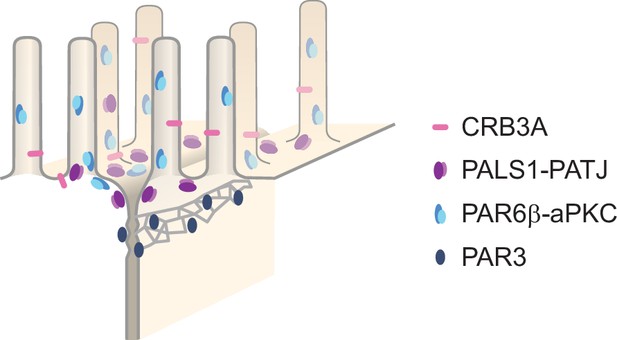
Super-resolution imaging uncovers the nanoscopic segregation of polarity proteins in epithelia
Figures
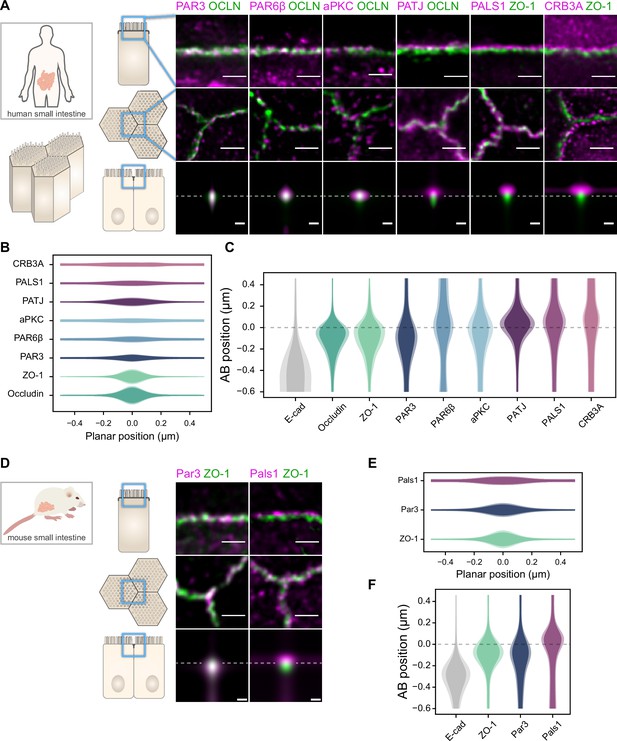
Polarity proteins localize in separate subdomains in the tight junction (TJ) region in human (A–C) and murine (D–F) small intestine biopsies.
(A, D) STED images of protein localization in the TJ area. TJ proteins in green, polarity proteins in magenta. Top row: apico-basal orientation; middle row: planar orientation; bottom row: estimates of average protein localization in the apico-basal orientation perpendicular to the junction, obtained by multiplying average localizations estimated in (B) and (C) for human biopsies and (E) and (F) for murine biopsies. Top row and middle row, scale bar 1 µm; bottom row, scale bar 200 nm. (B, E) Average localization of polarity proteins in the planar orientation, obtained by measuring the intensity profile of proteins perpendicular to the junction, using the TJ protein position as a reference. (C, F) Average localization of polarity proteins in the apico-basal orientation, obtained by measuring the intensity profile of proteins along the apico-basal orientation, using the TJ protein position as a reference. In (B, C, E, F), on a given position dark colors represent average intensity values, and lighter colors the average added with the standard deviation. We used three biological replicates for each human and mouse experiment (details in Figure 1—source data 1). Details of the analysis are specified in the ‘Materials and methods’ section.
-
Figure 1—source data 1
Details of the number of junctions used in each replicate.
- https://cdn.elifesciences.org/articles/62087/elife-62087-fig1-data1-v2.docx
-
Figure 1—source data 2
Estimate of the average density versus localization for each protein.
- https://cdn.elifesciences.org/articles/62087/elife-62087-fig1-data2-v2.zip
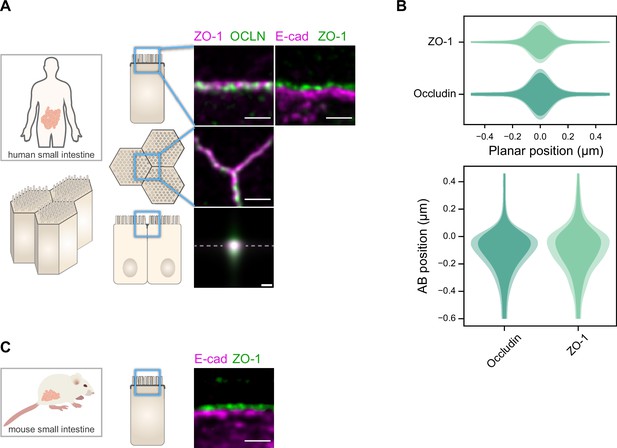
Localization of ZO-1 vs. Occludin in the human small intestine and E-cadherin vs. ZO-1 in human and mouse small intestine.
(A) Stimulated‐emission‐depletion (STED) images of the tight junction (TJ) proteins ZO-1 and Occludin, and E-cadherin in the TJ area of human small intestine samples. Top row: apico-basal orientation; middle row: planar orientation; bottom row: estimates of average protein localization in the apico-basal orientation perpendicular to the junction, obtained by multiplying average localizations estimated in (B) for human biopsies. Top row and middle row, scale bar 1 µm; bottom row, scale bar 200 nm. (B) Average localization of ZO-1 and Occludin in the planar orientation (top) and apico-basal orientation (bottom), obtained by measuring the intensity profile of proteins perpendicular to the junction, using the ZO-1 position as a reference. On a given position, dark colors represent average intensity values, and lighter colors are the average added with the standard deviation. Details of the analysis are specified in the ‘Materials and methods’ section. (C) STED images of ZO-1 and E-cadherin in the TJ area in the apico-basal orientation of murine small intestine samples. Scale bar: 1 µm.
-
Figure 1—figure supplement 1—source data 1
Details of the number of junctions used in each replicate.
- https://cdn.elifesciences.org/articles/62087/elife-62087-fig1-figsupp1-data1-v2.docx
-
Figure 1—figure supplement 1—source data 2
Estimate of the average density versus localization for each protein.
- https://cdn.elifesciences.org/articles/62087/elife-62087-fig1-figsupp1-data2-v2.zip
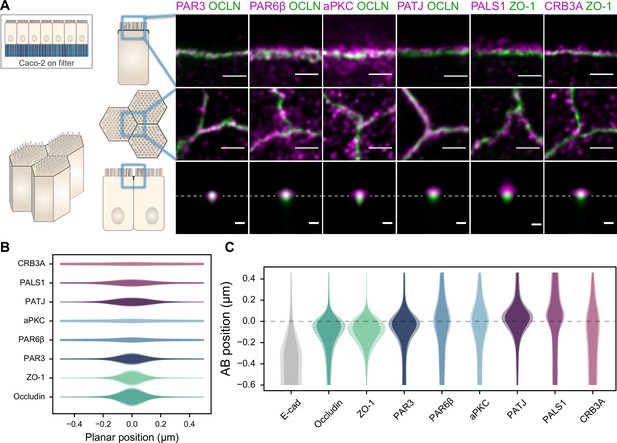
Polarity proteins localize in separate subdomains in the tight junction (TJ) region in Caco-2 cells.
(A) Stimulated‐emission‐depletion (STED) images of protein localization in the TJ area. TJ proteins in green, polarity proteins in magenta. Top row: apico-basal orientation (obtained from cryo-sectioning cells grown on filter); middle row: planar orientation; bottom row: estimates of average protein localization in the apico-basal orientation perpendicular to the junction, obtained by multiplying average localizations estimated in (B) and (C). Top row and middle row, scale bar 1 µm; bottom row, scale bar 200 nm. (B) Average localization of polarity proteins in the planar orientation obtained by measuring the intensity profile of proteins perpendicular to the junction, using the TJ protein position as a reference. (C) Average localization of polarity proteins in the apico-basal orientation obtained by measuring the intensity profile of proteins along the apico-basal orientation, using the TJ protein position as a reference. In (B, C), on a given position dark colors represent average intensity values, and lighter colors the average added with the standard deviation. We used three cell culture replicates (details in Figure 1—source data 1). Details of the analysis are specified in the ‘Materials and methods’ section.
-
Figure 2—source data 1
Details of the number of junction used in each replicate.
- https://cdn.elifesciences.org/articles/62087/elife-62087-fig2-data1-v2.docx
-
Figure 2—source data 2
Estimate of the average density versus localization for each protein.
- https://cdn.elifesciences.org/articles/62087/elife-62087-fig2-data2-v2.zip
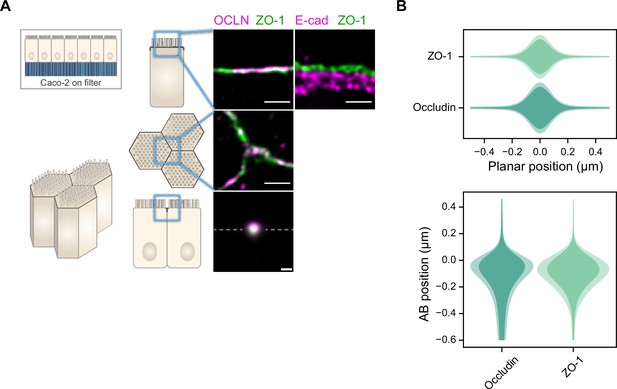
Localization of ZO-1, Occludin, and E-cadherin ZO-1 Caco-2 cells.
(A) Stimulated‐emission‐depletion (STED) images of tight junction (TJ) proteins ZO-1 and Occludin, and E-cadherin in the TJ area of Caco-2 cells. Top row: apico-basal orientation; middle row: planar orientation; bottom row: estimates of average protein localization in the apico-basal orientation perpendicular to the junction, obtained by multiplying average localizations estimated in (B). Top row and middle row, scale bar 1 µm; bottom row, scale bar 200 nm. (B) Average localization of ZO-1 and Occludin in the planar orientation (top) and apico-basal orientation (bottom), obtained by measuring the intensity profile of proteins perpendicular to the junction, using the ZO-1 position as a reference. On a given position, dark colors represent average intensity values, and lighter colors are the average added with the standard deviation. Details of the analysis are specified in the ‘Materials and methods’ section.
-
Figure 2—figure supplement 1—source data 1
Details of the number of junction for each replicate.
- https://cdn.elifesciences.org/articles/62087/elife-62087-fig2-figsupp1-data1-v2.docx
-
Figure 2—figure supplement 1—source data 2
Estimate of the average density versus localization for each protein.
- https://cdn.elifesciences.org/articles/62087/elife-62087-fig2-figsupp1-data2-v2.zip
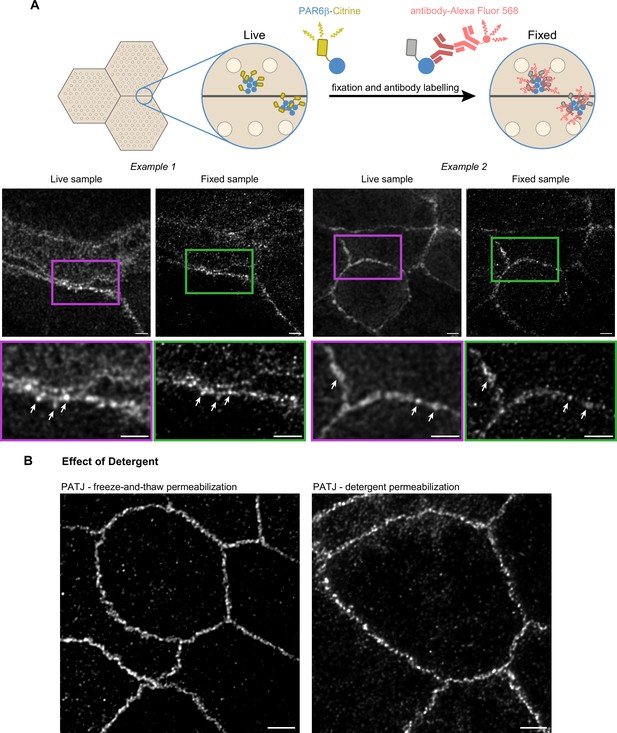
Confirmation of the cluster organization by alternative methods.
(A) Two examples of stimulated‐emission‐depletion (STED) images obtained on living Caco-2 cells expressing PAR6β-Citrine that were then fixed and immunolabeled and zoom on junctions (insets). Imaging of the same cells shows that clusters are observed in live and fixed conditions (arrows pointing at the same clusters in both conditions). (B) Images showing that permeabilization using freeze-and-thaw or detergent lead to very similar results, showing that detergents are not the cause of protein clustering. Scale bars: 2 µm. We obtained the same conclusions on three independent cell culture replicates.
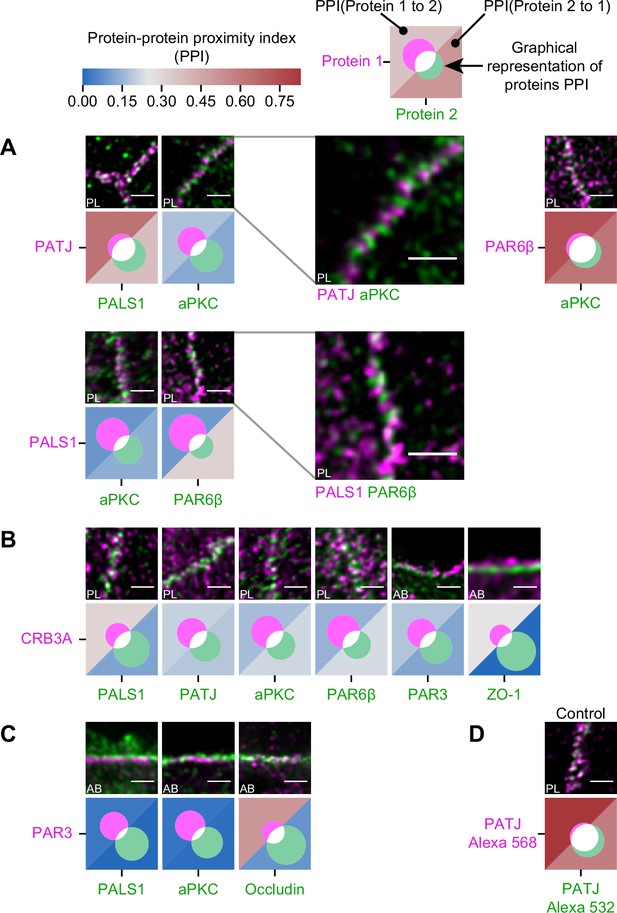
Proximity analysis of polarity proteins redefines protein complexes.
The analysis is carried out in Caco-2 cells, where we used the concept of protein–protein proximity index (PPI) introduced by Wu et al., 2010, indicating the proximity of two different protein populations. PPI of 0 indicates no proximity (or no colocalization), and PPI of 1 indicates perfect proximity (or perfect colocalization); intermediate values give an estimate of the fraction of a given protein being in close proximity (or colocalize) with another one. Here, the result of the proximity analysis is represented graphically with color-coded values and Venn diagrams as depicted on the top of the figure (details in ‘Materials and methods’). The analysis has been carried out on apico-basal (AB) or planar (PL) orientation images to minimize apparent colocalization due to overlapping in different planes; this is reported in the representative image of each experiment. (A) Proximity analysis for PATJ, PALS1, aPKC, and PAR6β and corresponding representative images. Zoomed images (PATJ/aPKC and PALS1/PAR6β) illustrate the segregation of these proteins. (B) Proximity analysis for CRB3A and the other polarity proteins. (C) Proximity analysis for PAR3 with PALS1, aPKC, and Occludin. (D) Control experiment with PATJ labeled with an Alexa 532 secondary antibody and an Alexa 568 tertiary antibody. We used three cell culture replicates for each protein pair (details in Figure 4—source data 1). The details of the analysis are specified in ‘Materials and methods.’ Scale bars: 1 µm.
-
Figure 4—source data 1
Details of the number of junctions for each repicate.
- https://cdn.elifesciences.org/articles/62087/elife-62087-fig4-data1-v2.docx
-
Figure 4—source data 2
PPI index for each protein couple.
- https://cdn.elifesciences.org/articles/62087/elife-62087-fig4-data2-v2.txt
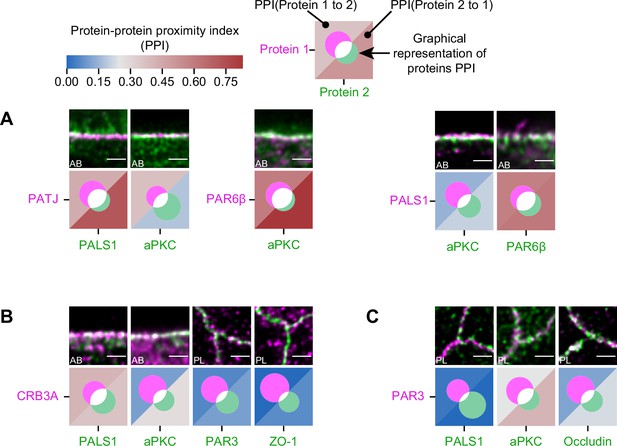
Protein–protein proximity index (PPI) analysis on the orientations can lead to artificially higher PPI because of higher protein overlap.
(A) Proximity analysis in the apico-basal orientation for PATJ, PALS1, aPKC, and PAR6β and corresponding representative images. (B) Proximity analysis in the apico-basal orientation for CRB3A and some of the other polarity proteins. (C) Proximity analysis for PAR3 with PALS1, aPKC, and Occludin in the planar orientation.
-
Figure 4—figure supplement 1—source data 1
Details of the number of junctions for each replicate.
- https://cdn.elifesciences.org/articles/62087/elife-62087-fig4-figsupp1-data1-v2.docx
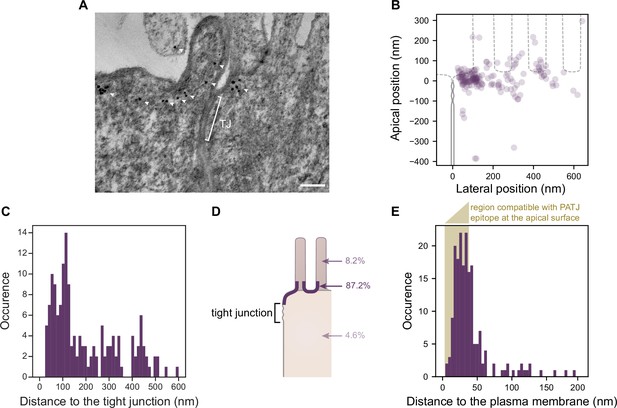
Electron tomography shows that PATJ localizes as clusters at the plasma membrane apically of the tight junction (TJ) in Caco-2 cells.
(A) Representative image of PATJ labeled with gold particles (arrowheads pointing at single particles or clusters of particles). The bracket with TJ indicates the TJ. Minimum intensity projection of a 150-nm-thick tomogram, scale bar: 100 nm. (B) Localization of gold particles labeling PATJ with respect to the TJ both in the apico-basal and lateral directions. (C) Distance between the center of gold particle labels and the TJ. (D) Summary of gold particles localization in the microvilli, in the vicinity of the plasma membrane and the cytoplasm. (E) Distance between gold particles and the apical surface. In amber, the region of distances compatible with PATJ epitope being at the apical surface, between 3 nm (radius of gold particles) and 37 nm (size of the primary and gold-labeled secondary antibody combination added with the presumed size of PALS1; Li et al., 2014). Tomograms of 300 nm in thickness of 12 junctions were used to extract the position of 169 gold particles labeling PATJ proteins. These junctions were obtained from one cell culture.
-
Figure 5—source data 1
Individual distance of gold particles versus the tight junction and the apical membrane.
- https://cdn.elifesciences.org/articles/62087/elife-62087-fig5-data1-v2.zip
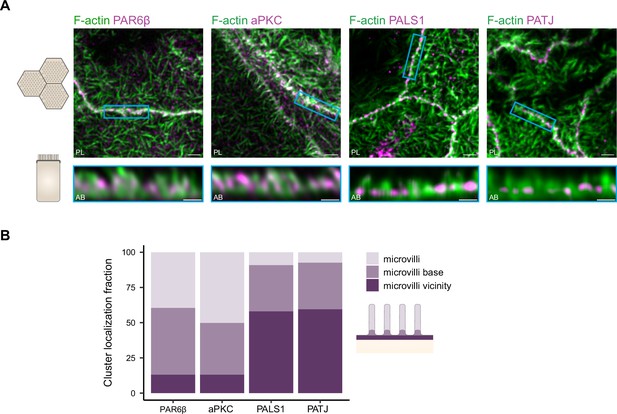
Organization of PAR6β, aPKC, PATJ, and PALS1, with respect to the actin cytoskeleton.
(A) 3D stimulated‐emission‐depletion (STED) imaging of cells labeled with Phalloidin and antibodies against polarity proteins with (top) top view and (bottom) side view on cell–cell junctions. Scale bars: top 2 µm, bottom 1 µm. (B) Localization analysis of PAR6β, aPKC, PATJ, and PALS1 vs. microvilli organization. We used three independent cell cultures. Detailed counts of clusters are given in Figure 6—source data 1.
-
Figure 6—source data 1
Contingency table of the number of counts of polarity proteins in the different microvilli regions.
- https://cdn.elifesciences.org/articles/62087/elife-62087-fig6-data1-v2.txt
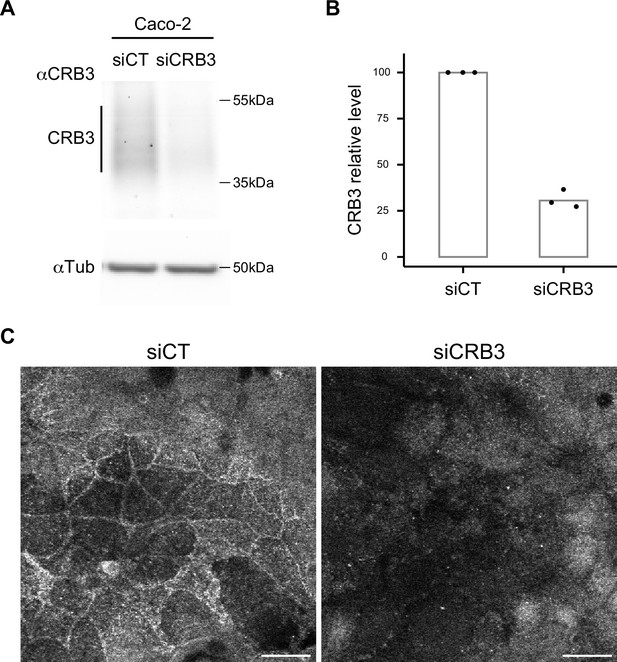
Characterization of α-CRB3 antibody.
(A) Immunoblot analysis of CRB3 expression level in CT (siCT) and CRB3 knock-down (siCRB3) Caco-2 cells with the rat monoclonal α-CRB3 antibody. α-Tubulin is used as a loading control. (B) Quantification of CRB3 in siCT and siCRB3 cells. (C) Confocal imaging of siCT and siCRB3 Caco-2 cells labeled with the rat monoclonal α-CRB3 antibody. Scale bars: 20 µm.
-
Appendix 1—figure 1—source data 1
Unnanotated blots in white light, fluorescence and annotated blots.
- https://cdn.elifesciences.org/articles/62087/elife-62087-app1-fig1-data1-v2.zip
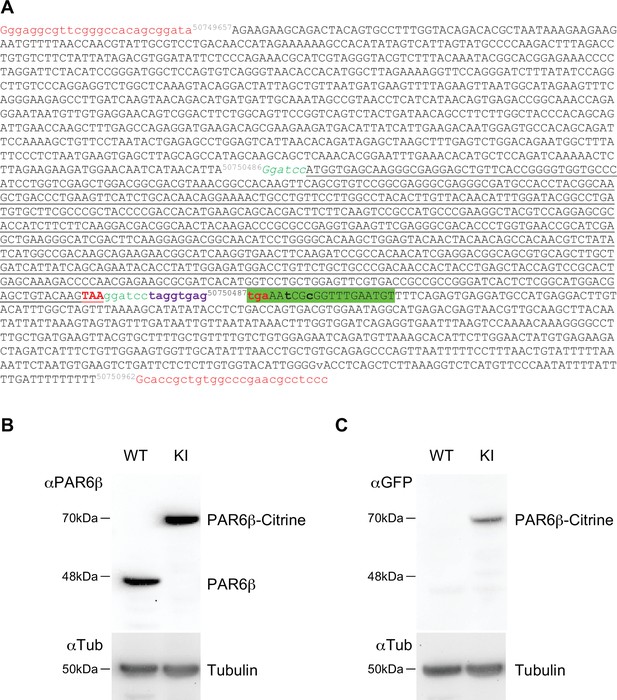
Characterization of the CRISPR/Cas9 Caco-2Par6β::Citrine cells.
(A) Donor Par6β-Citrine sequence: letters in red, universal guide sequence used by Cas9 to release the plasmid repair matrix into transfected cells; in black, sequence of homology arms; in green, GS peptide linker (and BamHI site to replace the Citrine sequence with another cDNA); in black underlined, Citrine sequence; in bold red, stop codon; in bold violet, a sequence including a STOP codon in each reading phase; in green highlight, sequence matching the gRNA. Mutations (in bold lowercase) were introduced in the noncoding part so that the gPARD6 guide could not lead to a cut (by Cas9) in the donor or modified genomic sequence after insertion. (B) Immunoblot analysis of PAR6β expression level in wild-type and Caco-2Par6β::Citrine Caco-2 cells. α-Tubulin is used as a loading control. (C) Immunoblot analysis of PAR6β-Citrine expression level in wild-type and Caco-2Par6β::Citrine cells Caco-2 cells. B and C deposits were independent.
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62087/elife-62087-transrepform1-v2.pdf
-
Source code 1
ImageJ marco toolsets to extract intensities from images and Python script to compute cross-correlation used in proximity analysis.
- https://cdn.elifesciences.org/articles/62087/elife-62087-code1-v2.zip
-
Appendix 1—figure 1—source data 1
Unnanotated blots in white light, fluorescence and annotated blots.
- https://cdn.elifesciences.org/articles/62087/elife-62087-app1-fig1-data1-v2.zip