Symbiont-mediated cytoplasmic incompatibility: What have we learned in 50 years?
Figures
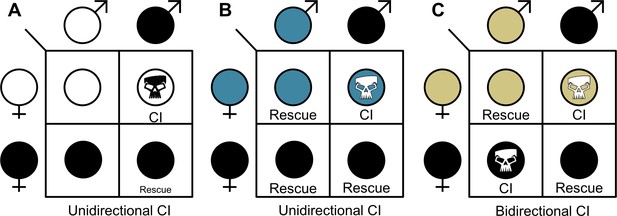
The three CI crossing relationships.
(A) Unidirectional CI results in embryonic lethality when symbiont-containing males are crossed with aposymbiotic females. Rescue of this embryonic lethality occurs if the female carries a compatible symbiont strain. (B) In some cases, unidirectional CI can emerge when one strain can rescue another strain, but the other strain does not reciprocate the rescue. (C) Bidirectional CI occurs when incompatible strains are present in a population. Rescue occurs if the female likewise harbors the same strain. Filled sex symbols indicate symbiotic hosts. Different colors represent different symbiont strains. Skull symbols represent embryonic death.
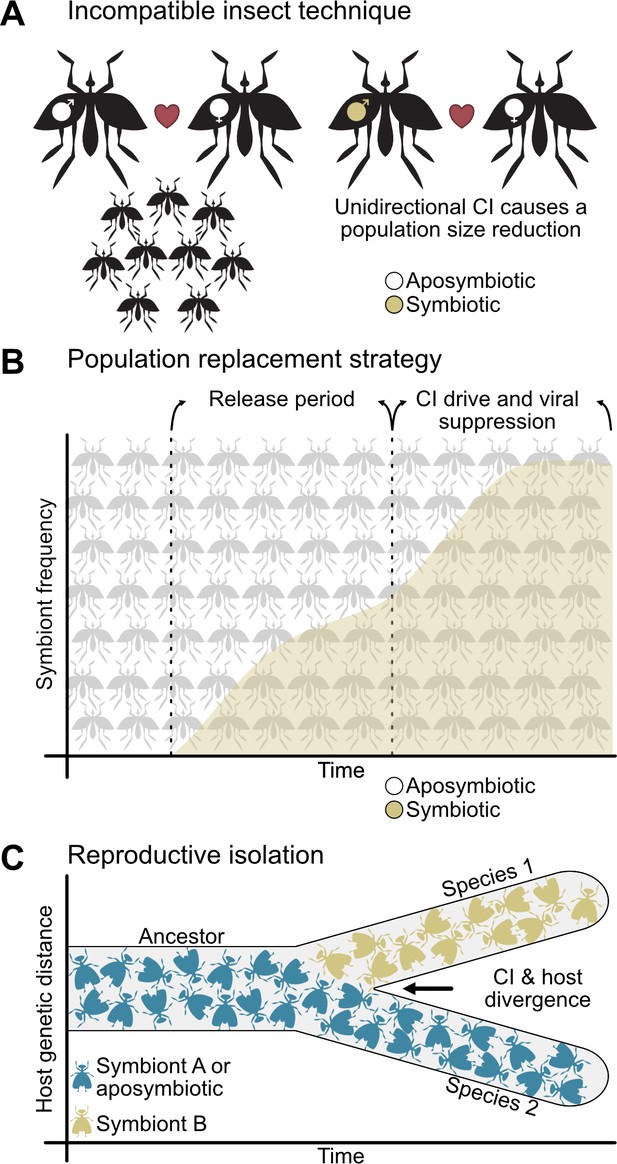
CI is important to vector control and reproductive isolation between species.
(A) The incompatible insect technique is used to reduce population sizes (Crawford et al., 2020; Laven, 1967). Typically, two aposymbiotic individuals will mate and produce viable offspring (left), but if males bearing CI-inducing symbionts are released into the population, then they will cause unidirectional CI when they mate with aposymbiotic females (right) or bidirectional CI when they mate with females harboring incompatible symbionts (not shown). This yields a reduction in egg hatching and population size. (B) The population replacement strategy involves the release of both males and females bearing CI-inducing and pathogen blocking symbionts (Hoffmann et al., 2011; O'Neill, 2018). After a period of releases, CI will spread the symbiont to high frequencies where it can block the replication of human diseases. (C) CI-inducing symbionts can cause reproductive isolation through unidirectional or bidirectional CI when different individuals, populations, or species have different incompatible symbiont states (Bordenstein et al., 2001; Breeuwer and Werren, 1990; Gebiola et al., 2017; Jaenike et al., 2006). This reproductive barrier reduces gene flow between hosts with different symbiont states, allowing for their divergence.
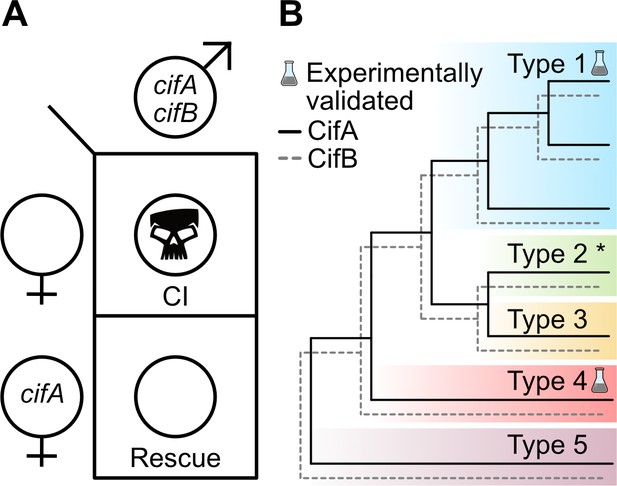
Two-by-One genetic model of cif-induced CI and Cif phylogeny.
(A) The Two-by-One genetic model of CI surmises that both cifA and cifB must be expressed in males to cause CI, and cifA must be expressed in females to rescue CI (Shropshire and Bordenstein, 2019). (B) CifA and CifB codiverge and are classified into at least five different phylogenetic Types (1-5) (Bing et al., 2020b; LePage et al., 2017; Lindsey et al., 2018; Martinez et al., 2020). To date, only Type 1 cifs from wMel and wPip, and Type 4 cifs from wPip, have been experimentally confirmed to cause and rescue CI (Beckmann et al., 2017; Chen et al., 2019; LePage et al., 2017; Shropshire et al., 2018; Shropshire and Bordenstein, 2019). Moreover, unpublished results from JDS and SRB suggest that Type 2 cifs from wRi are CI and rescue-capable (denoted with an asterisk).
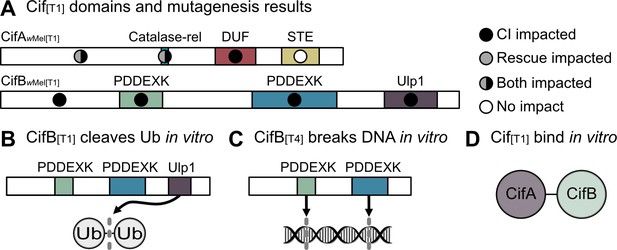
Biochemical characterization of Cif proteins.
(A) Annotated domains in the CifA and CifB proteins and the relative importance of conserved residues in each domain for CI (black circles), rescue (gray circles) or neither phenotype (white circles) as determined by transgenic expression of mutated proteins in aposymbiotic D. melanogaster (Beckmann et al., 2017; Shropshire et al., 2020). (B) CifB[T1] can cleave ubiquitin chains via its Ulp1 deubiquitinase domain in vitro (Beckmann et al., 2017). (C) CifB[T4] nuclease domains can cause DNA breaks in vitro (Chen et al., 2019). (D) CifA[T1] and CifB[T1] bind each other in vitro (Beckmann et al., 2017). Domain architecture is based on homology-based analyses and is of low predictive value (20–30% probability) for CifA (Lindsey et al., 2018), and CifB[T1] PDDEXK nuclease domains lack the canonical PD-(D/E)XK motif (Beckmann et al., 2017), but remain structurally homologous to other PDDEXK nucleases (Lindsey et al., 2018).
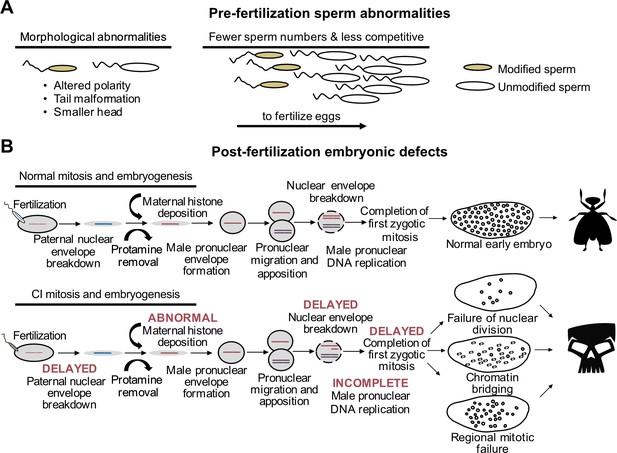
CI-associated defects occur pre- and post-fertilization.
(A) In males harboring Wolbachia, there are several types of sperm abnormalities when compared to their aposymbiotic counterparts. (B) When fertilized with sperm derived from Wolbachia-carrying males, embryonic nuclear defects result in the form of delayed paternal nuclear envelope breakdown, abnormal histone deposition and other early mitotic events. These defects then cause embryonic phenotypes observed in CI including chromatin bridging and regional mitotic failures.
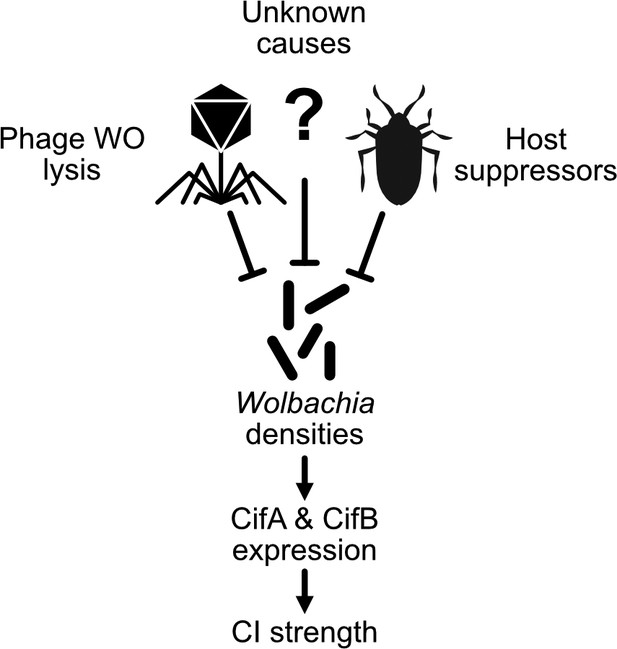
An expanding Wolbachia density model of CI strength variation.
The proximal cause of CI is likely CifA and CifB, whose transcriptional level has been connected with intensity in transgenic studies (LePage et al., 2017). Wolbachia densities have often correlated with factors that influence CI strength variation (Werren, 1997). In many cases, it remains unknown how these factors influence Wolbachia densities. Phage WO lysis (Bordenstein and Bordenstein, 2011) and host suppressors are well documented correlates or causes of density changes (Funkhouser-Jones et al., 2018; Poinsot et al., 1998; Walker et al., 2011).
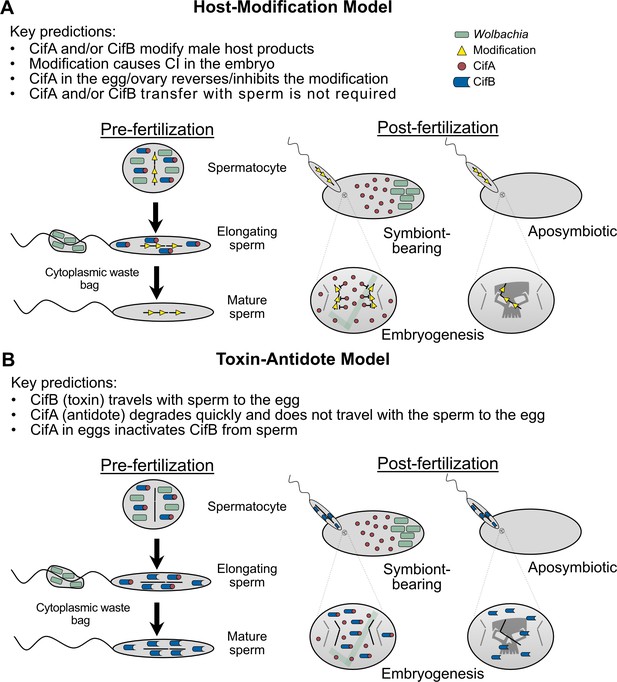
The Host-Modification and Toxin-Antidote models of CI mechanism.
(A) The Host-Modification (HM) model predicts that the Cif proteins impart a modification on male-derived products that result in CI unless CifA is available in the embryo to reverse or otherwise inhibit the male-derived modification (Shropshire et al., 2019; Werren, 1997). (B) The Toxin-Antidote (TA) model predicts that CifB is the primary toxin that is transferred to the embryo via the sperm, and that rescue occurs when CifA binds CifB in the embryo and inhibits its toxicity (Beckmann et al., 2019a; Hurst, 1991; Shropshire et al., 2019; Werren, 1997).
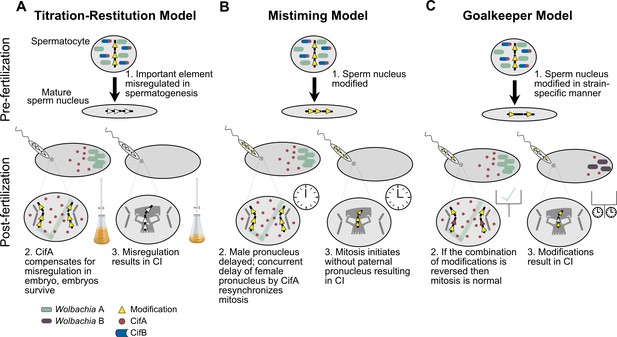
Extensions of the Host-Modification model.
(A) The Titration-Restitution Model posits that an element within mature sperm is either over- or under-expressed in males due to Cif protein expression, but this alteration is then remedied in the female as a result of CifA through a reconstitution of the required element (Werren, 1997). (B) The Mistiming Model posits that a modification in the male sperm causes a delay in the formation of the male pronucleus that results in CI if CifA does not cause a concurrent delay in the maternal pronucleus, resynchronizing mitosis between the two pronuclei (Tram and Sullivan, 2002). (C) The Goalkeeper Model expands on the Mistiming Model and posits that the male product modification occurs in a strain-specific quantity, and may involve multiple modifications that need to be remedied to rescue the lethality (Bossan et al., 2011).





