A feedback loop between the androgen receptor and 6-phosphogluoconate dehydrogenase (6PGD) drives prostate cancer growth
Figures
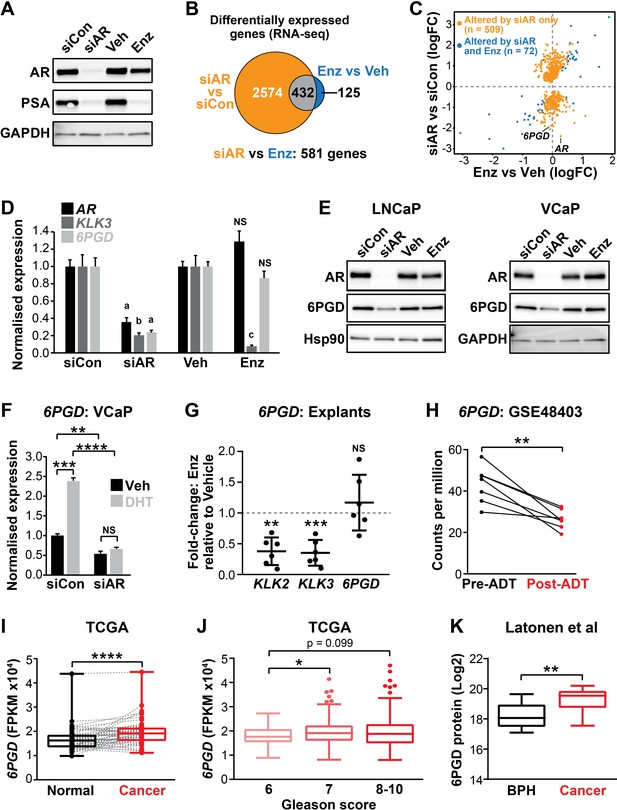
6PGD is an androgen receptor (AR)-regulated gene and is elevated in prostate cancer.
(A) Effect of siAR and enzalutamide (Enz) on the AR target, PSA. LNCaP cells were transfected with AR (siAR; 12.5 nM) or control (siCon) siRNA for 48 hr or treated with Enz (1 µM) or vehicle (Veh) for 24 hr, after which AR and PSA proteins were evaluated by immunoblotting. GAPDH was used as loading control. (B) Numbers of genes differentially expressed (false discovery rate [FDR] < 0.05) by siAR (versus siCon) or Enz (vs. Veh) are shown in the Venn diagram (at top). Below: an alternative analysis identified 581 genes differentially expressed (FDR < 0.05) by siAR versus Enz. (C) Scatterplot of genes affected by siAR and Enz. The 581 genes differentially expressed by siAR versus Enz are shown in blue (n = 72, genes differentially expressed by siAR versus siCon and Enz versus Veh) and yellow (n = 509), genes differentially expressed by siAR versus siCon but not by Enz versus Veh. (D) Validation of 6PGD expression in response to siAR and Enz by RT-qPCR. Gene expression was normalised to GUSB and L19 and represents the mean ± standard error of the mean (SEM) of three biological replicates; siCon and Veh were set to 1. Differential expression was evaluated using unpaired t tests (a, p<0.01; b, p<0.001; c, p<0.0001; NS, not significant). (E) 6PGD protein levels in response to siAR and Enz treatments were measured by immunoblotting in LNCaP (left) and VCaP (right) cells. HSP90 and GAPDH were used as loading controls. (F) RT-qPCR of 6PGD expression in response to DHT and siAR in VCaP cells. Cells were transfected with siRNAs for 24 hr, and then treated with 1 nM DHT for another 24 hr. Gene expression was normalised and graphed as in (D). Differential expression was evaluated by t tests (**p < 0.01; ***p < 0.001; ****p < 0.0001). (G) RT-qPCR of KLK2, KLK3, and 6PGD expression in response to Enz treatment (1 µM, 72 hr) in patient-derived explants. Gene expression was normalised to GAPDH, PPIA, and TUBA1B and is represented as fold-change relative to vehicle. Differential expression was evaluated by one-sample t tests (**p<0.01; ***p<0.001). (H) 6PGD mRNA expression in prostate tumours pre- and post-androgen deprivation therapy (ADT; GSE48403). A Wilcoxon matched-pairs signed-rank test was used to compare expression in the groups. (I) 6PGD expression is elevated in primary prostate cancer. The TCGA dataset comprises 52 patient-matched normal and cancer samples. Boxes show minimum and maximum (bottom and top lines, respectively) and mean (line within the boxes) values. A paired t test was used to compare expression in normal versus cancer. FPKM: fragments per kilobase of exon per million mapped reads. (J) 6PGD expression by Gleason grade in the TCGA cohort. Boxes show minimum and maximum (bottom and top lines, respectively) and mean (line within the boxes) values. Unpaired t tests were used to compare expression between the groups. (K) 6PGD protein expression in clinical prostate samples (benign prostatic hyperplasia [BPH] and tumours) was measured mass spectrometry. Boxes show minimum and maximum (bottom and top lines, respectively) and mean (line within the boxes) values. An unpaired t test was used to compare expression between the groups.
-
Figure 1—source data 1
Differentially-expressed genes in LNCaP prostate cancer cells treated with siAR or Enz.
- https://cdn.elifesciences.org/articles/62592/elife-62592-fig1-data1-v2.xlsx
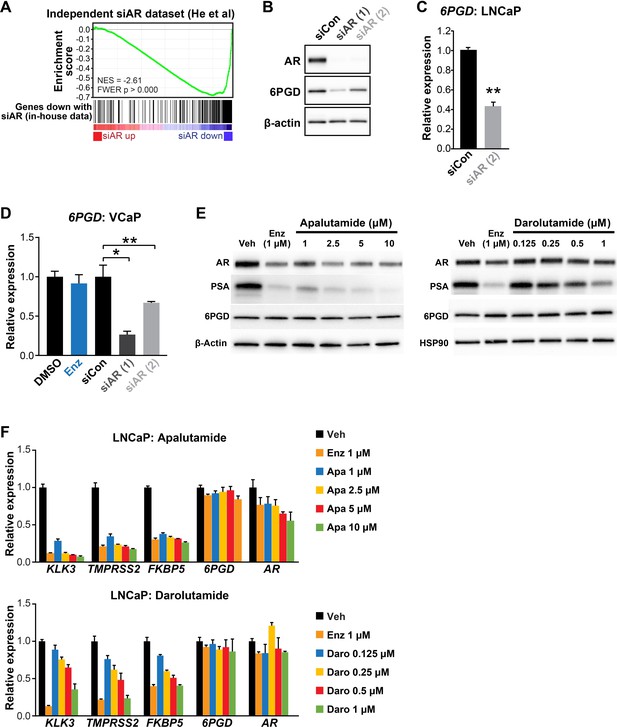
6PGD expression is decreased by AR knockdown but not by AR inhibition.
(A) Concordance between our siAR RNA-seq data and an independent dataset, as demonstrated by gene set enrichment analysis (GSEA; Subramanian et al., 2005). RNA-seq data from He and colleagues (He et al., 2014) was kindly provided by Nicholas Mitsiades (Baylor College of Medicine), and genes were ranked by fold-change in siAR treatment versus siControl. Genes downregulated by siAR versus siControl in our dataset (false discovery rate [FDR] < 0.01, n = 305) were used as the gene set of interest. Running enrichment scores are plotted (top graph) and normalised enrichment scores (NES) and p values are indicated. (B) Two distinct AR siRNAs (siAR [Vander Heiden et al., 2009] and siAR [Bader and McGuire, 2020]; 12.5 nM) reduce the expression of 6PGD at the protein level in LNCaP cells. Cells were transfected with 12.5 nM of each siRNA; after 48 hr, proteins were extracted and assessed by western blotting. (C) siAR (Bader and McGuire, 2020) reduces the expression of 6PGD mRNA in LNCaP cells. Transfection of siRNAs was performed as in (A). Differential expression was evaluated using an unpaired t test (**p <0.001). (D) siAR (Vander Heiden et al., 2009) and siAR (Bader and McGuire, 2020), but not enzalutamide (Enz, 1 µM), reduce the expression of 6PGD mRNA in VCaP cells. Transfection of siRNAs was performed as in (A). Cells were treated with DMSO or Enz for 24 hr. Differential expression compared to DMSO or siCon was determined using ANOVA and Dunnett’s multiple comparison tests (*p<0.05; **p<0.01). (E, F) Next-generation AR antagonists apalutamide and darolutamide inhibit AR target gene expression at the protein (E) and mRNA (F) level, but do not reduce expression of 6PGD protein or mRNA. Cells were treated for 24 hr with the indicated doses of each drug.
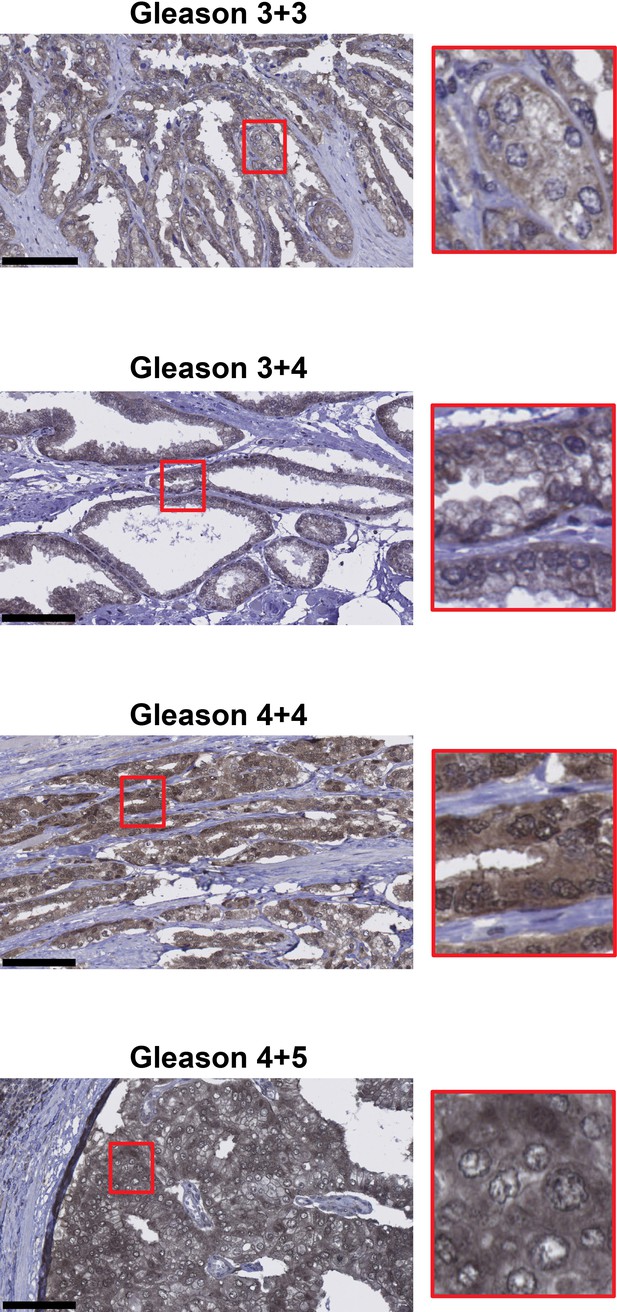
Representative images of 6PGD immunohistochemistry in patient tumours.
Gleason grades are shown. Scale bars represent 100 µm.
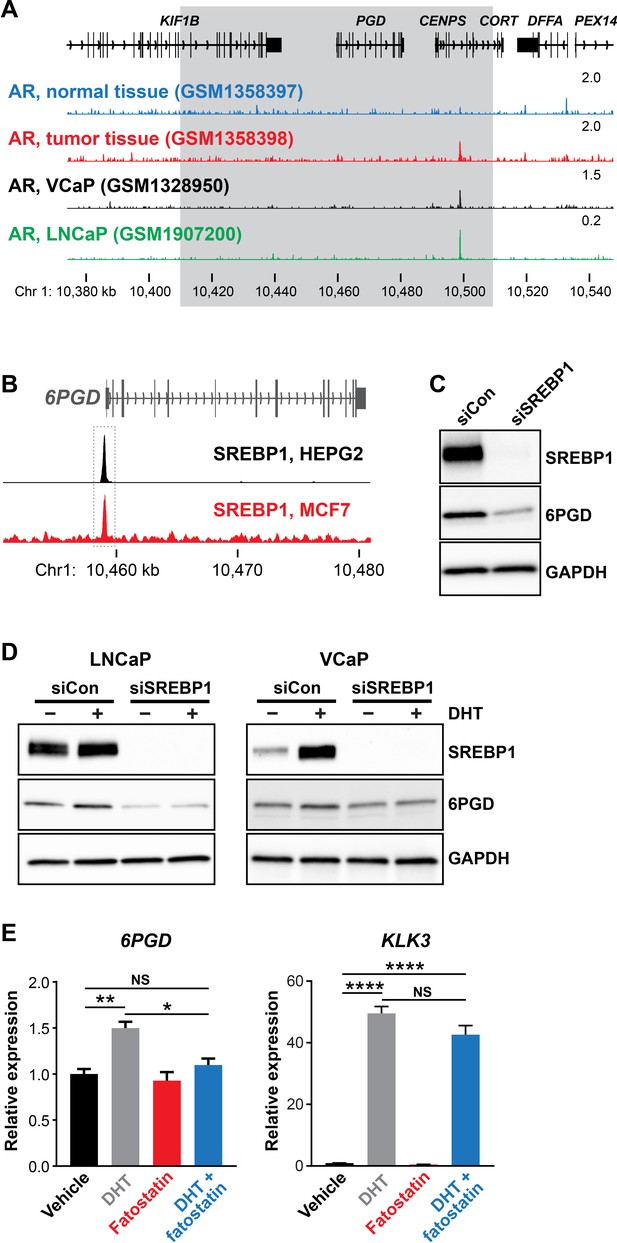
SREBP1 mediates induction of 6PGD downstream of the androgen receptor (AR).
(A) ChIP-seq data showing AR DNA binding near the 6PGD gene in non-malignant and prostate tumour samples (Pomerantz et al., 2015) and the LNCaP (Barfeld et al., 2017) and VCaP (Asangani et al., 2014) cell line models. The grey box indicates a region ±50 kb of the 6PGD transcriptional start site. (B) ChIP-seq data showing SREBP1 DNA binding at the 6PGD promoter in HEPG2 and MCF7 cells. Data is from ENCODE (ENCODE Project Consortium, 2012; HEPG2: ENCFF000XXR; MCF7: ENCFF911YFI). (C) Effect of siSREBP1 on 6PGD protein. LNCaP cells were transfected with siRNA (siSREBP1; 12.5 nM) or control (siCon) for 72 hr after which SREBP1 and 6PGD protein levels were evaluated by immunoblotting. GAPDH was used as loading control. (D) Effect of siSREBP1 on 6PGD induction by DHT. LNCaP (left) or VCaP (right) cells were transfected with siRNA (siSREBP1; 12.5 nM) or control (siCon) in charcoal-stripped FBS media for 72 hr and then treated with 10 nM DHT for another 24 hr. SREBP1 and 6PGD protein levels were evaluated by immunoblotting. GAPDH was used as loading control. (E) RT-qPCR of 6PGD expression in response to DHT and fatostatin in LNCaP cells. Cells were serum starved in charcoal-stripped FBS media for 72 hr and then treated with Veh or 10 nM DHT ±10 µM fatostatin for another 24 hr. Gene expression was normalised to GUSB and L19 and represents the mean + SEM of three biological replicates. Treatment effects were evaluated using ANOVA and Dunnett’s multiple comparison tests (*p<0.05; **p<0.01; ****p<0.0001; NS, not significant).
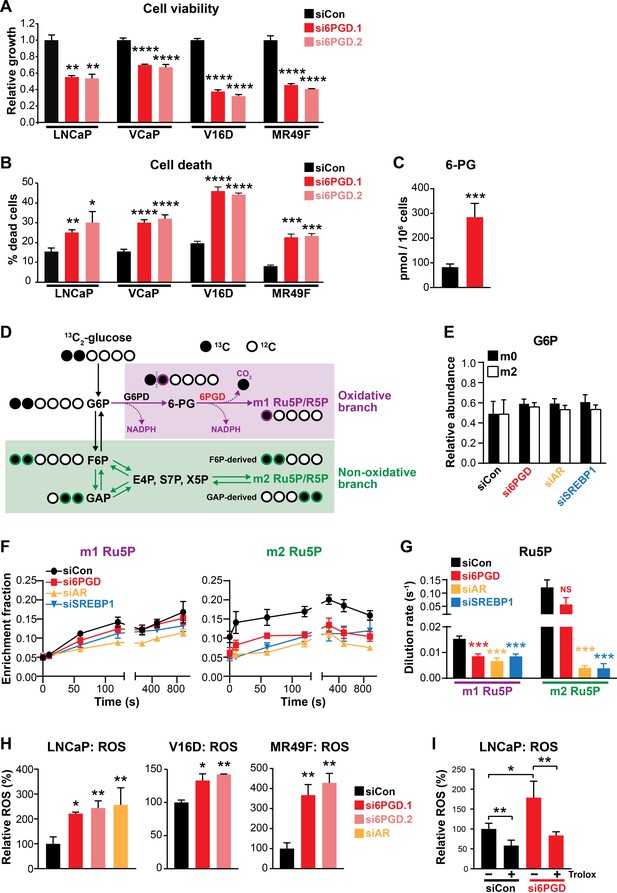
An AR-SREBP1-6PGD axis influences prostate cancer cell growth and activity of the pentose phosphate pathway.
(A, B) Knockdown of 6PGD with two distinct siRNAs (si6PGD.1 and si6PGD.2) reduced viability (A) and increased cell death (B) of four prostate cancer cell lines, as assessed using Trypan blue exclusion assays. LNCaP and VCaP cells were evaluated 3 days post-transfection; V16D and MR49F cells were evaluated 5 days post-transfection. Error bars are standard error of the mean (SEM) of triplicate samples and are representative of three independent experiments. Treatment effects were evaluated using ANOVA and Dunnett’s multiple comparison tests (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001). (C) Knockdown of 6PGD causes accumulation of intracellular 6PG in LNCaP cells, as determined by mass spectrometry. Results are representative of two independent experiments. Error bars are SEM of triplicate samples. Treatment effect was evaluated using an unpaired t test (p<0.001). Colour key is as in (A). (D) Schematic demonstrating flux of 1,2-13C2 glucose through the PPP and incorporation into Ru5P and R5P. Unlabelled 12C carbon is shown as open circles, whereas 13C is shown as filled circles. The oxidative and non-oxidative branches of the PPP are indicated in purple and green, respectively. 6PG: 6-phosphogluconate; E4P: erythrose 4-phosphate; F6P: fructose 6-phosphate; G6P: glucose 6-phosphate; GAP: glyceraldehyde 3-phosphate; R5P: ribose 5-phosphate; Ru5P: ribulose 5-phosphate; S7P: sedoheptulose 7-phosphate; X5P: xylulose 5-phosphate. (E) Isotopic steady-state G6P enrichments of LNCaP cells fed with 1,2-13C2 glucose and natural glucose at 1:1 ratio show control and treatments cells were labelled to a similar extent. Error bars are standard deviation (SD). (F) Accumulation of singly (left, m1) and doubly (right, m2) labelled Ru5P produced via the oxidative and non-oxidative branches, respectively, of the PPP. Error bars are SD. (G) Dilution rate (turnover rate) calculated from the accumulation of singly and doubly labelled Ru5P (data from E) using the continuous stirred-tank reactor (CSTR) equation. For statistical analysis of treatment effects, refer to Materials and methods (***p<0.001; NS, not significant). Error bars are SD. (H) Knockdown of 6PGD and androgen receptor (AR) causes increased levels of reactive oxygen species (ROS) in LNCaP, V16D, and MR49F cells. Data was normalised to siCon, which was set to 100%. Error bars are SEM of triplicate samples. Treatment effects were evaluated using ANOVA and Dunnett’s multiple comparison tests (*p<0.05; **p<0.01). (I) ROS production in LNCaP cells in response to si6PGD is reversed by the antioxidant. Trolox data was normalised to siCon in the absence of Trolox, which was set to 100%. Error bars are SEM of triplicate samples. Treatment effects were evaluated using ANOVA and Tukey’s multiple comparison tests (*p<0.05; **p<0.01). Colour key is as in (A).
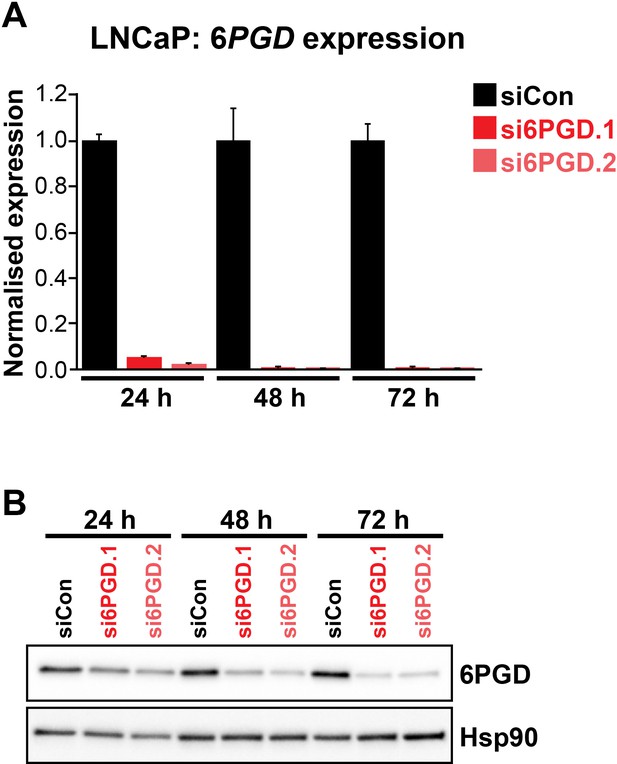
Two distinct 6PGD siRNAs (si6PGD.1 and si6PGD.2) effectively reduce 6PGD expression in LNCaP cells.
Cells were transfected with 12.5 nM of each siRNA for 72 hr, after which 6PGD mRNA was measured by RT-qPCR (A) or 6PGD protein was measured by immunoblotting (B).
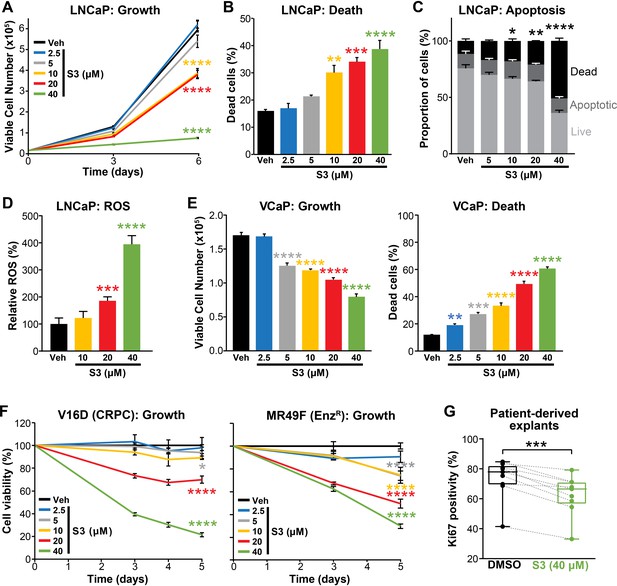
Pharmacological inhibition of 6PGD suppresses prostate cancer growth and increases reactive oxygen species (ROS).
(A, B) The 6PGD inhibitor, S3, dose-dependently decreased viability (A) and increased death (B) of LNCaP cells, as determined by Trypan blue exclusion assays. Dead cells were counted at day 6. Data represent the mean of triplicate samples and are representative of three independent experiments. Error bars are SEM. Growth (day 6) and death for each dose was compared to vehicle using ANOVA and Dunnett’s multiple comparison tests (****p<0.0001). Veh: vehicle. (C) S3 causes apoptosis of LNCaP cells, as determined using flow cytometry-based Annexin V/7-AAD assays. Cells were assessed 72 hr after treatment. Data represent the mean ± SE of triplicate samples and are representative of four independent experiments. Dead cell proportions were compared to vehicle using ANOVA and Dunnett’s multiple comparison tests (*p<0.05; **p<0.01; ****p<0.0001). (D) S3 causes increased levels of ROS in LNCaP cells. Data was normalised to Veh, which was set to 100%. Effects were evaluated using ANOVA and Dunnett’s multiple comparison tests (***p<0.001; ****p<0.0001). (E) S3 dose-dependently decreased viability (left) and increased death (right) of VCaP cells, as determined by Trypan blue exclusion assays. Live and dead cells were counted 4 days after treatment. Data represent the mean ± SE of triplicate samples and are representative of three independent experiments. Effects were evaluated using ANOVA and Dunnett’s multiple comparison tests (**p<0.01; ***p<0.001; ****p<0.0001). (F) S3 suppresses the growth of castration-resistant prostate cancer (CRPC) cells (V16D) and enzalutamide-resistant CRPC cells (MR49F), as determined using CyQuant Direct Cell Proliferation Assay. Fluorescence from day 0 was set to 100%. Data represent the mean ± SEM of triplicate samples and are representative of two independent experiments. Effects (at day 5) were evaluated using ANOVA and Dunnett’s multiple comparison tests (*p<0.05; ****p<0.0001). (G) S3 inhibits the proliferation of prospectively collected human tumours grown as patient-derived explants (PDEs). PDEs (from n = 9 patients) were treated for 72 hr. Ki67 positivity, a marker of proliferation, was determined using immunohistochemistry. Boxes show minimum and maximum (bottom and top lines, respectively) and mean (line within the boxes) values. A paired t test was used to compare Ki67 positivity in treated versus vehicle-treated control samples (***p<0.001).
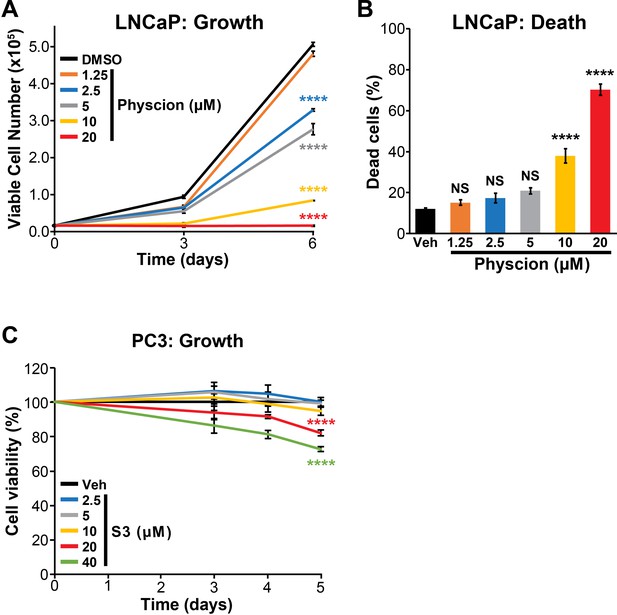
Physcion effectively suppresses growth (A) and causes death (B) of LNCaP cells.
Live and dead cells were measured (A, at the indicated timepoints; B, at day 6) using Trypan blue exclusion assays. Physion’s effects on growth and death compared to vehicle (Veh) were determined using ANOVA and Dunnett’s multiple comparison tests (****p<0.0001; NS, not significant). (C) Effect of S3 on growth of PC3 cells. Cell viability assessed by CyQuant Direct Cell Proliferation Assay. Fluorescence at day 0 was set to 100%. The effect of S3 on growth compared to vehicle (Veh) was determined using ANOVA and Dunnett’s multiple comparison tests; only 20 µM and 40 µM doses were significantly different to Veh (****p<0.0001).
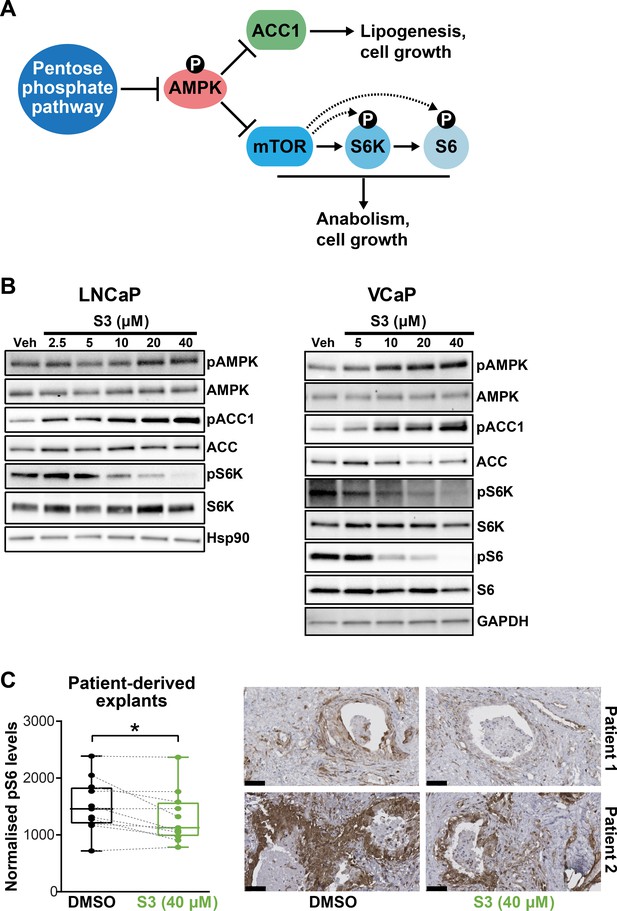
Targeting 6PGD activates AMPK and represses ACC1 and mTOR pathways.
(A) Schematic showing key metabolic pathways downstream of the pentose phosphate pathway (PPP). By suppressing AMPK signalling, the PPP can enhance the activity of ACC1 and mTOR and subsequently various growth-promoting anabolic processes. (B) S3 activates AMPK and inhibits ACC1 and mTOR signalling. LNCaP (left) and VCaP (right) cells were treated for 24 hr with the indicated doses of S3 prior to analysis of indicated proteins by immunoblotting. (C) S3 inhibits mTOR signalling, as indicated by reduced pS6, in patient-derived explants (PDEs). PDEs (from n = 11 patients) were treated for 72 hr. The levels of pS6 were measured using immunohistochemistry (IHC). Boxes (graph on left) show minimum and maximum (bottom and top lines, respectively) and mean (line within the boxes) values. A paired t test was used to compare Ki67 positivity in treated versus vehicle-treated control samples (***p<0.001). Representative IHC images are shown on the right (scale bars represent 50 µm).
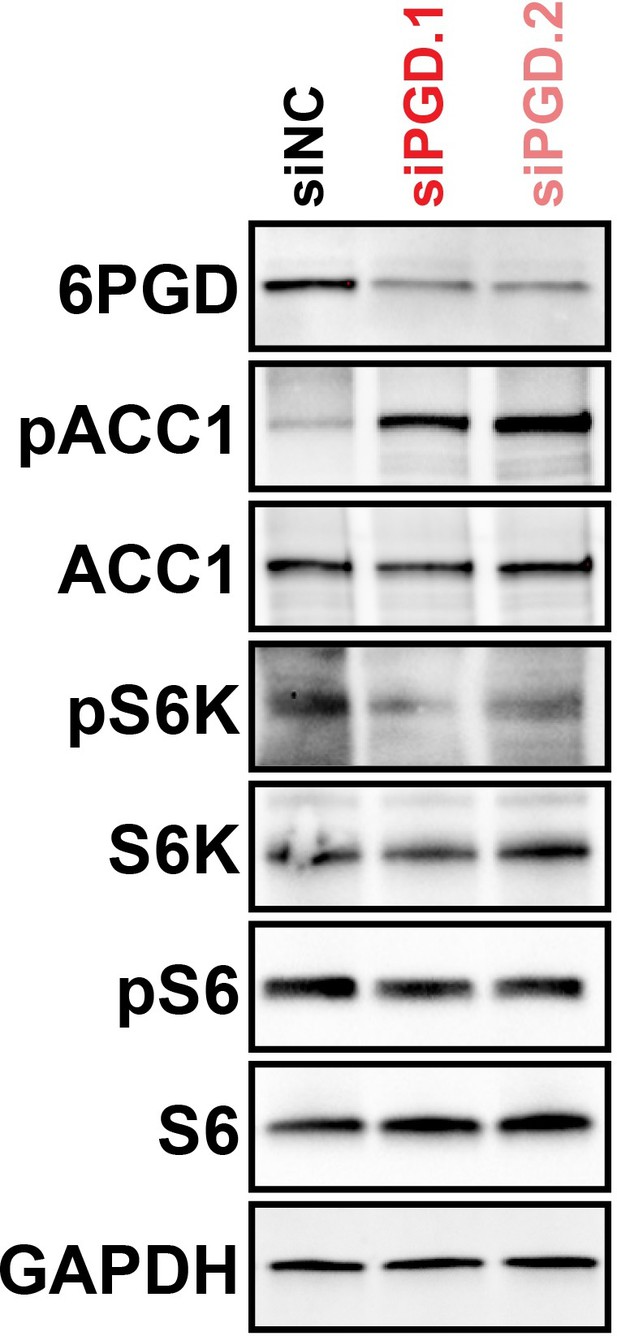
6PGD knockdown inhibits ACC1 and mTOR signalling, as determined by increased levels of pACC1 and decreased levels of pS6/pS6K, respectively.
VCaP cells were transfected with two distinct 6PGD siRNAs (siPGD.1 and siPGD.2) for 48 hr prior to analysis of indicated proteins by immunoblotting.
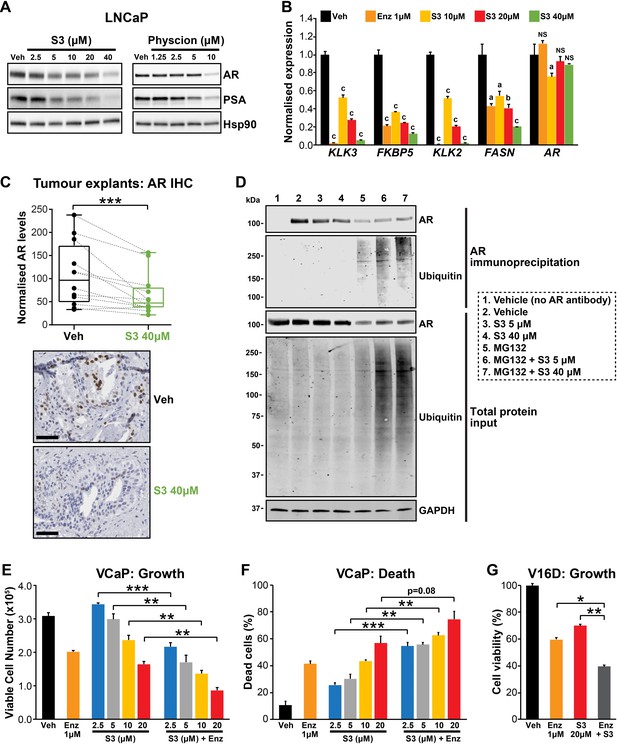
Targeting the androgen receptor (AR)/PGD feedback loop in prostate cancer.
(A) Protein levels of AR and its target PSA in response to S3 (24 hr of treatment) and physcion (48 hr of treatment) in LNCaP cells, as determined by immunoblotting. HSP90 was used as a loading control. (B) AR target gene expression in response to S3 treatment in LNCaP cells, as determined by RT-qPCR. Gene expression was normalised to GUSB and L19 and represents the mean + SEM of three biological replicates; Veh was set to 1. Differential expression was evaluated using ANOVA and Dunnett’s multiple comparison tests (a, p<0.01; b, p<0.001; c, p<0.0001; NS, not significant). (C) S3 reduces AR protein levels in patient-derived explants (PDEs). AR levels in tumours from 14 patients were measured by immunohistochemistry (IHC; left). Boxes show minimum and maximum (bottom and top lines, respectively) and mean (line within the boxes) values. A paired t test was used to compare AR levels in treated versus control samples (***p<0.001). Representative IHC images are shown on the right (scale bars represent 50 µm). (D) S3 enhances AR ubiquitylation. LNCaP cells were treated with indicated concentrations of S3 ±10 µM MG132, or 10 µM MG132 alone, for 24 hr prior to AR immunoprecipitation. Both immunoprecipitates and total protein inputs (1/30 of immunoprecipitates) were subjected to immunoblotting analysis for the indicated proteins. (E, F) Anti-cancer effects of combined Enz and S3 treatment in VCaP cells. Live (E) and dead (F) cells were measured by Trypan blue exclusion assays 4 days after treatment. Data represent the mean + SEM of triplicate samples and are representative of three independent experiments. (G) Anti-cancer effects of combined Enz and S3 treatment in V16D cells. Live cells (F) were measured as in (D) after 3 days of treatment; data are representative of three independent experiments.
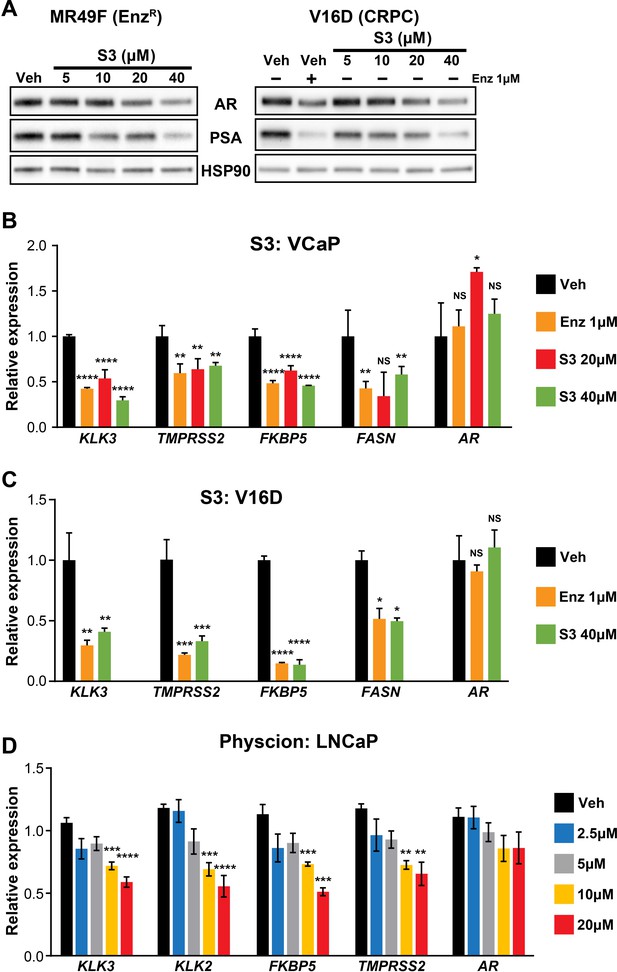
6PGD inhibitors suppress AR expression and activity.
(A) S3 decreases androgen receptor (AR) and PSA protein levels in MR49F (left) and V16D (right) cells. Protein was extracted from cells at 24 hr and assessed by western blotting. HSP90 is shown as a loading control. EnzR: enzalutamide-resistant. (B, C) S3 suppresses AR target gene expression in VCaP (B) and V16D (C) cells after 24 hr treatment. Expression is shown relative to GUSB and L19; vehicle (Veh) was set to 1. (D) Physcion suppresses AR target gene expression in LNCaP cells after 24 hr treatment. Expression is shown relative to GUSB and L19. Differential expression compared to vehicle (B–D) was determined using ANOVA and Dunnett’s multiple comparison tests (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; NS, not significant).
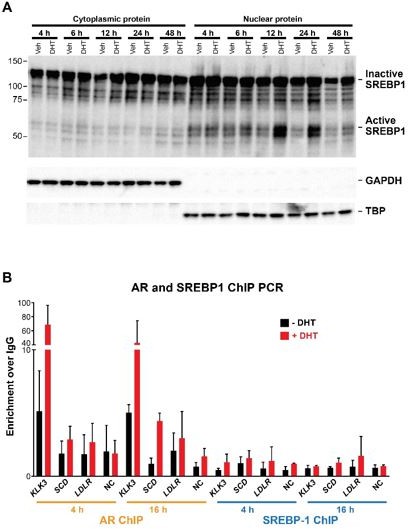
SREBP1 activation and ChIP PCR analysis for AR/SREBP1 in prostate cancer cells.
(A) LNCaP cells were cultured in the absence or presence of DHT (10nM) for varying timepoints up to 48 hours. Nuclear and cytoplasmic protein extracts were evaluated for the active (cleaved) form of SREBP1. (B) DHT-induced recruitment of AR or SREBP1 to canonical AR/SREBP1 binding sites after 4 or 16 hrs culture.

Influence of ectopic 6PGD overexpression on AR protein levels in stably transduced LNCaP cells at passages 6/9.
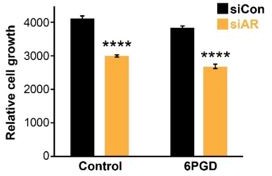
Effect of 6PGD overexpression on response of LNCaP prostate cancer cells to siAR treatment.
Cells stably transduced with control or 6PGD-overexpressing vectors were transfected with siCon or siAR and viable cell number measured after 6 days in culture. **** P<0.001 c.f. siCon.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo-sapiens) | LNCaP | ATCC | ATCC CRL-1740 (RRID:CVCL_1379) | |
| Cell line (Homo-sapiens) | VCaP | ATCC | ATCC CRL-2876 (RRID:CVCL_2235) | |
| Cell line (Homo-sapiens) | PC3 | ATCC | ATCC CRL-7934 (RRID:CVCL_0035) | |
| Cell line (Homo-sapiens) | 22RV1 | ATCC | ATCC CRL-2505 (RRID:CVCL_1045) | |
| Cell line (Homo-sapiens) | V16D | PMID:27046225 | Kind gift from Prof. Amina Zoubeidi | |
| Cell line (Homo-sapiens) | MR49F | PMID:27046225 | Kind gift from Prof. Amina Zoubeidi | |
| Transfected construct (Homo sapiens) | Negative control siRNA | Ambion; Thermo Fisher Scientific | AM4637 | |
| Transfected construct (Homo sapiens) | siAR | Thermo Fisher Scientific | Silencer Select 4390824 | |
| Transfected construct (Homo sapiens) | siAR | Thermo Fisher Scientific | Silencer Select 4390825 | |
| Transfected construct (Homo sapiens) | siAR | Thermo Fisher Scientific | Custom 4399665 | |
| Transfected construct (Homo sapiens) | si6PGD | Thermo Fisher Scientific | 4427038 | |
| Tansfected construct (Homo sapiens) | siSREBP1 | Dharmacon | ON-TARGETplus 6720 | |
| Antibody | ACC-1 [C83B10] (rabbit monoclonal) | Cell Signaling Technology, Inc | 3676 (RRID:AB_2219397) | Western blot: (1:1000) |
| Antibody | pACC-1 [Ser79] (rabbit polyclonal) | Cell Signaling Technology, Inc | 3661 (RRID:AB_330337) | Western blot: (1:1000) |
| Antibody | β-Actin (AC-15) (mouse monoclonal) | Sigma Aldrich | A5441 (RRID:AB_476744) | Western blot: (1:1000) |
| Antibody | AR-N20 (rabbit polyclonal) | Santa Cruz Biotechnology Inc | sc-816 (RRID:AB_1563391) | Western blot: (1:1000) |
| Antibody | AR (rabbit monoclonal) | Abcam | ab108341 (RRID:AB_10865716) | Immunohistochemistry: (1:200) |
| Antibody | AR (mouse monoclonal) | Santa Cruz Biotechnology Inc | sc-7305 (RRID:AB_626671) | Immunoprecipitation: (0.2 µg) |
| Antibody | AMPKα (rabbit polyclonal) | Cell Signaling Technology, Inc | 2532 (RRID:AB_330331) | Western blot: (1:1000) |
| Antibody | pAMPKα [Thr172] 40H9 (rabbit monoclonal) | Cell Signaling Technology, Inc | 2535 (RRID:AB_331250) | Western blot: (1:1000) |
| Antibody | GAPDH (HuCAL recombinant) | BioRad | 12004168 | Western blot: (1:1000) |
| Antibody | Hsp90 (rabbit polyclonal) | Cell Signaling Technology, Inc | 4874 (RRID:AB_2121214) | Western blot: (1:1000) |
| Antibody | Ki67 (mouse monoclonal) | Agilent Technologies | M724001-2 (RRID:AB_2631211) | Immunohistochemistry: (1:200) |
| Antibody | P70 S6 Kinase (49D7) (rabbit monoclonal) | Cell Signaling Technology, Inc | 2708 (RRID:AB_390722) | Western blot: (1:1000) |
| Antibody | pP70 S6 Kinase [Thr389] (rabbit polyclonal) | Cell Signaling Technology, Inc | 9205S (RRID:AB_330944) | Western blot: (1:2000) |
| Antibody | 6PGD (rabbit polyclonal) | ThermoFisher Scientific | PA5-21376 (RRID:AB_11153623) | Western blot: (1:1000) |
| Antibody | 6PGD (rabbit polyclonal) | Sigma Aldrich | HPA031314 (RRID:AB_10610278) | Immunohistochemistry: (1:800) |
| Antibody | PSA (rabbit polyclonal) | ProteinTech Group | 10679-1-AP (RRID:AB_2134244) | Western blot: (1:1000) |
| Antibody | S6 (5G10) (rabbit monoclonal) | Cell Signaling Technology, Inc | 2217 (RRID:AB_331355) | Western blot: (1:1000) |
| Antibody | pS6 [Ser235/236] (rabbit polyclonal) | Cell Signaling Technology, Inc | 2211 (RRID:AB_331679) | Western blot: (1:1000) Immunohistochemistry: (1:200) |
| Antibody | Ubiquitin (mouse monoclonal) | Genesearch | 3936 | Western blot: (1:1000) |
| Antibody | Goat Anti-Rabbit (Biotinylated) | Agilent Technologies | E043201-8 | Immunohistochemistry: (1:400) |
| Sequence-based reagent | AR For | This paper | qRT-PCR primers | CAACTCCTTCAGCAACAGCA |
| Sequenced-based reagent | AR Rev | This paper | qRT-PCR primers | TCGAAGTGCCCCCTAAGTAA |
| Sequence-based reagent | FKBP5 For | This paper | qRT-PCR primers | AAAAGGCCAAGGAGCACAAC |
| Sequenced-based reagent | FKBP5 Rev | This paper | qRT-PCR primers | TTGAGGAGGGGCCGAGTTC |
| sSquence-based reagent | GAPDH For | This paper | qRT-PCR primers | TGCACCACCAACTGCTTAGC |
| Sequenced-based reagent | GAPDH Rev | This paper | qRT-PCR primers | GGCATGGACTGTGGTCATGAG |
| Sequence-based reagent | GUSB For | This paper | qRT-PCR primers | CGTCCCACCTAGAATCTGCT |
| sSquenced-based reagent | GUSB Rev | This paper | qRT-PCR primers | TTGCTCACAAAGGTCACAGG |
| Sequence-based reagent | KLK2 For | This paper | qRT-PCR primers | GGTGGCTGTGTACAGTCATGGAT |
| Sequenced-based reagent | KLK2 Rev | This paper | qRT-PCR primers | TGTCTTCAGGCTCAAACAGGTTG |
| Sequence-based reagent | KLK3 For | This paper | qRT-PCR primers | ACCAGAGGAGTTCTTGACCCCAAA |
| Sequenced-based reagent | KLK3 Rev | This paper | qRT-PCR primers | CCCCAGAATCACCCGAGCAG |
| Sequence-based reagent | L19 For | This paper | qRT-PCR primers | TGCCAGTGGAAAAATCAGCCA |
| Sequenced-based reagent | L19 Rev | This paper | qRT-PCR primers | CAAAGCAAATCTCGACACCTTG |
| Sequenced-based reagent | PGD For | This paper | qRT-PCR primers | CACAGCAGGGTTCTCCAGTT |
| Sequenced-based reagent | PGD Rev | This paper | qRT-PCR primers | GTCAGTGGTGGAGAGGAAGG |
| Sequenced-based reagent | PPIA For | This paper | qRT-PCR primers | GCATACGGGTCCTGGCAT |
| Sequenced-based reagent | PPIA Rev | This paper | qRT-PCR primers | ACATGCTTGCCATCCAACC |
| Sequenced-based reagent | TMPRSS2 For | This paper | qRT-PCR primers | GACCAAGAACAATGACATTGCG |
| Sequenced-based reagent | TMPRSS2 Rev | This paper | qRT-PCR primers | GTTCTGGCTGCAGCATCATG |
| Sequenced-based reagent | TUBA1B For | This paper | qRT-PCR primers | CCTTCGCCTCCTAATCCCTA |
| Sequenced-based reagent | TUBA1B Rev | This paper | qRT-PCR primers | CCGTGTTCCAGGCAGTAGA |
| Chemical compound, drug | Dihydrotestosterone | Sigma Aldrich | Cas#: 521-18-6 | |
| Chemical compound, drug | Enzalutamide | Selleck Chemicals | Cat#: S1250 | |
| Chemical compound, drug | Apalutamide | Selleck Chemicals | Cat#: S2840 | |
| Chemical compound, drug | Darolutamide | Selleck Chemicals | Cat#: S7559 | |
| Chemical compound, drug | S3 | Sigma Aldrich | Cat#: R164046 | |
| Chemical compound, drug | Physcion | Sigma Aldrich | Cat#: 93893 | |
| Chemical compound, drug | Trolox | Selleck Chemicals | Cat#: S3665 | |
| Chemical compound, drug | 1,2-13C2 glucose | Sigma Aldrich | Cat#: 453188 | |
| Commercial assay or kit | RNeasy Mini extraction kit | Qiagen | Cat#: 74104 | |
| Commercial assay or kit | iScriptTM cDNA Synthesis kit | Bio-Rad | Cat#: 1708890 | |
| Commercial assay or kit | NEXTflex Rapid Illumina Directional RNA-Seq Library Prep Kits | Perkin-Elmer | Cat#: NOVA-5138 | |
| Commercial assay or kit | CyQuant Assay Cell Proliferation Assays | Thermo Fisher Scientific | Cat#: C7026 | |
| Commercial assay or kit | CellROX Orange Flow Cytometry Assay Kits | Life Technologies | Cat#: C10493 | |
| Other | Lipofectamine RNAiMAX transfection reagent | Thermo Fisher Scientific | 13778075 | |
| Software, algorithm | GraphPad Prism | GraphPad Software, Inc. | Prism V7 RRID:SCR_002798 | |
| Software, algorithm | R | R Core Team (2019) | R version 3.6.2 RRID:SCR_001905 | |
| Software, algorithm | ImageJ analysis software | NIH | ImageJ RRID:SCR_003070 | |
| Software, algorithm | TraceFinder v5.0 | Thermo Fisher Scientific | OPTON-30688 |
Additional files
-
Supplementary file 1
Primers used for quantitative reverse transcription PCR (qRT-PCR).
- https://cdn.elifesciences.org/articles/62592/elife-62592-supp1-v2.docx
-
Supplementary file 2
Antibodies (primary and secondary) used for western blotting and immunohistochemistry (IHC).
- https://cdn.elifesciences.org/articles/62592/elife-62592-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62592/elife-62592-transrepform1-v2.docx
-
Source data 1
Western blot source data, including all individual Western blot images and a summary file.
- https://cdn.elifesciences.org/articles/62592/elife-62592-supp3-v2.zip





