A natural variant of the essential host gene MMS21 restricts the parasitic 2-micron plasmid in Saccharomyces cerevisiae
Figures
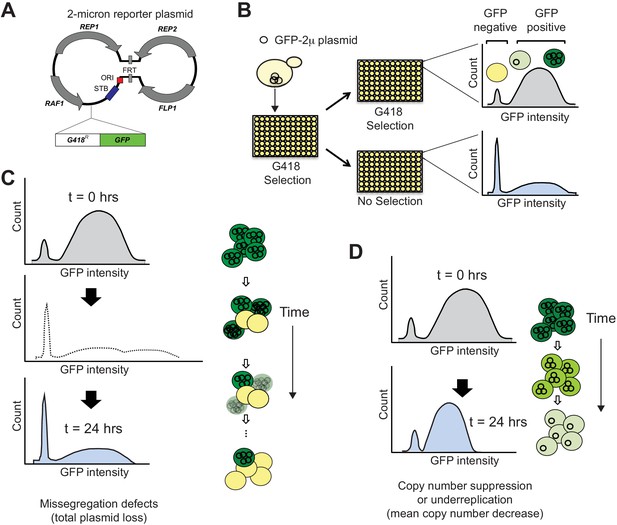
SCAMPR, a novel method to measure 2μ plasmid stability and dynamics.
(A) Schematic of GFP-reporter 2μ plasmid. The endogenous 2μ plasmid encodes an origin of replication (ori), four protein-coding genes (REP1, REP2, RAF1, FLP1) and their interacting DNA loci (STB, and FRT). The GFP-2μ reporter plasmid described here utilizes the full 2μ genome with an additional integrated G418-resistance and GFP expression cassette. (B) A Single Cell Assay for Measuring Plasmid Retention (SCAMPR) utilizes the dual reporter cassette: G418 resistance to ensure plasmid retention while under selection and GFP to facilitate screening of plasmid-positive cells. Cells with the reporter plasmid are kept on G418 selection to ensure the plasmid is present at t = 0 and either released to media without selection or passaged with continued G418 selection. Comparing the GFP intensities of the cell populations with and without G418 selection after 24 hr reveals the plasmid retention dynamics and population heterogeneity of the host genetic background (Figure 1—figure supplement 1). SCAMPR can therefore distinguish between alternate mechanisms of plasmid instability, illustrated in (C) and (D), or the relative contribution of both mechanisms. (C) Gross segregation defects in which plasmids are not distributed to both daughter cells would cause an increase in GFP-negative cells, as well as an increase in ‘super-green’ cells that retain twice as many two plasmids (light shading, dotted histogram). However, we infer that these cells would either be lost or not proliferate due to growth defects associated with high plasmid copy number. As a result of this selection, we expect to see a rapid increase in GFP-negative cells but no dramatic change in the median expression of (surviving) GFP-positive cells. (D) Plasmid instability caused by under-replication or copy number suppression would not cause a precipitous decline in GFP-positive cells as in (C) but would instead lead to a reduction in the median GFP intensity of the GFP-positive cells.
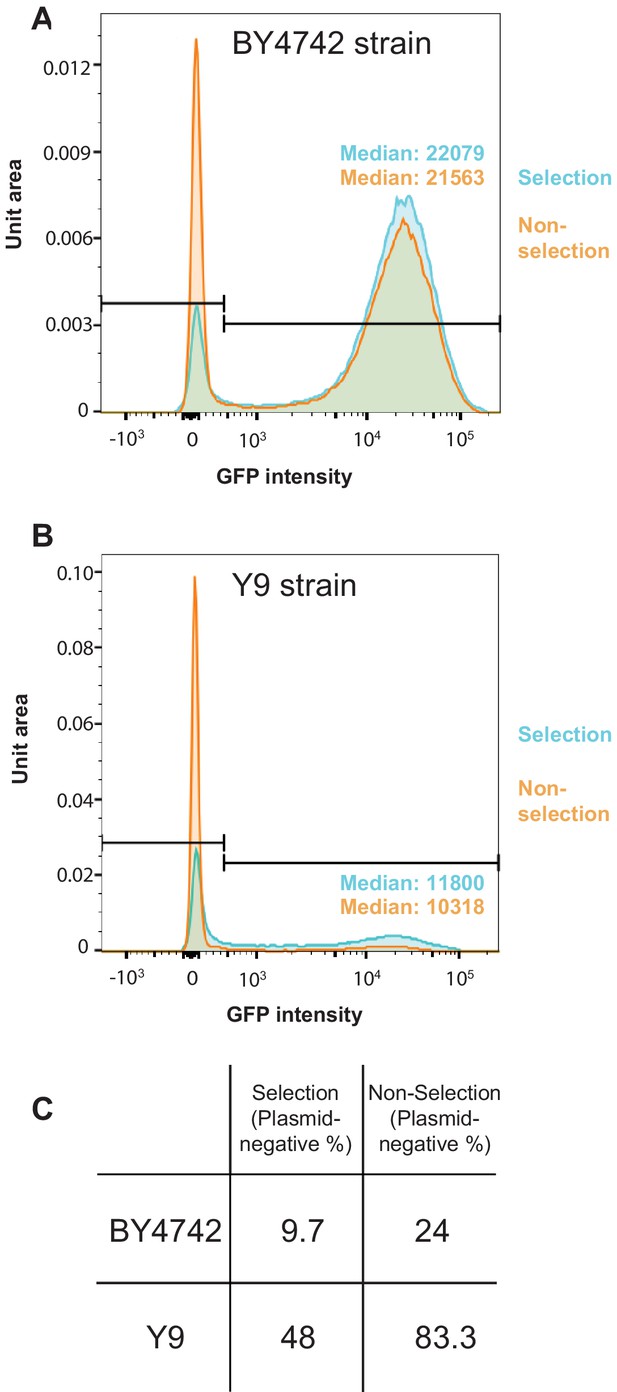
SCAMPR analysis for permissive and non-permissive S. cerevisiae strains.
(A) SCAMPR analysis in the laboratory BY4742 strain reveals that GFP intensity for the 2μ reporter plasmid is roughly normally distributed across single cells. Upon relaxation of G418 selection, there is an increase in the number of cells lacking 2μ plasmid from ~10% to~24%, although the median GFP intensity of plasmid-bearing cells remains almost unchanged. (B) In non-permissive Y9 strains, there is an increase of plasmid-lacking cells from 48% to 83% upon relaxation of G418 selection. Again, the median GFP intensity of plasmid-bearing cells remains largely unchanged. From these analyses, we conclude that plasmid instability in Y9 cells occurs via mis-segregation defects. (C) Table summarizing the plasmid-negative cell fractions in BY4742 and Y9 cells, with and without G418 selection.
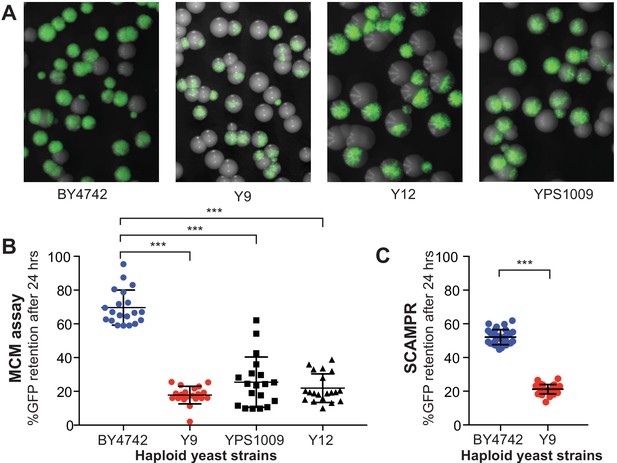
Plasmid instability is a heritable trait in three natural S. cerevisiae isolates.
(A) A colony sectoring assay qualitatively measures GFP-2μ reporter plasmid loss on solid media. Whereas the majority of colonies in the BY4742 background express GFP, only a small fraction of cells in colonies from wild isolates Y9, Y12, and YPS1009 express GFP. (B) The MCM assay quantifies the frequency of 2μ loss events in different yeast strains. Haploid cells from three wild isolates (Y9, Y12, YPS1009) have significantly lower plasmid retention than haploid cells from the laboratory BY4742 strain. ***p<0.0001, Kruskal-Wallis test. (C) SCAMPR assays confirm that a significantly smaller fraction of Y9 strain haploid cells retain the GFP-2μ reporter plasmid after 24 hr, relative to haploid BY4742 cells. ***p<0.0001, Kruskal-Wallis test.
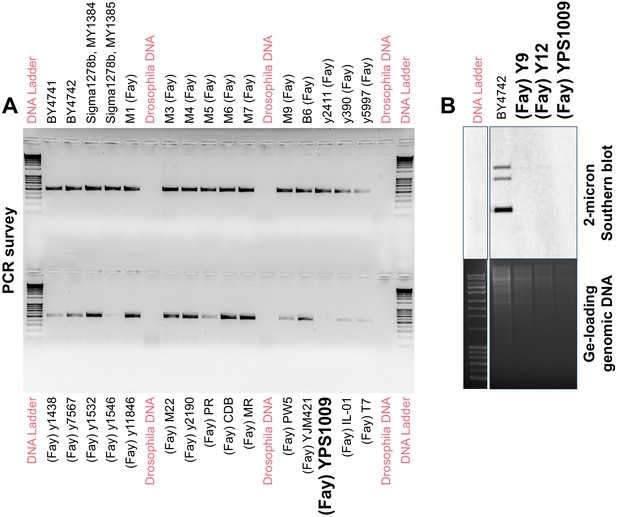
Three natural S. cerevisiae isolates lack endogenous 2μ plasmids.
(A) Representative PCR analysis shows that most of the 52 natural isolates tested harbor endogenous 2μ plasmids, except for three strains (one indicated). Drosophila melanogaster DNA was included as a negative control template. Representative gel shown for the presence of REP1 (Materials and methods) (B) Southern blot analysis confirms the absence of endogenous 2μ plasmids in the three natural isolates Y9, Y12, and YPS1009 as compared to the BY4742 positive control.
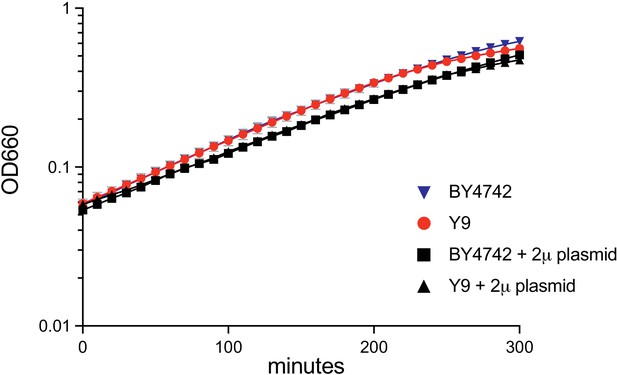
BY4742 and Y9 show similar growth rates.
BY4742 and haploid Y9 show similar growth dynamics in defined liquid media, both without the GFP-2μ reporter plasmid (blue triangle and red circle respectively), or with the reporter plasmid in the presence of G418 selection (black square and black triangle respectively).
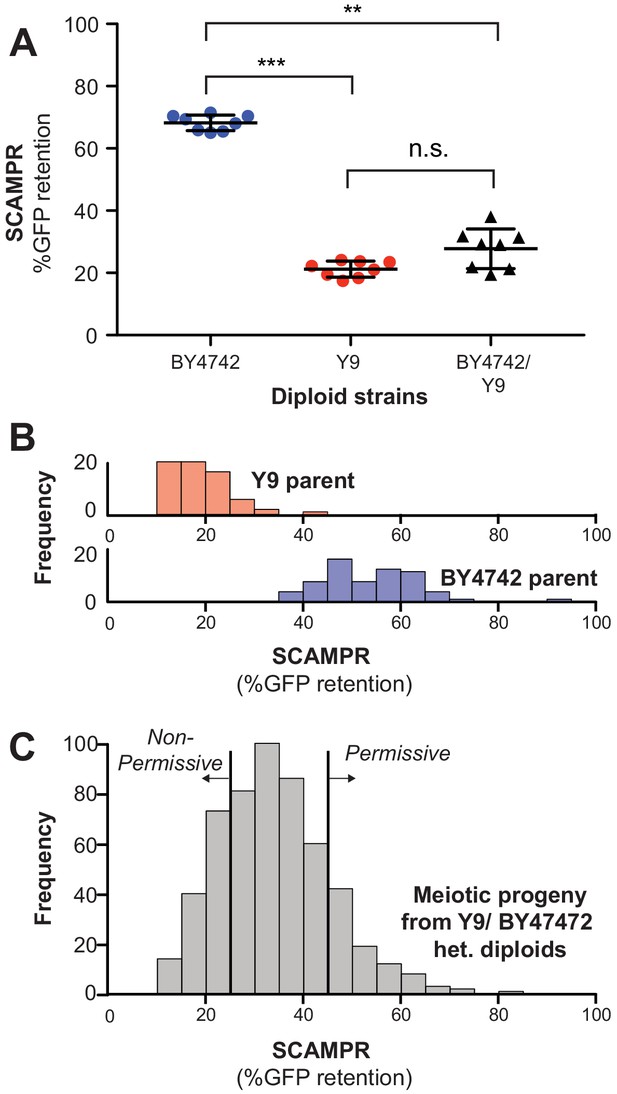
Genetic architecture and dominance of the Y9 plasmid instability phenotype.
(A) Compared to homozygous BY4742 diploids, heterozygous BY4742/Y9 diploid cells display low plasmid retention after 24 hr, similar to homozygous Y9 diploids. This suggests that the plasmid instability of Y9 cells is a dominant trait. All strains were analyzed with the SCAMPR plasmid retention assay. **p<0.001, ***p<0.0001, Kruskal-Wallis test; n.s. = not significant. (B–C) Phenotype distribution across ~600 random progeny strains (C) shows that most have an intermediate phenotype between that of the parental haploids (B). All strains were analyzed in triplicate with the SCAMPR assay. We selected the bottom ~20% (‘non-permissive’) and top ~20% (‘permissive’) of strains from this distribution for bulk sequencing and segregant analysis.
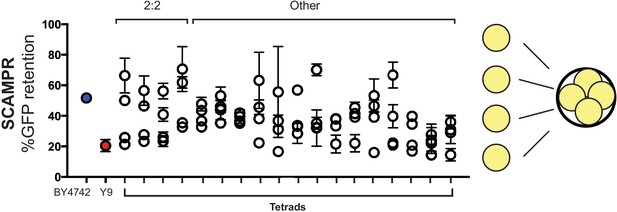
Tetrads dissected from meiosis of B4742/Y9 heterozygous diploids reveal a range of plasmid stability phenotypes.
All four spores dissected from meiotic tetrads were individually assayed by SCAMPR. While some tetrads display a 2:2 segregation pattern of plasmid instability/stability consistent with a single Mendelian locus, others suggest a more complex inheritance pattern. This pattern is consistent with at least 2–3 independently segregating loci in the Y9 genome that inhibit 2μ plasmid stability. Values plotted are the mean of SCAMPR measurements of three replicates per progeny.
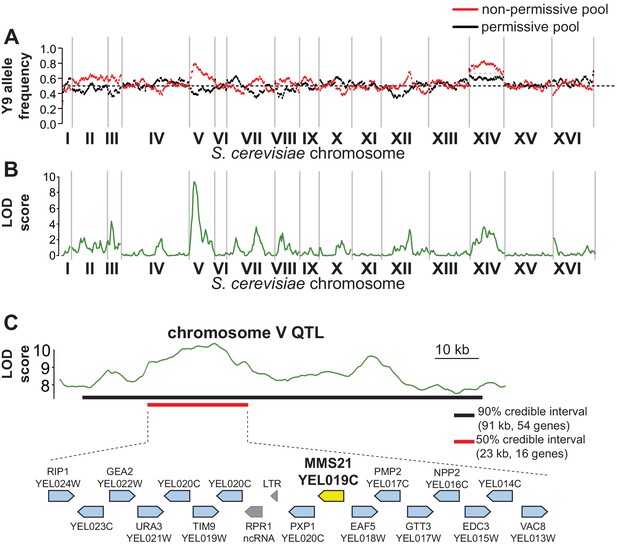
QTL mapping identifies a plasmid instability locus on Y9 chromosome V.
(A) We plotted the mean Y9 SNP allele frequency in 20 kb windows for the ‘non-permissive’ (red) and ‘permissive’ (black) pools of meiotic haploid progeny from BY4742/Y9 heterozygous diploid parents. Associations with a plasmid instability locus would show an increased representation of Y9 alleles in the non-permissive pool and a decreased representation of the BY4742 haplotype in the permissive pool (dotted line indicates equal representation). The increased representation of Y9 alleles on chromosome XIV in both pools is a result of a segregating disomy in the Y9 parent that we show does not affect the plasmid instability phenotype (Figure 4—figure supplement 2). (B) Based on the allele frequencies of individual SNPs, we used MULTIPOOL to calculate LOD scores for association with the plasmid instability phenotype. We observe a highly significant LOD score (10.00) on chromosome V. The peak is fairly sharp and reaches maximal LOD score at chrV:122.3–122.7 kb (sacCer3 coordinates). All loci have allele frequencies skewed in the expected direction; the restrictive pool is enriched for Y9 alleles. (C) MULTIPOOL 90% (54 genes, 91.2 kb, chrV:92.4–183.6 kb) and 50% (16 genes, 23.1 kb, chrV:107.3–130.4 kb) credible intervals for the chromosome V QTL. Among the 16 genes in the 50% credible interval is MMS21, which encodes an essential SUMO E3-ligase.
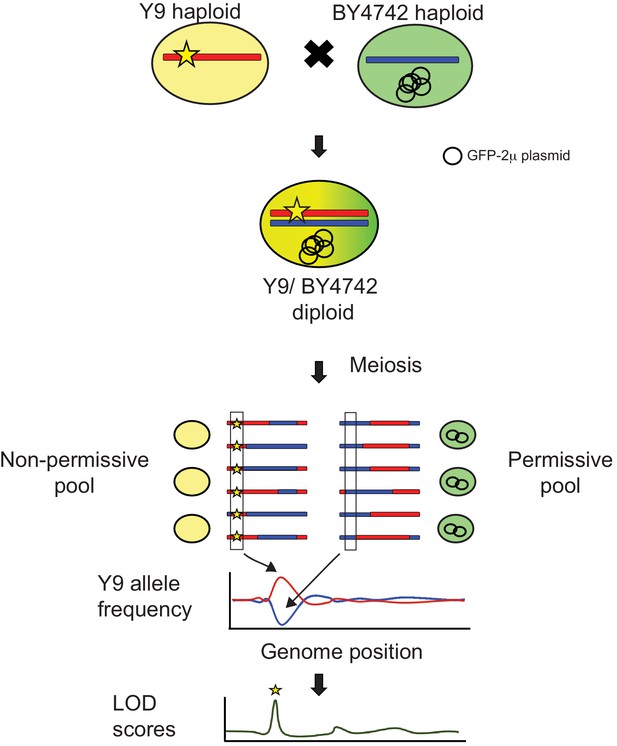
Schematic of QTL mapping by bulk segregant analysis (Magwene et al., 2011).
We crossed non-permissive Y9 and permissive BY4742 haploid cells to create heterozygous diploids. We expect any Y9 alleles associated with plasmid instability (yellow star) to be concentrated in pools of meiotic progeny that show plasmid instability with SCAMPR analyses. Therefore, by determining where the Y9 allele frequency is elevated in the non-permissive pool and depleted in the permissive pool, we can identify genetic loci that are likely to significantly contribute to the plasmid instability phenotype. We use allele frequencies as input to the MULTIPOOL algorithm to calculate LOD scores that indicate statistical likelihoods of each genetic locus contributing to the plasmid instability phenotype (Edwards and Gifford, 2012).
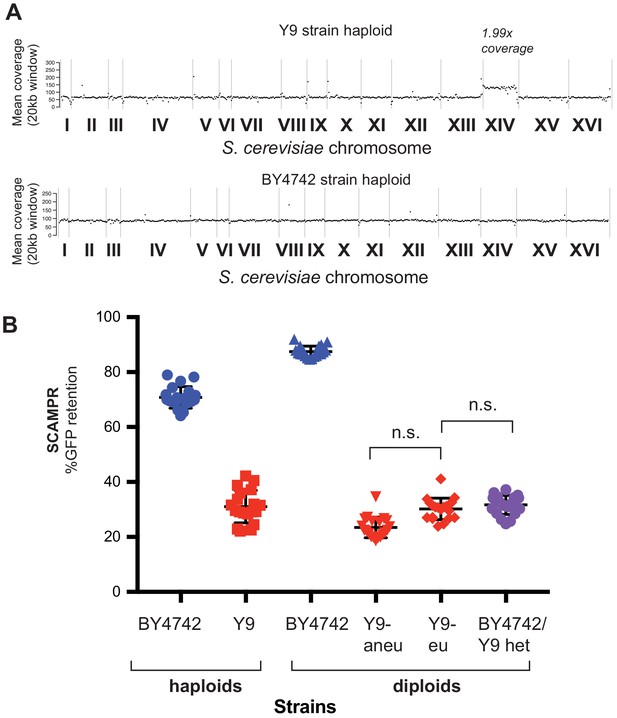
Aneuploidy of chromosome XIV in Y9 strain.
(A) Whole genome sequencing reveals ~2X coverage of chromosome XIV in the sequenced Y9 haploid strain, indicating that this parent is disomic for this chromosome. This disomy segregates among the meiotic progeny from BY4742/Y9 heterozygous diploids. (B) Y9-derived diploid strains that were euploid or aneuploid for chromosome XIV were identified by qPCR and phenotyped (Pavelka et al., 2010). We find no difference in plasmid instability phenotypes, indicating that disomy of chromosome XIV is not likely to contribute to this trait.
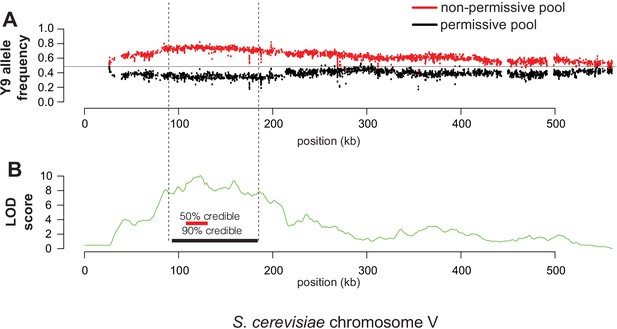
Y9 chromosome V is most strongly associated with plasmid instability.
(A) Over-representation of chromosome V Y9 alleles in the non-permissive pool (red) and under-representation in the permissive pool (black). Each datapoint is an individual SNP. (B) Based on the allele frequencies shown in (A), we calculated LOD scores and 50% (chrV:107.3–130.4 kb), 90% (chrV:92.4–183.6 kb) credible intervals for association with the plasmid instability phenotype. The highest LOD score in the region is approximately 10.00 at chrV:122 kb.
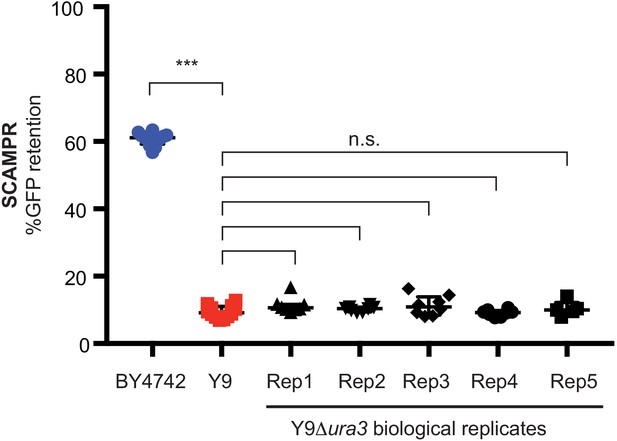
Deletion of URA3 from Y9 haploid cells does not affect their plasmid instability phenotype.
Independently derived biological replicates (Rep1 through 5) of Δura3 Y9 cells are not different from wildtype Y9 in terms of their plasmid instability phenotypes. ***p<0.0001, Kruskal-Wallis test, n.s. = not significant.
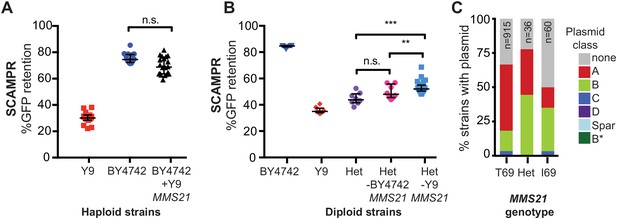
A single SNP in Y9 MMS21 contributes to the plasmid instability phenotype.
(A) Introduction of the Y9 MMS21 allele into BY4742 haploid cells is not sufficient to significantly lower plasmid instability. (B) However, removal of the Y9 MMS21 allele but not the BY4742 MMS21 allele increases plasmid stability in BY4742/Y9 heterozygous diploids, showing that the Y9 allele of MMS21 plays an important role in the Y9 plasmid instability phenotype. **p<0.001, Kruskal-Wallis test, n.s. = not significant. (C) Plasmid prevalence (by plasmid class) for each MMS21 Thr9Ile genotype within 1011 sequenced S. cerevisiae strains. Plasmid data and genotypes from Peter et al., 2018. Strains with the Y9 MMS21 allele (I69) have a lower frequency of harboring 2μ plasmids in general, and A-type 2μ plasmids, in particular. However, this effect can be confounded by the phylogenetic relatedness of these strains.
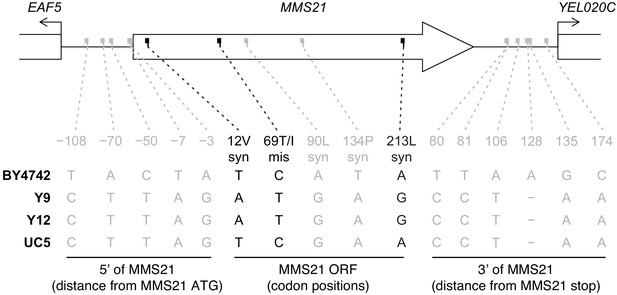
Comparative analysis of MMS21 and flanking regions.
We show all differences between Y9 and BY4742 in the MMS21 ORF and flanking intergenic regions (chrV:120199–121571, − strand). We also show genotypes for Y12 (closely-related non-permissive strain) and UC5 (closely-related permissive strain), and show SNPs where genotype is congruent (in black) or incongruent (in gray) with plasmid status. ‘syn’=synonymous, ‘mis’=missense.
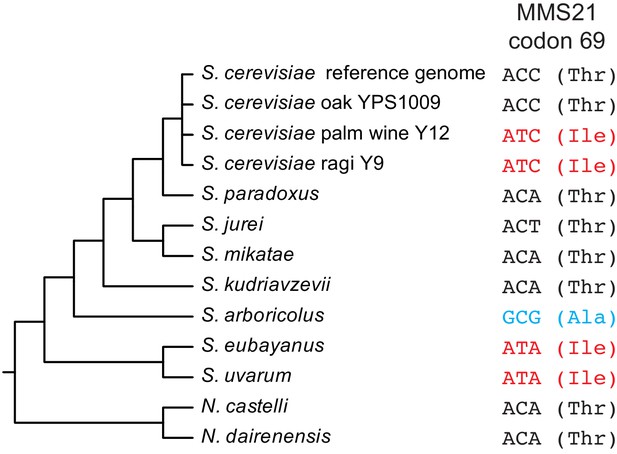
Sequence of MMS21 codon 69 across the Saccharomyces sensu stricto clade, as well as selected S. cerevisiae strains and two outgroup Naumovozyma species.
Species cladogram was adapted from previous studies (Naseeb, 2018; Borneman and Pretorius, 2014). This analysis shows that the Thr69 allele of MMS21 is ancestral in S. cerevisiae but is not universally conserved in closely-related species.
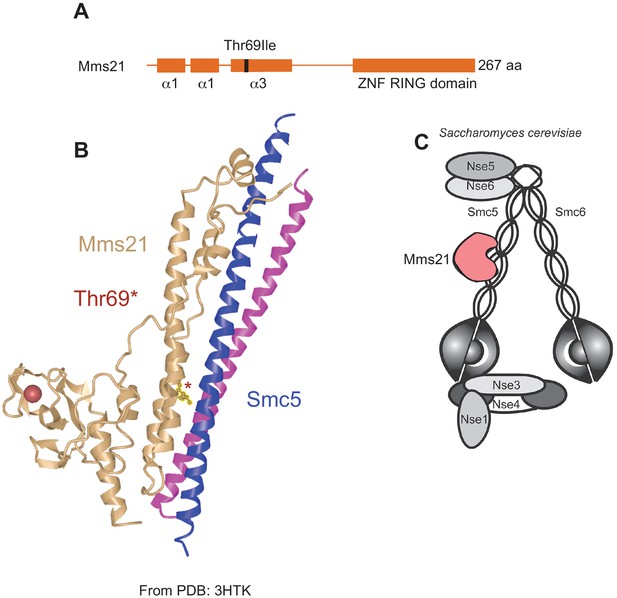
The Thr69Ile polymorphism is located at the Mms21-Smc5 binding interface.
(A) The Thr69Ile polymorphism occurs in the third alpha-helix in the Mms21 N-terminal domain. The C-terminal domain of MMS21 encodes the SUMO E3-ligase associated RING domain. (B) Schematic of co-crystal structure of Mms21 with the coiled coil domain of Smc5 (PDB: 3HTK, adapted from Figure 1, Duan et al., 2009) shows that the Thr69 residue (*) in the N-terminal domain is at the direct binding interface between the two proteins. (C) Cartoon of Smc5/6 complex in S. cerevisiae showing the Mms21-Smc5 interaction, reproduced from Figure 1 (right panel), Stephan et al., 2011.
© 2015, Stephan et al. https://creativecommons.org/licenses/by-nc-nd/3.0/ Reproduced from Figure 1 (right panel), Stephan et al., 2011, under the terms of a CC-BY-NC-ND 3.0 license. This panel is not available under the terms of the CC-BY 4.0 license and any further reproduction must adhere to the terms of the original license.
Additional files
-
Supplementary file 1
Natural S. cerevisiae isolates screened for the presence or absence of endogenous 2μ plasmids.
- https://cdn.elifesciences.org/articles/62337/elife-62337-supp1-v2.xlsx
-
Supplementary file 2
Engineered S. cerevisiae strains used in this study.
- https://cdn.elifesciences.org/articles/62337/elife-62337-supp2-v2.xlsx
-
Supplementary file 3
Missense polymorphisms in the 90% credible QTL interval for plasmid instability.
The table lists missense polymorphisms shared between the non-permissive Y9 and Y12 strains, but distinct from the closely-related plasmid-permissive strain, UC5, and the permissive laboratory strain, BY4742.
- https://cdn.elifesciences.org/articles/62337/elife-62337-supp3-v2.xlsx
-
Supplementary file 4
Missense polymorphisms in Smc5/6 complex members and other SUMO ligases.
All non-synonymous differences between Y9 and BY4742 strains in all components of the Smc5/6 complex (Smc5, Smc6, Nse1-6; Nse2 is a synonym of Mms21) and in SUMO ligases Siz1 and Siz2.
- https://cdn.elifesciences.org/articles/62337/elife-62337-supp4-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62337/elife-62337-transrepform-v2.docx





