Principles of dengue virus evolvability derived from genotype-fitness maps in human and mosquito cells
Figures
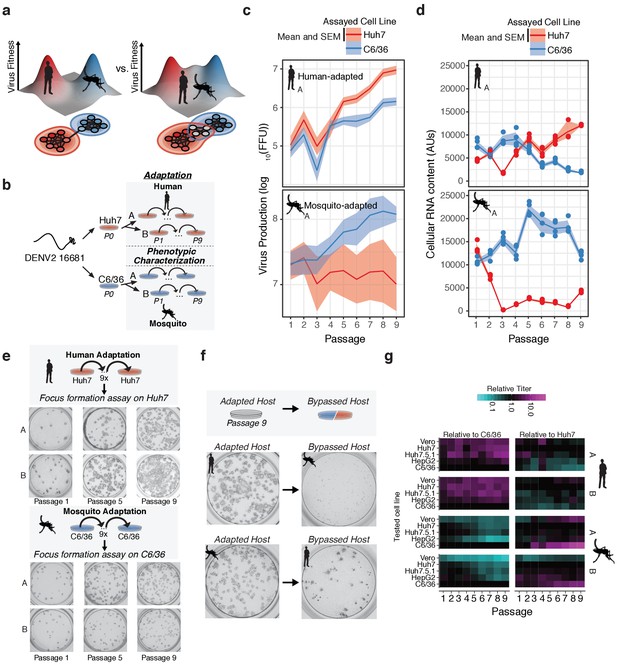
Dengue navigates distinct fitness landscapes in its alternative hosts.
(a) Two potential models of the genotype-fitness landscape and mutational network in alternative arboviral hosts. The relative topography of the viral genotype-fitness landscape determines the extent of evolutionary trade-offs associated with transmission, and the paths through the mutational network toward host adaptation, and the proportion of genotypes viable in the alternative host environment (gray nodes). (b) Outline of our in vitro DENV evolution experiment. Dengue virus RNA (Serotype 2/16881/Thailand/1985) was electroporated into mosquito (C6/36) or human cell lines (Huh7), and the resulting viral stocks were passaged at fixed population size (MOI = 0.1, 5 × 105 FFU/passage) for nine passages in biological duplicates. After passage, samples of virus from each passaged population were characterized for phenotypic measures of fitness. (c) Viral production assays comparing mosquito-adapted (top panel) and human-adapted (bottom panel) DENV populations. Adapted populations show increased virus production on their adapted hosts. Biological replicate A is shown for all experiments. (d) Analysis of viral RNA content by qRT-PCR. Cellular DENV RNA is significantly decreased when adapted lines are propagated on the by-passed, alternative host. Lines and shading represent the mean and standard deviation of four technical replicates, respectively. Biological replicate A is shown for all experiments. (e) Focus forming assays of the adapted lineages performed on each population over the course of passage. Passages 1, 5, and 9 are shown for all lineages. Focus size increased markedly throughout passage on the adapted host. (f) Focus forming assays of the P9 virus on the adapted (left) and by-passed (right) host. Changes in focus size and morphology suggest evolutionary trade-offs between the alternative hosts. (g) Heatmap showing the virus production of the passaged lineages in human, primate, and mosquito cell lines.
-
Figure 1—source data 1
Virus titer data from focus forming assays on multiple cell lines.
- https://cdn.elifesciences.org/articles/61921/elife-61921-fig1-data1-v2.txt
-
Figure 1—source data 2
Table of intracellular RNA content measurements.
- https://cdn.elifesciences.org/articles/61921/elife-61921-fig1-data2-v2.txt
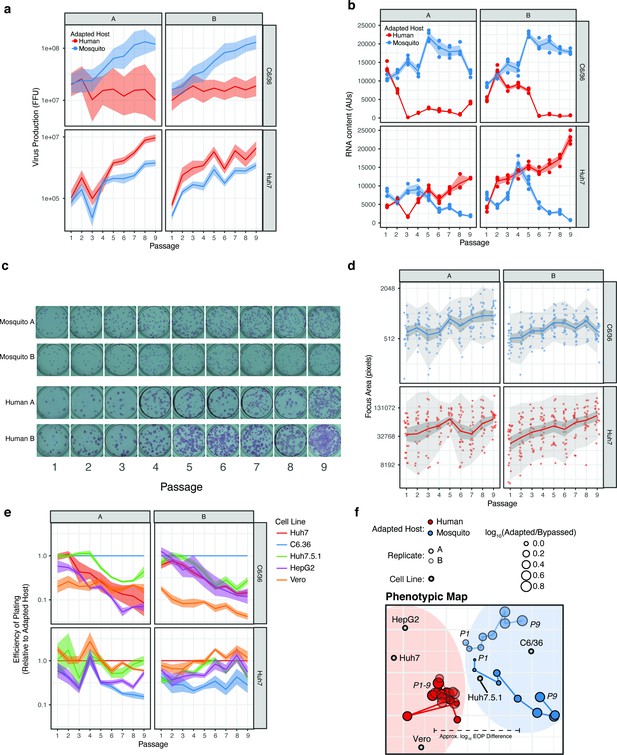
Phenotypic characterization of passaged viral populations.
(a) Quantification of virus production. Vital titers were quantified by focus forming assay for both replicates. (b) Intracellular RNA content determined by qRT-PCR. (c) Efficiency of Plating (EOP) data represented in the embedding in Figure 1g. EOP was determined based on comparison to the adapted cell line. (d) Tropic cartography of DENV in vitro host adaptation. The large number of comparisons in 2D space were visualized using an embedding technique that summarizes the relative EOP as an approximate distance in two dimensions. This approach is similar to the technique of antigenic cartography used to describe antigenic evolution from pairwise measurements of antibody neutralizing titers (Smith et al., 2004). The movement of the sequenced viral populations (Figure 1g, red and blue lines) relative to the placement of the cell lines (gray circles) reflects the change in relative titer between both cell lines. Human-adapted viruses exhibited similar titers in human- and primate-derived cell lines, resulting in their clustering separately from the titers in mosquito-derived C6/36 cells. The movement of C6/36-adapted populations (blue line) toward the C6/36 cell line reflects the significant mosquito-specific adaptation in these populations. A two-dimensional embedding of the relative titer (mean of four replicates), or efficiency of plating (EOP), of the adapted populations (red and blue trajectories) relative to five assayed cell lines (gray points). Movement corresponds to a change in EOP over passage on a log scale. The size of the red and blue points indicates the ratio of each population’s titer on the adapted and bypassed cell lines to illustrate the change in EOP over passage on the adapted lines.
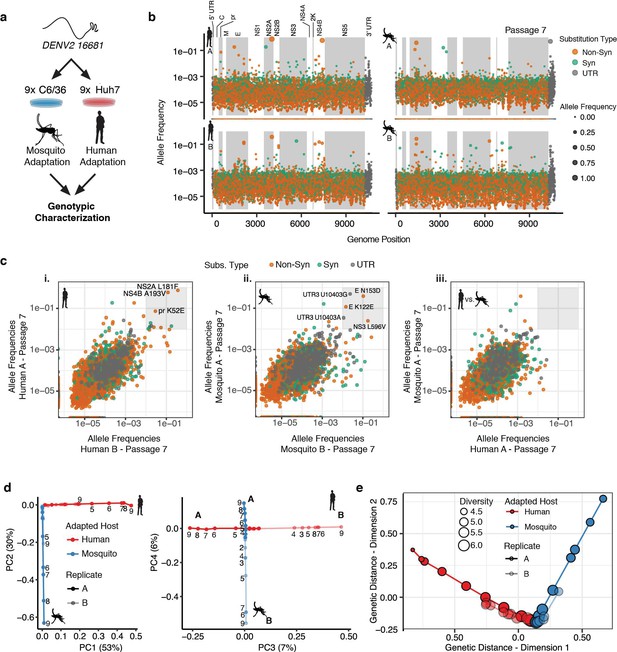
Adapting viral lineages show host-specific patterns of genetic variance.
(a) Adapted viral populations were subject to genotypic characterization by ultra-deep sequencing using the CirSeq procedure. (b) Plots of allele frequency across the viral genomes for all four viral populations at passage 7. Alleles are colored by mutation type (Nonsynonymous, Orange; Synonymous, Green; Mutations in the untranslated region (UTR), Dark Gray). Shaded regions denote mature peptide boundaries in viral ORF. (c) Scatter plots comparing allele frequencies between adapted populations of human- and mosquito-adapted dengue virus. Replicate host-adapted populations share multiple high-frequency non-synonymous mutations, but populations from alternative hosts do not (gray square, >10%). (d) Dimension reduction of the allele frequencies by principal components analysis summarizes the host-specific patterns of variance (left), and the replicate-specific differences in genetic variability over passage (right). (e) A two-dimensional embedding of the pairwise genetic distances between the sequenced viral populations (Weir-Reynolds Distance) by multidimensional scaling. The viral populations (red- and blue-hued trajectories) project out from the founding genotype in orthogonal and host-specific directions.
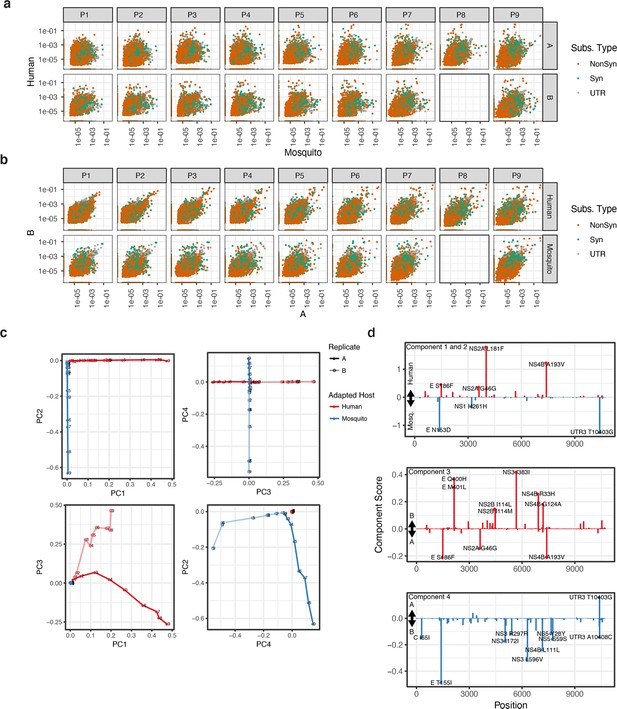
Genotypic characterization of passaged DENV2 populations.
(a) Scatter plots showing pairwise comparisons for all allele frequencies in each passage for Human and Mosquito-adapted populations in replica lineages A and B. (b) Scatter plots showing pairwise comparisons for all allele frequencies in replica lineages A and B for each passage for Human and Mosquito-adapted populations. (c) PC loadings of genetic diversity in sequenced populations. Graphs compare the contribution of principal components: The first and second components are host specific, while the third and fourth capture replicate specific differences between the populations. (d) PC scores of individual allele variants. Each score represents the contribution of the allele to the specific pattern of variance captured in the component. Major alleles are highlighted and labeled.
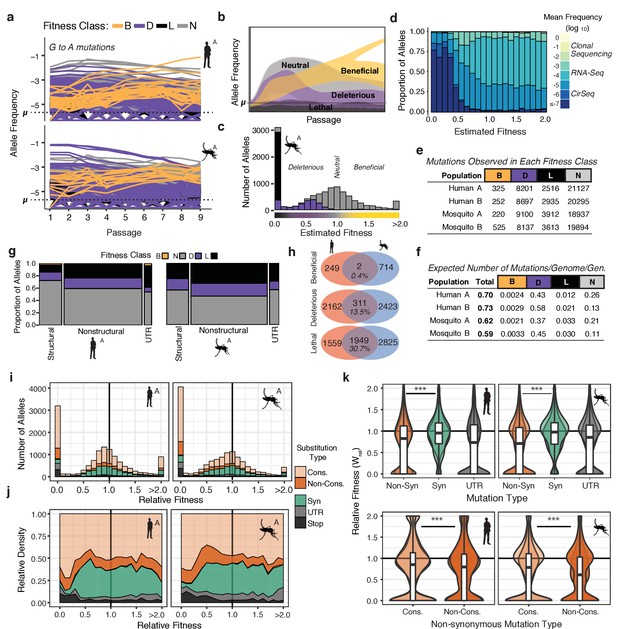
The distribution of fitness effects reveals patterns of evolutionary constraint.
(a) The frequency trajectories for G-to-A mutations in the adapting populations determined by CirSeq. Colors represent the classification of each allele as beneficial, deleterious, lethal, or neutral (not statistically distinguishable from neutral behavior) (b) Schematic illustrating the expected frequency behavior of specific fitness classes relative to their corresponding mutation rate, µ. Changes in allele frequency between passages are used to estimate the fitness effects of individual alleles in the population (see Materials and methods). (c) Histogram showing the distribution of mutational fitness effects (DMFE) of DENV passaged in mosquito cells. The data shown are from mosquito A and represent the high confidence set of alleles (see text). The fitness classifications of alleles in each bin, based on their 95% confidence intervals, is indicated by the fill color. (d) The relative density of each mutation type across the fitness spectrum illustrates the sequencing depth necessary to observe regions of the fitness spectrum. Fill color represents the average frequency of the mutation over passage. (e) Tabulation of all alleles by fitness class. (f) Estimate of the genomic mutation rate per genome per generation ('Total'), and fitness class-specific mutation rates ('B', 'D', 'N', and 'L', Supplementary file 2). (g) Area plot showing the fitness effects associated with mutations in structural, non-structural, and UTR regions of the DENV genome. The relative width of the columns indicates the number of alleles in each class, the relative height of the colored regions indicates the proportion of alleles of a given class. (h) Venn diagrams showing the number of mutations identified as beneficial, deleterious, or lethal in the high confidence set alleles (see Materials and methods). These counts include alleles identified in either A or B replica populations. (i) Histograms of the DMFE broken down by mutation type. (j) Density plot of the relative density of mutation types across the DMFE to emphasize the local enrichment of specific classes. (k) Violin plots showing the relative fitness of nonsynonymous and synonymous mutations, and those in the viral UTRs. Overlapping plots are shown for replicates A and B for each host. Boxplots are computed based on the DMFE of both replica lineages in each host. Nonsynonymous mutations can further be partitioned into conservative and non-conservative classes.
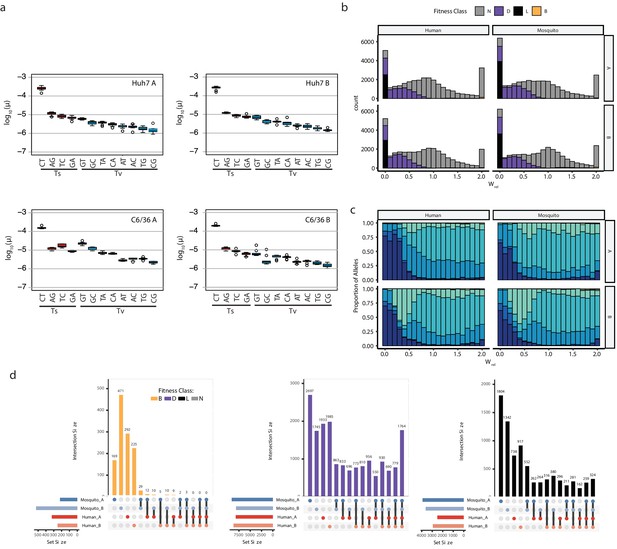
Details of mutation rates, distributions of mutational fitness effects, and fitness class assignments.
(a) Box plots of the nine individual mutation rate estimates obtained for each passaged population, indicating transitions (‘Ts’) and Transversions (‘Tv’). (b) Distribution of mutational fitness effects for all possible alleles in the population. The bars in the histogram are shaded to show the proportion of alleles called as Beneficial, Neutral, Deleterious or Lethal, according to their 95% CI. (c) Filled Histogram showing the sequencing depth required to observe alleles of a given fitness class for all populations. (d) UpSet Plots (Lex et al., 2014) comparing shared alleles of individual fitness classes between experimental sets. These comparisons reveal the stochastic nature of beneficial mutations, which are largely unique to the individual populations. Deleterious and lethal mutations act more deterministically and have more universal effects on fitness across the different host environments.
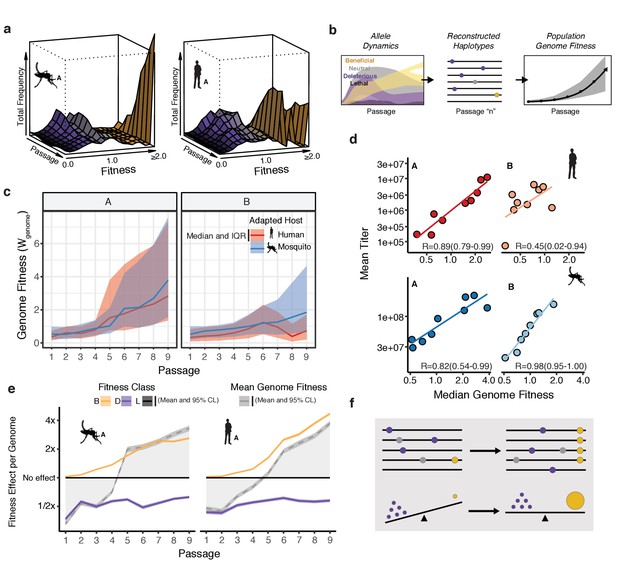
Connecting global evolutionary dynamics and population fitness.
(a) Surface plot of the 'fitness wave' illustrating the change in frequency of alleles, colored by fitness bin, throughout a passage experiment. The height of the surface represents the sum of frequencies of alleles in a given bin. Deleterious and neutral mutations (purple and gray regions) make up a large proportion of the population early in the experiment. They are largely, but not entirely, driven out in later passages as beneficial mutations (yellow) increase in frequency. (b) The shift in allele fitness effects circulating in the population suggests a net increase in the fitness of genotypes within the adapting populations. To estimate the population-level change in fitness during adaptation, we used the allele frequencies in each passage (left panel) to reconstruct haplotypes for that passage (e.g. passage 'n', middle panel). The potential genotypes from each sequenced population were inferred based on empirical allele frequencies to determine the probability of a sequence identity at each site for each estimated genotype. We then computed the fitness of the resulting genotype as the product of the fitness effects across the genome. Sampling a large population of representative genotypes from the populations, we were able to generate a distribution of genome fitness values for each sequenced population (right panel). (c) Median and interquartile range of genotypic fitness () of 50,000 reconstructed genotypes sampled from the empirical allele frequencies in each sequenced population. (d) Correlation plots comparing the median genotypic fitness () of the reconstructed populations versus mean virus titer from focus forming assays (N=4). (e) Line plots showing the mean effect of beneficial (yellow), deleterious (purple), and lethal (black) mutations on mean fitness of the population (gray line). The shaded area represents the 95% confidence interval of the mean from 50,000 reconstructed genomes.
Changes in allele composition.
(a) Fitness Wave representations of the allele fitness dynamics of all of the experimental populations. (b) Line plot showing the change in expected fitness of a randomly drawn allele in the population over time. (c) Line plots showing the mean number of mutations per genome of each fitness class in the sampled genomes used to estimate population fitness.
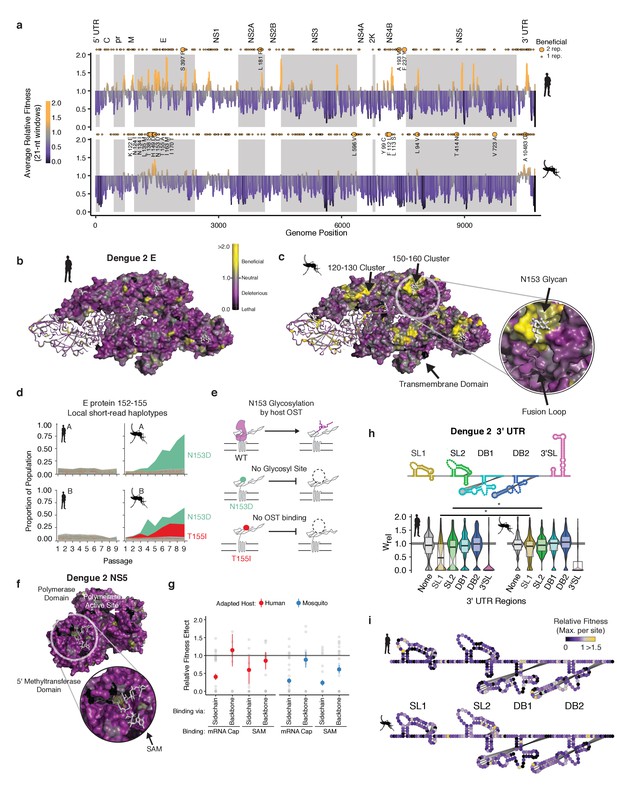
Structural analysis reveals hotspots of viral adaptation.
(a) Bar plot of the mean fitness effect of alleles in 21 nt windows across the DENV genome. Fitness estimates from both replicates are used to compute means. Synonymous alleles are removed to emphasize the fitness effects of coding changes. Yellow points above each line denote the locations of beneficial mutations (95% CI >1). Larger, labeled yellow points denote beneficial mutations identified in both replicates of host cell passage. (b) The empirical fitness estimates displayed on a trimer of envelope and M proteins (PDB: 3J27) in an antiparallel arrangement, similar to that found on the mature virion (Zhang et al., 2013). To emphasize rare sites of positive selection, the color of each residue represents the maximum of the lower confidence intervals of fitness effect estimates at that site. Human-adapted populations show significant negative selection on the envelope protein surface, with no residues showing significant positive selection (yellow color). (c) Mosquito-adapted DENV exhibit two patches of pronounced positive selection on the exterior face of the virion (labeled 120–130 and 150–160). These clusters are absent from the human-adapted populations. Cluster 150–160 (Zoom), consists of a loop containing a glycosylation site at N153. This loop and glycan enclose the viral fusion loop of the anti-parallel monomer prior to activation and rearrangement in the endosome after entry. (d) Plots of the frequency of local read haplotypes for the area overlapping N153 and T155. Mutations at N153 (N153D) and T155 (T155I) are positively selected in mosquitoes, but never occur together on individual reads. (e) Schematic describing the phenotypically equivalent effects of the N153D and T155I mutations. These mutations block recognition and modification by the host oligosaccharyltransferase (OST), which initiates glycosylation. (f) CirSeq also reveals patterns of negative selection. Patches of significant evolutionary constraint can be seen around the methyltransferase active site highlighted by numerous positions with lethal fitness effects (Zoom). (g) Comparison of fitness effects of non-synonymous mutations targeting residues in NS5-MT that interact with the 5 (h) Insights into host-specific RNA structural constraints. Violin plot comparing the fitness effects of mutations in the stem-loop (SL) and dumb-bell (DB) structures of the 3′ UTR RNA of DENV2 shown in the schematic. Fitness effects of mutations in the conserved structures reveal differences in fitness effects associated with SLI and SLII in human- and mosquito-adapted dengue virus populations. (i) Nucleotide-resolution map of fitness effects on the viral 3′ UTR reveals regions of SL1 and 2 that are under tighter constraints in human passage.
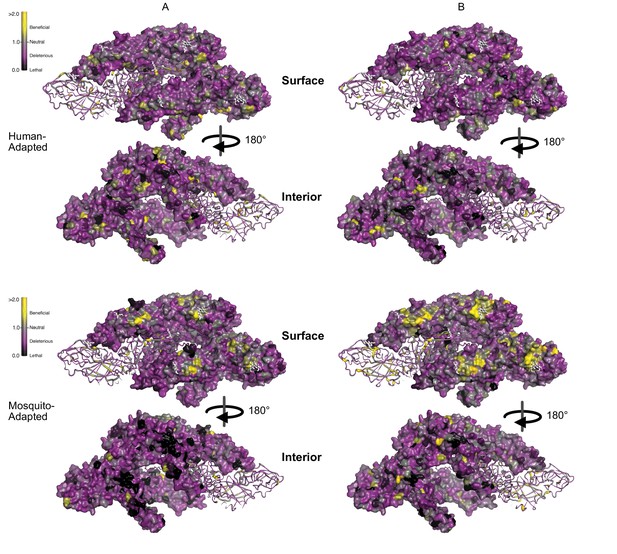
Panels showing the surface and interior views of the DENV E and M proteins.
All replicates are shown. To emphasize rare sites of significant positive selection, the color of each residue represents the maximum of the lower confidence intervals of fitness effect estimates at that site.
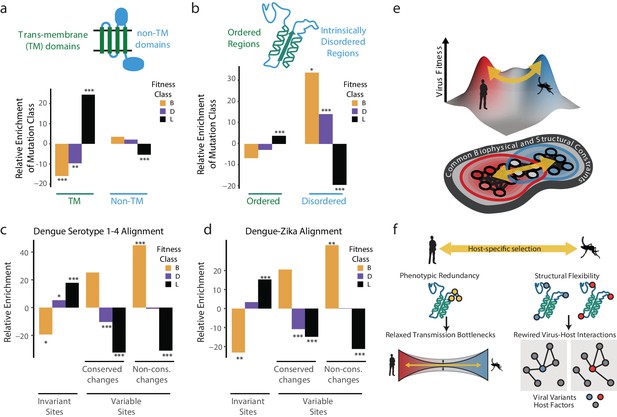
Biophysical and biological themes in DENV adaptation.
(a, b) Distribution of mutations in regions with different biophysical characteristics. (a) The relative enrichment of each type of mutations (beneficial, deleterious, and lethal) in transmembrane (TM) regions versus non-TM regions (Figure 2a) and disordered versus ordered regions (Figure 2b). Relative enrichment is computed as the normalized difference in occurrence of this type of mutation in the specific region tested, and its occurrence across the entire polyprotein. Significance values (FDR-corrected, Fisher exact values) are shown (* = p<0.05, ** = p<0.01, *** = p<0.001). (c) Cross-dengue conservation. Distribution of mutations in regions with different levels of conservation across dengue virus strains. The relative enrichment of each type of mutations (beneficial, deleterious, and lethal) in residues that are identical (‘Invariant’), similar (‘Conserved’) or dissimilar (‘Variable’) across the four dengue strains. Relative enrichment was computed as the normalized difference in occurrence of this type of mutation in the specific region tested, and its occurrence across the entire polyprotein. Significance values (FDR-corrected, Fisher exact values) are shown (* = p<0.05, ** = p<0.01, *** = p<0.001). (d) Zika-Dengue conservation. Distribution of mutations in regions with different levels of conservation between DENV and ZIKV virus. The relative enrichment of each type of mutations (beneficial, deleterious, and lethal) in residues that are identical, similar, or dissimilar between the two viruses. Relative enrichment was calculated as the normalized difference in occurrence fraction of this type of mutation in this specific region and its occurrence across the entire polyprotein. Significance values (FDR-corrected, Fisher exact values) are shown (* = p<0.05, ** = p<0.01, *** = p<0.001). (e) Visualization of a simplified landscape of dengue host adaptation. The landscape is shaped by common biophysical and functional constraints that operate similarly in both hosts, defining the outline of the fitness landscape. Positive selection of host-specific phenotypes drives host adaptation. (f) Host adaptation is associated with trade-offs that form a bottleneck to transmission. This bottleneck is relaxed by phenotypic redundancy and structural flexibility at key hotspots of adaptation.
-
Figure 6—source data 1
Pooled count data used for computing Fisher’s exact test enrichment.
- https://cdn.elifesciences.org/articles/61921/elife-61921-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Fitness values and classifications used in analysis in Figure 6.
- https://cdn.elifesciences.org/articles/61921/elife-61921-fig6-data2-v2.csv
Videos
Animation of the allele frequencies in the adapting populations over nine passages.
Colors: Orange, non-synonymous mutations; Green, synonymous mutation; and Gray, mutations in the UTR.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Dengue Virus) | Dengue Virus, Serotype 2, Thailand, 16681 | PMID:9143286 | Plasmid pD2/IC-30P | |
| Cell Line (Homo sapiens) | HepG2 | PMID:233137 | RRID:CVCL_0027 | |
| Cell Line (Aedes albopictus) | C6/36 | PMID:690610 | RRID:CVCL_Z230 | |
| Cell line (Homo sapiens) | Huh7 | PMID:30894373 | RRID:CVCL_7927 | |
| Cell Line (Homo sapiens) | Huh7.5.1 | PMID:15939869 | RRID:CVCL_E049 | |
| Cell Line (Chlorocebus sabaeus) | Vero cells | ISSN: 0047–1852 | RRID:CVCL_Z230 | |
| Antibody (Mouse anti-DENV Envelope) | Anti-E antibody | Genetex | GTX127277 |
Additional files
-
Supplementary file 1
Table of fitness estimates, confidence intervals, annotations, mutation classifications, and computational statistics for all data presented here.
- https://cdn.elifesciences.org/articles/61921/elife-61921-supp1-v2.csv.zip
-
Supplementary file 2
Class-specific mutation rate and error estimates for each passaged population.
- https://cdn.elifesciences.org/articles/61921/elife-61921-supp2-v2.txt
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61921/elife-61921-transrepform-v2.docx





