GH31, alpha-glucosidase | Fungus | 4 | 39.1 | |
| 157 | GH31, alpha-glucosidase | Ant | 4 | 7.9 | |
| 108 | GH35, beta-galactosidase | Fungus | 4 | 13.6 | |
| 154 | GH37, trehalase | Ant | 4 | 19.8 | |
| 90 | GH43, arabinan-endo-1,5-alpha-L-arabinosidase | Fungus | 4 | 21.8 | |
| 11 | GH51, arabinofuranosidase | Fungus | 4 | 85.7 | |
| 46 | GH53, arabinogalactanase | Fungus | 4 | 36.2 | |
| 77 | GH55, exo-1,3-beta-glucanase | Fungus | 4 | 34.5 | |
| 7 | GH78, α-L-rhamnosidase | Fungus | 4 | 14.0 | |
| 63 | GH79, betaglucoronidase | Fungus | 4 | 35.0 | |
| 62 | GH88, glucuronyl hydrolase | Fungus | 4 | 3.0 | |
| 76 | GH92, exo-alpha-mannosidase? | Fungus | 4 | 16.6 | |
| 68 | GH92, exo-alpha-mannosidase? | Fungus | 3 | 3.8 | |
| 57 | CE8, pectinesterase | Fungus | 4 | 69.0 | |
| 40 | CE12, rhamnogalacturonan acetylesterase | Fungus | 4 | 5.8 | |
| 67 | CE12, Rhamnogalacturonan acetylesterase | Fungus | 4 | 27.6 | |
| 208 | PL1, pectate lyase | Fungus | 4 | 34.6 | |
| 122 | PL4, rhamnogalacturonan lyase | Fungus | 4 | 9.6 | |
| Lipidolytic enzymes | 124 | Lipase, triacylglycerol lipase | Fungus | 4 | 2.5 |
| 65 | Neutral/alkaline nonlysosomal ceramidase | Fungus | 4 | 6.2 | |
| 138 | Pancreatic lipase-related protein | Ant | 4 | 6.5 | |
| 44 | Phosphatidylglycerol/phosphatidylinositol transfer protein | Fungus | 3 | 2.8 | |
| 32 | Alpha/beta hydrolase, cephalosporin esterase | Fungus | 4 | 10.1 | |
| 18 | Alpha/beta hydrolase, triacylglycerol lipase, carotenoid ester lipase | Fungus | 4 | 5.5 | |
| 27 | Carboxylesterase; alpha-beta hydrolase; lipase | Fungus | 4 | 1.5 | |
| Miscellaneous proteins | 212 | Fruit-body specific protein D | Fungus | 4 | 44.0 |
| 31 | Plant expansin, papain inhibitor | Fungus | 3 | 1.7 | |
| 129 | Regucalcin | Ant | 4 | 5.8 | |
| 113 | SnodProt1, cerato platanin | Fungus | 4 | 114.7 | |
| 10 | Ubiquitin | Fungus | 4 | 1.7 | |
| 136 | Epididymal secretory protein E1 | Ant | 4 | 6.4 | |
| 158 | CEN-like protein 2, OV-16 antigen | Fungus | 4 | 4.8 | |
| 106 | Hypothetical protein | Fungus | 4 | 39.2 | |
| 195 | Hypothetical protein | Fungus | 3 | 4.5 | |
| 119 | Hypothetical protein, symbiosis related protein, MAPK? | Fungus | 4 | 52.8 |
Additional files
-
Supplementary file 1
Data for all fecal fluid proteins identified in the study.
- https://cdn.elifesciences.org/articles/61816/elife-61816-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61816/elife-61816-transrepform-v2.docx





