Calcium depletion challenges endoplasmic reticulum proteostasis by destabilising BiP-substrate complexes
Figures
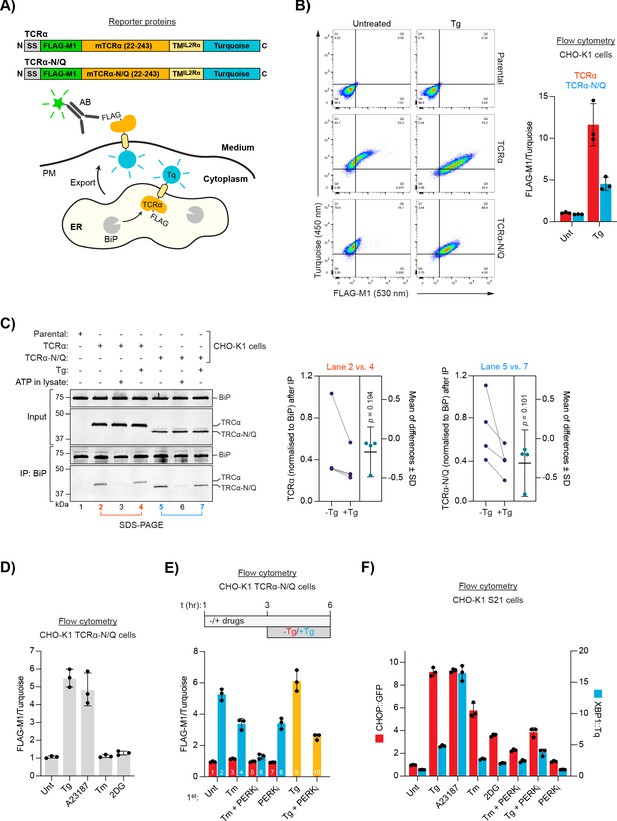
ER Ca2+ depletion-dependent trafficking of TCRα-based reporter proteins.
(A) Schematic representation of reporter proteins and detection principles. Signal sequence (SS); FLAG-M1 epitope tag; mouse T-cell antigen receptor α chain (mTCRα); transmembrane domain of the interleukin-2 receptor α subunit (TMIL2Rα); monomeric Turquoise (Tq); endoplasmic reticulum (ER); plasma membrane (PM). (B) Flow cytometry analysis of TCRα cell surface exposure. CHO-K1 cell lines stably expressing TCRα or glycosylation-deficient TCRα-N/Q were left untreated or exposed to Thapsigargin (Tg) for 3 hr before analysis. Dot plots of a representative experiment are shown, along vertical and horizontal guides to facilitate comparisons between the different plots. Graph: reporter surface exposure is plotted as the ratio between the FLAG-M1 and Turquoise median fluorescence signal (derived from all the cells scanned, relative to a data point of TCRα in untreated cells arbitrarily set to 1). Bars represent mean ± SD of the median signal from three independent experiments. Figure 1—source data 1. (C) Co-immunoprecipitation (IP) of TCRα reporter proteins with BiP from stable CHO-K1 cell lines (as in ‘B') analysed by SDS-PAGE and immunoblotting. Where indicated cells were treated with Tg for 3 hr prior to lysis. ATP was depleted from samples during lysis to stabilise BiP-substrate interactions except from the sample marked with ‘ATP’ to which additional ATP was added instead. Graph: quantification of TCRα and TCRα-N/Q signals normalised to BiP after IP from untreated (-Tg) and Tg-treated (+Tg) cells and mean of differences ± SD from four independent experiments. A two-tailed, paired, parametric t-test was performed. (D–E) Flow cytometry analysis as in ‘B’. (D) Cells were treated for 3 hr, as indicated, before analysis [untreated (Unt); tunicamycin (Tm); 2-deoxyglucose (2DG)]. Figure 1—source data 2. (E) Cells were exposed to the compounds indicated. After 3 hr, the medium was replaced by medium containing the same compounds without (red) or with additional Tg (blue) for further 3 hr before analysis. Samples 9 and 10 (yellow bars) contained the same amount of Tg before and after medium exchange. PERK inhibitor (PERKi). Bars: mean ± SD from three independent experiments. Figure 1—source data 3. (F) Flow cytometry analysis of a UPR reporter CHO-K1 cell line (S21) carrying a predominantly PERK-controlled CHOP::GFP marker and a predominantly IRE1-controlled XBP1::Turquoise marker. The cells were treated for 6 hr, as indicated. Median fluorescence signals relative to untreated cells are shown. Bars: mean ± SD from three independent experiments. Figure 1—source data 4.
-
Figure 1—source data 1
Source data for the flow cytometry experiment shown in Figure 1B.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Source data for the flow cytometry experiment shown in Figure 1D.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig1-data2-v2.xlsx
-
Figure 1—source data 3
Source data for the flow cytometry experiment shown in Figure 1E.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig1-data3-v2.xlsx
-
Figure 1—source data 4
Source data for the flow cytometry experiment shown in Figure 1F.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig1-data4-v2.xlsx
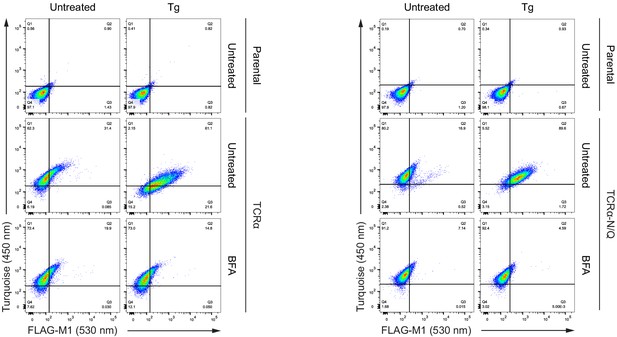
Brefeldin A blocks exposure of the T-cell antigen receptor α (TCRα) reporter on the cell surface.
Dot plots of a representative flow cytometry analysis of CHO-K1 cells stably expressing TCRα or TCRα-N/Q after treatment with thapsigargin (Tg) and brefeldin A (BFA) for 3 hr, as indicated.
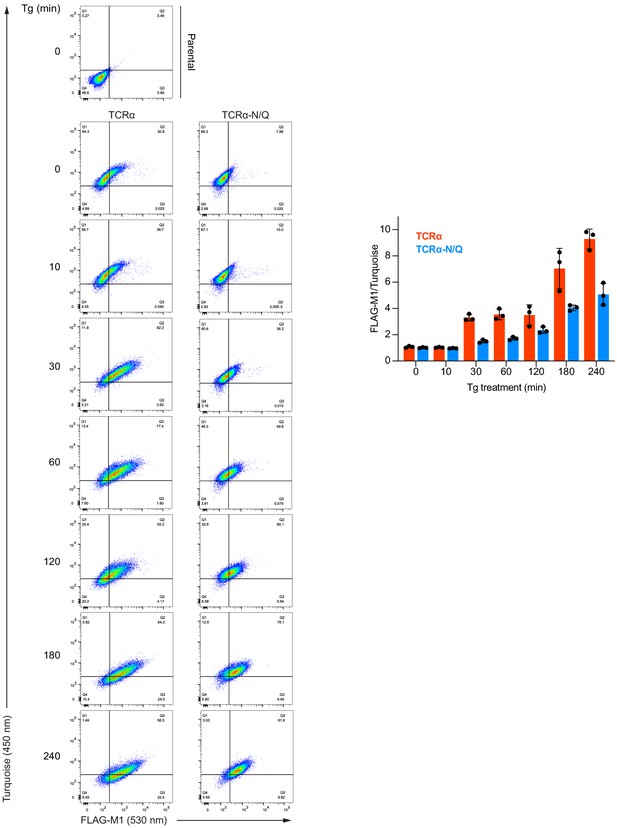
Time course of exposure of the T-cell antigen receptor α (TCRα) reporter on the cell surface during ER Ca2+ depletion.
Dot plots of a representative flow cytometry analysis of CHO-K1 cells stably expressing TCRα or TCRα-N/Q after treatment with thapsigargin (Tg) for indicated periods. Graph: reporter surface exposure is given as the ratio between the FLAG-M1 and turquoise median fluorescence signal relative to untreated cells. Bars: mean ± SD bars are shown from three independent experiments. Source data are provided in Supplementary file 1.
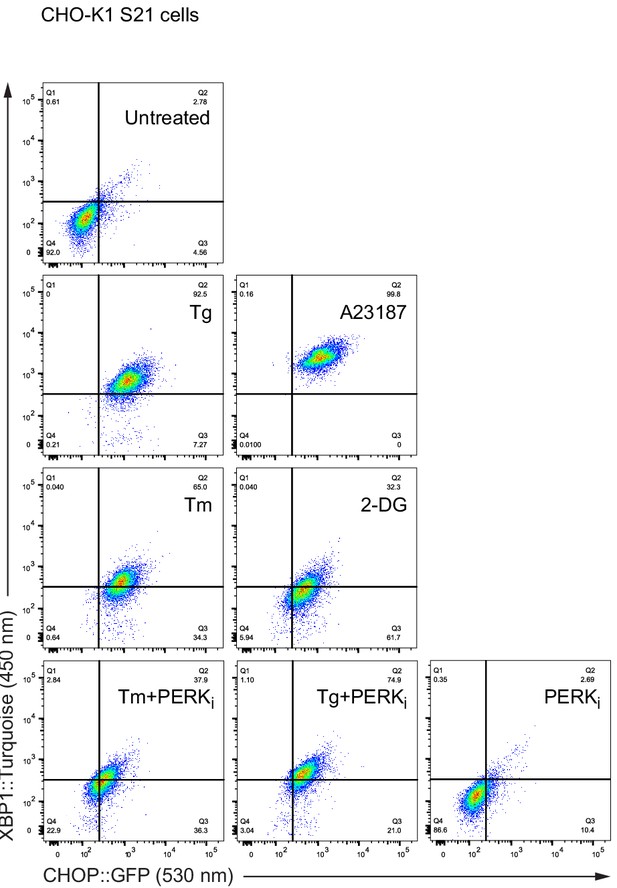
Endoplasmic reticulum (ER) Ca2+ depletion and ER stress induce the unfolded protein response (UPR).
Dot plots of a representative flow cytometry experiment using CHO-K1 CHOP::GFP XBP1::Tq cells (S21). The cells were treated for 6 hr, as indicated. Related to Figure 1F.
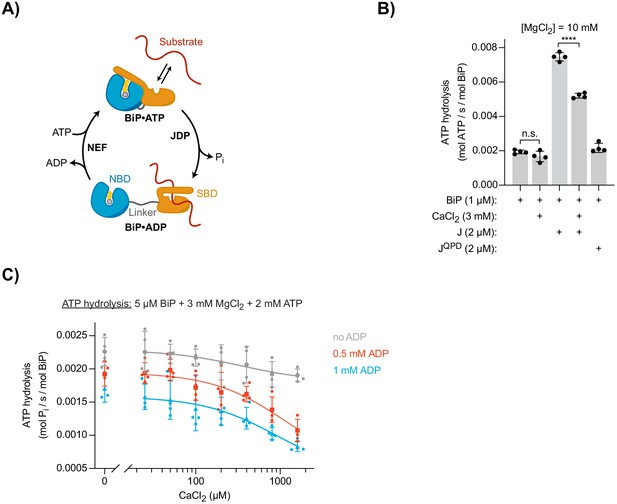
Effect of Ca2+ on BiP’s ATPase activity.
(A) Schematic representation of BiP’s chaperone cycle. Unfolded substrate protein (red); nucleotide binding domain (NBD); substrate binding domain (SBD); interdomain linker (grey); orthophosphate (Pi); J-domain protein (JDP); nucleotide exchange factor (NEF). (B) ATP hydrolysis by purified BiP measured with a NADH-based ATPase assay. All samples contained 10 mM MgCl2 and 1 µM BiP. Where indicated CaCl2 (3 mM) and wild-type (J) or mutant J-domain (JQPD) were added (both at 2 µM). Bars: mean ± SD from four independent experiments. ****p<0.0001, 95% CI −0.002605 to −0.001912; two-tailed, unpaired, parametric t-test. Figure 2—source data 1. (C) ATPase activity of BiP measured by detection of ATP hydrolysis product (Pi) accumulation using a malachite green-based assay. All samples contained 3 mM MgCl2 and 2 mM ATP. CaCl2 was titrated (0–1.6 mM). Where indicated samples contained 0.5 mM (red) and 1 mM (blue) ADP. Bold symbols represent mean values ± SD derived from individual data points (staggered dots). Best-fit lines are shown. Figure 2—source data 2.
-
Figure 2—source data 1
Source data and calculated rates for the ATPase experiment shown in Figure 2B and Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Source data and calculated rates for the ATPase experiment shown in Figure 2C.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig2-data2-v2.xlsx
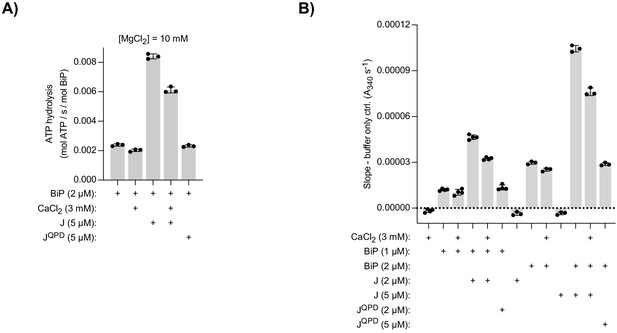
Effect of Ca2+ on BiP’s ATPase activity measured by a NADH-based assay.
ATP hydrolysis by purified BiP measured with a NADH-based ATPase assay that detects ADP production. All samples contained 10 mM MgCl2. Where indicated CaCl2 (3 mM) and wild-type (J) or mutant J-domain (JQPD) were added. (A) Same as Figure 2B but with higher protein concentrations. Bars: mean ± SD from three independent experiments. (B) Slopes of time-dependent NADH signals (absorbance at 340 nm; A340) of experiments shown in ‘A’ and Figure 2B. The value of the ‘buffer only’ control reaction was subtracted in each experiment. Bars: mean ± SD from at least three independent experiments. Note: the reaction velocity correlates with the BiP concentration.
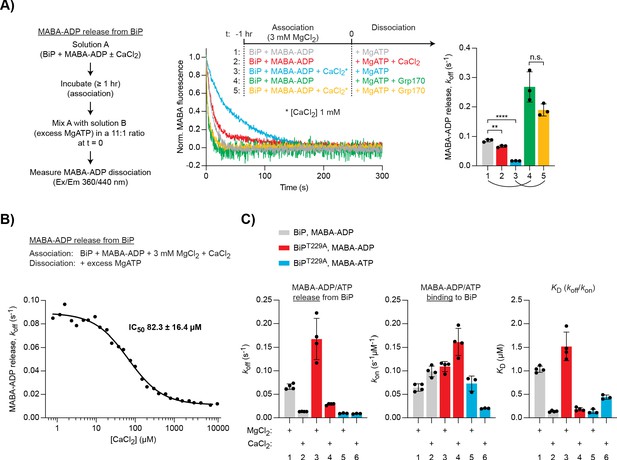
Differential effect of Ca2+ on ATP and ADP binding to BiP.
(A) Representative plot of fluorescence against time of pre-formed complexes of MABA-ADP and BiP (each 1.3 µM) challenged at t = 0 with excess of MgATP (1.5 mM) to reveal nucleotide release. All solutions contained 3 mM MgCl2. CaCl2 (1 mM) and Grp170 (1.3 µM) were added at t = −1 hr or t = 0 as indicated. Graph: mean MABA-ADP dissociation rate constants (koff) ± SD from three independent experiments (the curved lines connecting conditions 1 and 4 as well as 3 and 5 emphasise the effect of Ca2+ on the activity of Grp170). Figure 3—source data 1. (B) MABA-ADP dissociation rates from BiP plotted against [CaCl2] of a representative experiment. The experiment was performed as in ‘A’. All samples contained MgCl2 (3 mM). CaCl2 was present at increasing concentrations during nucleotide binding (solution A). The half maximal inhibitory concentration (IC50) of CaCl2 (mean ± SD) was calculated from three independent experiments. **p=0.0036, 95% CI −0.02899 to −0.01086; ****p<0.0001, 95% CI −0.07659 to −0.06222; two-tailed, unpaired, parametric t-test. Figure 3—source data 2. (C) Effect of Ca2+ on affinities of nucleotides for BiP. The dissociation rates of MABA-ADP and MABA-ATP from wildtype or ATPase-impaired T229A mutant BiP were measured as in ‘A’, whereby either MgCl2 or CaCl2 (1 mM) were present throughout the experiment (solutions A and B). The association rate constants (kon) were measured upon addition of MABA-labelled nucleotides to BiP in the presence of either divalent cation. The dissociation constants (KD) were calculated based on the rate constants (koff/kon). Nucleotide-free proteins were used. Bars: mean ± SD. All the data points from ≥3 independent experiments are shown. Source data and a summary of the calculated values are presented in Figure 3—source data 3.
-
Figure 3—source data 1
Source data and calculated rates for the MABA-ADP release experiment shown in Figure 3A.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Source data and calculated rates for the MABA-ADP release experiment shown in Figure 3B.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig3-data2-v2.xlsx
-
Figure 3—source data 3
Source data and summary of calculated values for the nucleotide binding experiments shown in Figure 3C.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig3-data3-v2.xlsx
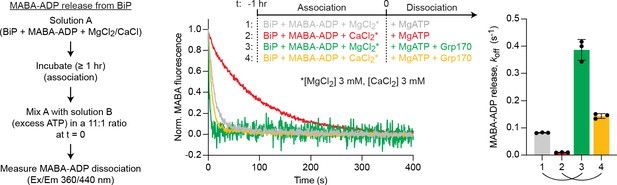
Dissociation BiP•MABA-ADP complexes formed in the presence of either Mg2+ or Ca2+.
Representative plot of fluorescence against time of complexes of MABA-ADP and BiP (each 1.3 µM) formed in the presence of either MgCl2 or CaCl2 (each 3 mM). Excess of MgATP (1.5 mM) without or with Grp170 (1.3 µM) was added at t = 0 to reveal nucleotide release. Graph: mean MABA-ADP dissociation rate constants (koff) ± SD from three independent experiments. Final concentrations after mixing are given. Note: the stronger stimulatory effect of Grp170 compared to Figure 3A may be due to variations in the experimental conditions. Source data are provided in Supplementary file 2.
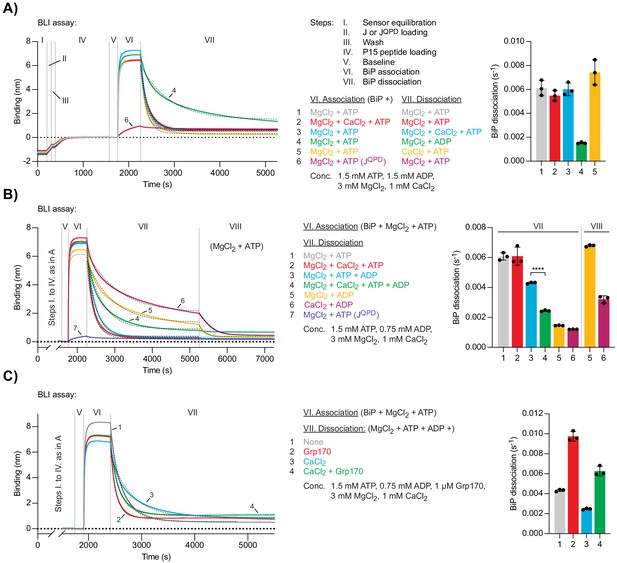
Ca2+ affects ATP-dependent dissociation of substrates from BiP.
(A) A representative plot of the Bio-Layer interferometry (BLI) signal against time. The individual steps of the experiment (I-VII) are indicated: following an initial equilibration step (I) biotinylated wild-type or QPD mutant J-domain protein was immobilised corresponding to an interference signal difference of ∼0.4 nm (II). After a wash step (III) the sensors were saturated with biotinylated P15 BiP substrate peptide (IV). After a stable baseline signal was established (V) the sensors were transferred into solutions containing BiP to measure association in the presence of ATP (VI). Dissociation of BiP from the sensor was measured in protein-free solutions containing either ADP or ATP (VII). The composition of solutions in steps VI and VII as well as the concentrations of the variable components are indicated. (A) MgCl2 and/or CaCl2 were present where indicated. Dashed lines represent single exponential fit curves. Graph: mean dissociation rate constants ± SD from three independent experiments. Figure 4—source data 1. (B) Same as ‘A’ but with identical solutions for all sensors in step VI and varying solution compositions in step VII. In the additional step (VIII) the sensors were introduced into solutions containing ATP and MgCl2. ****p<0.0001, 95% CI −0.00198822 to −0.00174311, two-tailed, unpaired, parametric t-test. Figure 4—source data 2. (C) Same as ‘B’ (reactions 3 and 4) with Grp170 present in step VII where indicated. Figure 4—source data 3.
-
Figure 4—source data 1
Source data and calculated rates for the BLI experiment shown in Figure 4A.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Source data and calculated rates for the BLI experiment shown in Figure 4B.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Source data and calculated rates for the BLI experiment shown in Figure 4C.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig4-data3-v2.xlsx
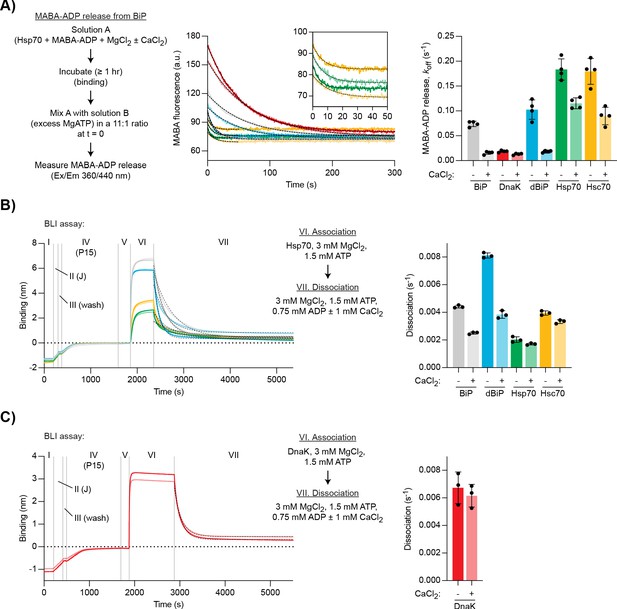
Ca2+ selectively stabilises BiP•ADP compared with cytoplasmic Hsp70s.
(A) Representative plot of fluorescence against time of complexes of MABA-ADP and indicated full-length Hsp70 chaperones (each 1.3 µM) formed without or with CaCl2 (1 mM) (dBiP, Drosophila BiP). All solutions contained MgCl2 (3 mM). Excess of MgATP (1.5 mM) was added at t = 0 to reveal MABA-ADP release. Final concentrations are given. Graph: mean MABA-ADP dissociation rate constants ± SD from four independent experiments. Figure 5—source data 1. (B–C) A representative plot of the Bio-Layer interferometry (BLI) signal against time. The experiments were performed as in Figure 4A with the indicated Hsp70s. Dissociation of Hsp70s from the sensor was measured in protein-free solutions (VII). The composition of solutions in steps VI and VII as well as the concentrations of the variable components are indicated. (B) Analysis of eukaryotic Hsp70s. Dashed lines represent single exponential fit curves. Graph: mean dissociation rate constants ± SD from three independent experiments. Figure 5—source data 2. (C) Same as ‘B’ but with E. coli DnaK. Figure 5—source data 3.
-
Figure 5—source data 1
Source data and calculated rates for the MABA-ADP release experiment shown in Figure 5A.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Source data and calculated rates for the BLI experiment shown in Figure 5B.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig5-data2-v2.xlsx
-
Figure 5—source data 3
Source data and calculated rates for the BLI experiment shown in Figure 5C.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig5-data3-v2.xlsx

Structural analysis of Ca2+ and ADP binding to Hsp70 nucleotide binding domains (NBDs).
(A) Ribbon diagram of the aligned crystal structures of BiP (yellow), Hsp70 (blue), and Hsc70 (pink) NBDs in complex with Ca2+ and ADP. The subdomains IA-IIB are indicated. Inset: coordination of Ca2+ ions (green) by water (small spheres) and ADP in the nucleotide binding cleft. (B) Nucleotide and metal ion ligands of BiP(NBD)•Ca2+•ADP (as in ‘A’) and bovine Hsc70(NBD) •Mg2+•ADP (PDB 1BA1) or human Hsp70-2(NBD)•Mg2+•ADP•Pi (PDB 3I33) based on nucleotide alignment. ADP, inorganic phosphate (Pi), Ca2+ (green sphere), and Mg2+ (grey sphere) are shown. (C) MABA-ADP dissociation rates from the indicated NBDs plotted against [CaCl2] of a representative experiment. The experiment was performed as in Figure 3A. All samples contained MgCl2 (3 mM). CaCl2 was present at increasing concentrations during NBD•MABA-ADP complex formation. Bars: the half maximal inhibitory concentration (IC50) of CaCl2 (mean ± SD) was calculated from three independent experiments. Figure 6—source data 1. (D) Melting temperatures (Tm) of the indicated NBDs (at 5 µM) were measured by differential scanning fluorimetry (DSF) in the presence of MgCl2 or CaCl2 (each 6 mM) without or with ADP (4 mM). Top: representative melt curves with their negative first derivatives (RFU, relative fluorescence units). Left bar graph: mean Tm ± SD of three independent experiments. Right bar graph: difference in Tm (ΔTm) between MgADP and CaADP containing samples. Figure 6—source data 2.
-
Figure 6—source data 1
Source data and calculated rates for the MABA-ADP release experiment shown in Figure 6C.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Source data and calculated melting temperatures for the DSF experiment shown in Figure 6D.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig6-data2-v2.xlsx
Structural details of nucleotide binding domains (NBDs) and their ligands.
(A) Polder OMIT maps (grey mesh) of BiP, Hsp70, and Hsc70 NBD ligands omitting ADP (sticks), Ca2+ (green sphere), and water molecules (red spheres) contoured at 3.80 RMSD and including density within 1.6 Å of all atoms shown. (B) Ribbon diagram of NBDs of (i) BiP(NBD)•Ca2+•ADP aligned with (ii) BiP(NBD)apo (PDB 3LDN) (RMSD i vs. ii 0.75 over 336 Cα) and (iii) BiP(28-549)apo (PDB 6HAB) (RMSD i vs. iii 0.776 over 311 Cα). The nucleotide is shown in surface representation and the Ca2+ ion (green sphere) is indicated. (C) Nucleotide and metal ion ligands of BiP(NBD)•Ca2+•ADP and DnaK•Mg2+•ATP (PDB 4B9Q) based on nucleotide alignment. ADP, ATP, Ca2+ (green sphere), and Mg2+ (grey sphere) are shown. (D) Nucleotide and metal ion ligands of human Hsp70-2(NBD)•Mg2+ADP•Pi (PDB 3I33) and human Hsp70(NBD)•Ca2+•ADP•Pi (PDB 1S3X) based on nucleotide alignment. ADP, Ca2+ (green sphere), and Mg2+ (grey sphere) are shown.
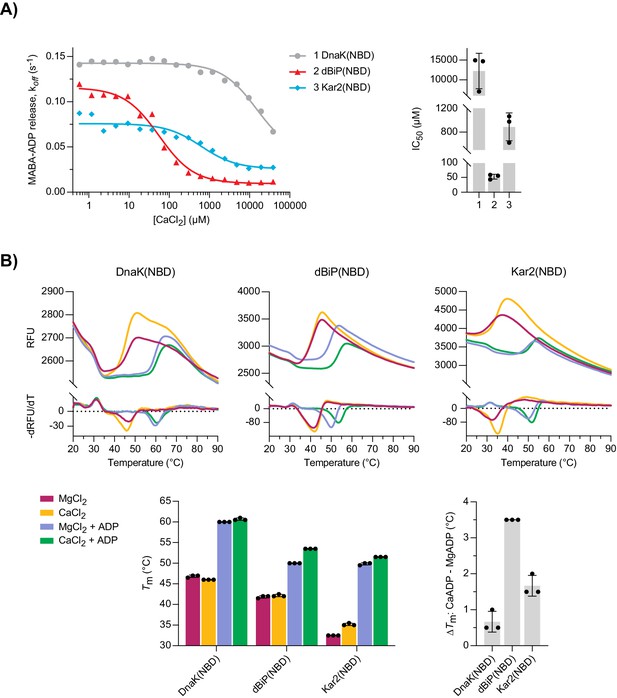
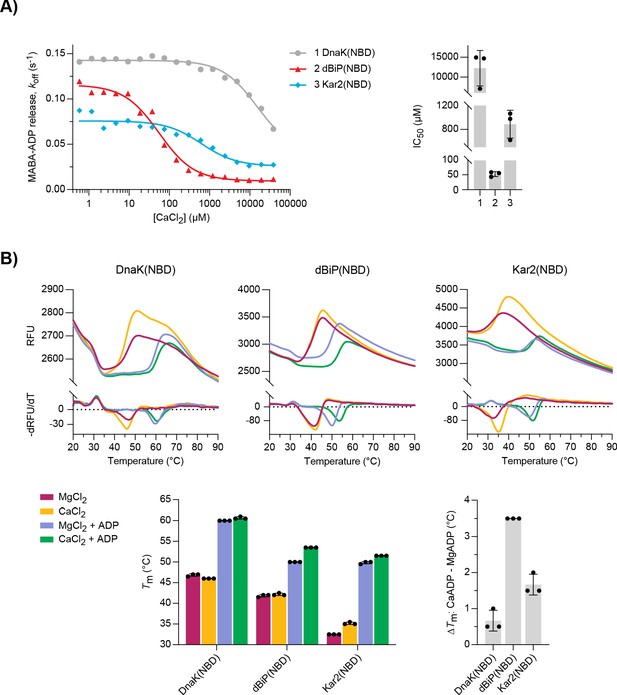
Characterisation of nucleotide binding domains (NBDs) of Drosophila BiP, DnaK, and Kar2.
(A) MABA-ADP dissociation rates from the indicated NBDs plotted against [CaCl2] of a representative experiment. The experiment was performed as in Figure 3A. All samples contained MgCl2 (3 mM). CaCl2 was present at increasing concentrations during NBD•MABA-ADP complex formation. Graph: the half maximal inhibitory concentration (IC50) of CaCl2 (mean ± SD) was calculated from three independent experiments. Source data are provided in Supplementary file 3. (B) Melting temperatures (Tm) of the indicated NBDs (at 5 µM) were measured by differential scanning fluorimetry (DSF) in the presence of MgCl2 or CaCl2 (each 6 mM) without or with ADP (4 mM). Top: representative melt curves with their negative first derivatives (RFU, relative fluorescence units). Left bar graph: mean Tm ± SD of three independent experiments. Right bar graph: difference in Tm (ΔTm) between MgADP- and CaADP-containing samples. Source data are provided in Supplementary file 4.
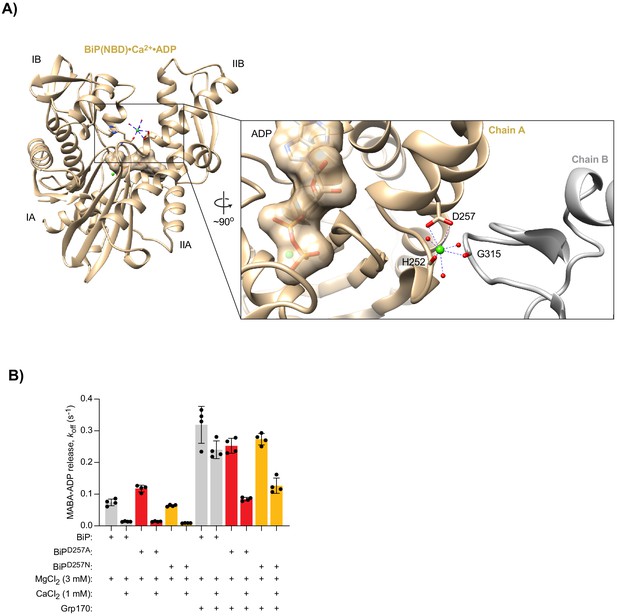
The surface-exposed second Ca2+- binding site in BiP’s NBD does not significantly contribute to the effect of Ca2+ on ADP release.
(A) Ribbon diagram showing coordination of a surface-bound Ca2+ ion (green sphere) in the BiP(NBD) structure, involving contacts to the crystallographic neighbouring molecule (chain B; grey). Relevant water molecules (red spheres) as well as ADP (surface representation) and Ca2+ within the nucleotide binding cleft are indicated. A Ca2+ ion was also identified at the corresponding surface position in structures of Hsp70(NBD) and Hsc70(NBD). In neither case does this Ca2+ ion participate in coordination of the nucleotide. (B) Dissociation of pre-formed complexes of MABA-ADP (1.3 µM) and wild-type BiP or BiP mutants (each 1.3 µM) after exposure to excess of MgATP (1.5 mM) without or with Grp170 (1.3 µM). All solutions contained 3 mM MgCl2. Where indicated CaCl2 (1 mM) was present during complex formation. Mean MABA-ADP dissociation rate constants (koff) ± SD from four independent experiments. Source Data.
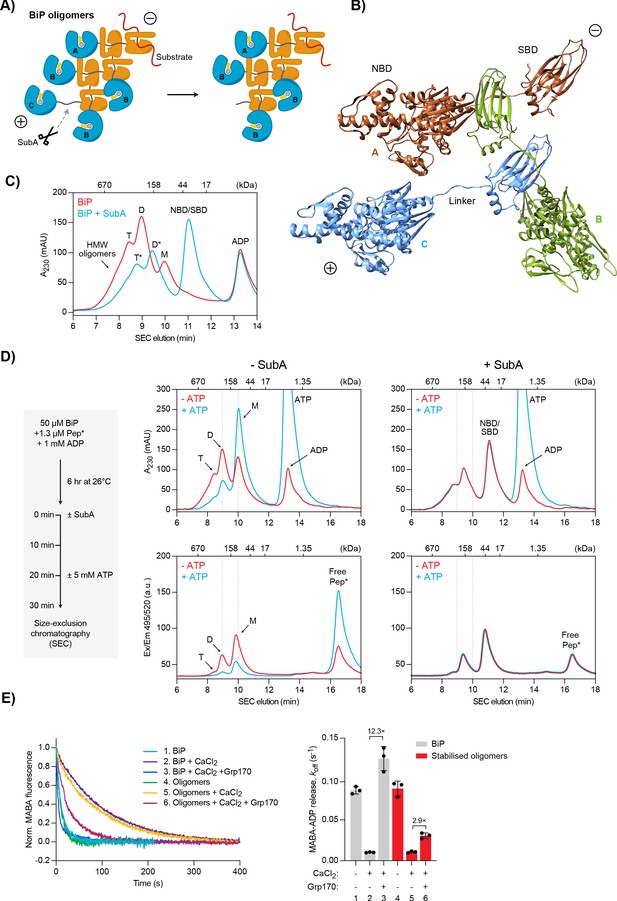
Effect of Ca2+ on BiP oligomers in vitro.
(A) Schematic representation of BiP oligomers. Unfolded substrate protein (red) and the distinct types of BiP protomers (A, B, and C) are shown. Oligomers are directional assemblies with minus (-) and plus (+) ends. The cleavage site for the BiP linker-specific protease SubA (scissors) is only accessible on (+)-end protomer (C). The predicted inert BiP oligomer lacking the C protomer nucleotide binding domain (NBD; a result of SubA cleavage) is shown on the right. (B) Ribbon diagram of apo BiPT229A-V461F oligomers formed by typical substrate interactions between protomers. Three protomers (labelled A-C according to ‘A’) are presented in different colours. Note the unusual C- to N-terminal engagement of BiP’s interdomain linker as a substrate bound to the substrate binding domain (SBD) of an adjacent BiP molecule. (C) Size-exclusion chromatography (SEC) elution profile of BiP. BiP was incubated with ADP (1 mM) and where indicated treated with SubA before SEC. Monomeric (M), dimeric (D), and trimeric BiP (T) is indicated. Heterogenous high-molecular weight (HMW) BiP complexes elute early as a shoulder. Note that the peaks of proteolytic cleavage products originating from trimers and dimers (*) are shifted to later elution times. Also note the disappearance of BiP monomers and a new peak of the individual NBD and SBD after treatment with SubA. Figure 7—source data 1. (D) SEC elution profile as in ‘C’ of BiP and substrate peptide. BiP was incubated with trace amounts of fluorescently labelled substrate peptide (NR; Pep*) in the presence of ADP at indicated concentrations. Where indicated the samples were exposed at t = 0 to SubA for 30 min before SEC. Where indicated excess of ATP was added 10 min before SEC. Peptide bond absorbance at 230 nm (A230) and the fluorescence signal of the labelled peptide were recorded separately. Figure 7—source data 2. (E) Representative plot of fluorescence against time of complexes of MABA-ADP and BiP or purified, stabilised BiP oligomers (each 1.3 µM) formed in the presence of either MgCl2 or CaCl2 (each 3 mM). Excess of ATP (1.5 mM) without or with Grp170 (1.3 µM) was added at t = 0 to reveal nucleotide release. Graph: mean MABA-ADP dissociation rate constants (koff) ± SD from three independent experiments. Final concentrations are given. Figure 7—source data 3.
-
Figure 7—source data 1
Source data for the SEC traces shown in Figure 7C.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig7-data1-v2.xlsx
-
Figure 7—source data 2
Source data for the SEC traces shown in Figure 7D.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig7-data2-v2.xlsx
-
Figure 7—source data 3
Source data and calculated rates for the MABA-ADP release experiment shown in Figure 7E.
- https://cdn.elifesciences.org/articles/62601/elife-62601-fig7-data3-v2.xlsx
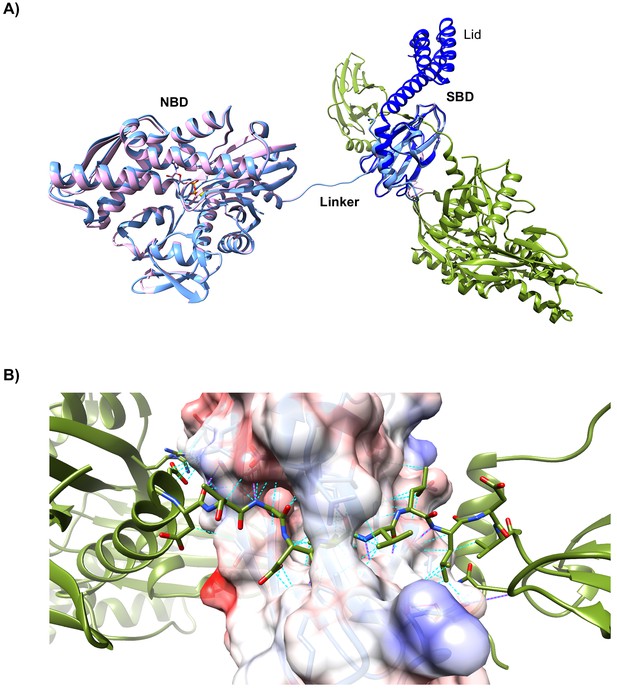
Structural features of oligomeric BiP.
(A) Superposition of the asymmetric unit of a lid-truncated, ADP-bound oligomeric BiP (pink) onto the biological (dimeric) assembly of oligomeric apo-BiP protomers (light blue and green, as in Figure 7B) reveals a highly similar conformation. ADP is shown in stick form with its coordinating K+ ion (yellow sphere) in the nucleotide binding domain (NBD). The isolated substrate binding domain (SBD) of DnaK, including the C-terminal lid structure in the closed conformation (dark blue; PDB 1DKZ), is also aligned and reveals only minor conformational differences to the lid-truncated SBD of oligomeric BiP. (B) Close-up view of the interdomain linker of one BiP protomer (green) bound to the SBDβ subdomain of another BiP protomer as in ‘A’. The surface of the linker-binding SBD is shown with translucent coulombic surface colouring. All residues participating in trans-protomer interactions are shown as sticks. Hydrogen bonds are highlighted with solid magenta pseudo bonds and van der Waals interactions are represented by cyan dashed pseudo bonds. Note that the majority of the linker side chains occupy the hydrophobic surface of the substrate- binding groove and most polar bonds are to mainchain amide atoms of the β-sheet scaffold of the SBD. Moreover, the protomer whose SBD-bound linker is shown forms a number of additional interactions with the surface of the SBDβ, in which it is engaged, outside of the canonical substrate- binding groove. These peripheral contacts may contribute to the orientation and stabilisation of the oligomeric BiP structure.
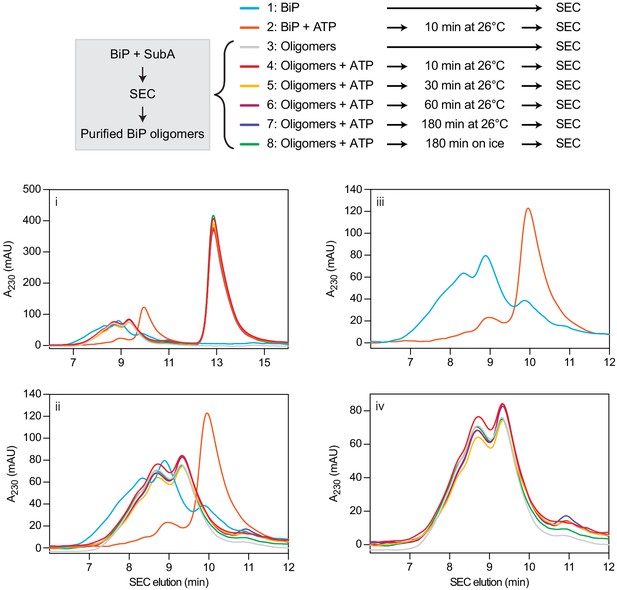
Stability of purified BiP oligomers after cleavage with SubA assessed by size-exclusion chromatography.
Size-exclusion chromatography (SEC) elution profile of BiP or purified BiP oligomers. BiP oligomers were isolated after cleavage by SubA and frozen. Thawed samples were exposed to ATP for indicated times before SEC analysis. As a control, uncleaved BiP was loaded. Note that purified BiP oligomers were stable in the presence of ATP. Source data are provided in Supplementary file 6. Panels ‘ii – iv’ represent different detailed views of the chromatogram shown in ‘i’.
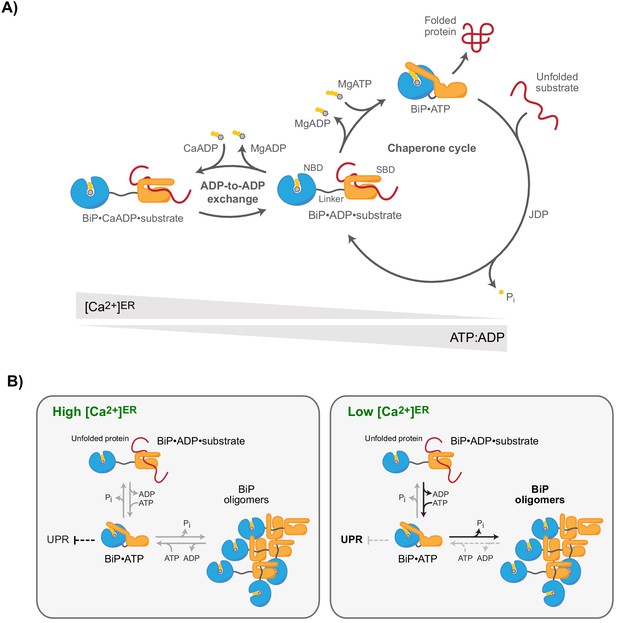
Ca2+-dependent alternative fates of BiP during nucleotide exchange.
(A) High [Ca2+]ER stabilises BiP•ADP•substrate complexes. The interaction of BiP with its substrates is driven by an ATPase-dependent chaperone cycle, whereby ATP binding triggers substrate release and ATP hydrolysis (to ADP and orthophosphate; Pi) induces high-affinity substrate binding (right cycle). ADP competes with ATP for binding to BiP during post-hydrolysis nucleotide exchange, causing ADP-to-ADP exchange cycles that delay substrate release. Because Ca2+ enhances the affinity of ADP for BiP, the substrate interaction-stabilising ADP-bound state of BiP is kinetically favoured by high [Ca2+]ER, whereas a decline in [Ca2+]ER accelerates ADP-to-ATP exchange and substrate release. A decrease in the ATP:ADP ratio also promotes ADP-to-ADP exchange and extension of BiP-substrate interactions. (B) Observed redistribution of BiP upon ER Ca2+ depletion. At high [Ca2+]ER (resting state; left panel), the concentration of unfolded proteins governs the formation of BiP-substrate interactions and competing oligomerisation. BiP oligomers are conformationally locked (functionally inactive) assemblies formed by ATP hydrolysis-dependent substrate interactions amongst BiP proteins. BiP also participates (both indirectly and by direct interaction with UPR signal transducers) in repression of UPR signalling. The restricted ability of oligomeric BiP to respond allosterically to ATP binding (and the resistance of oligomeric BiP to NEF-stimulated ADP release; not shown) kinetically stabilises oligomers at the expense of heterodimeric BiP-substrate complexes in the Ca2+-depleted ER (right panel). Faster substrate release from BiP at low [Ca2+]ER and lower BiP availability (due to enhanced sequestration into oligomers) likely favours de-repression of UPR signalling.
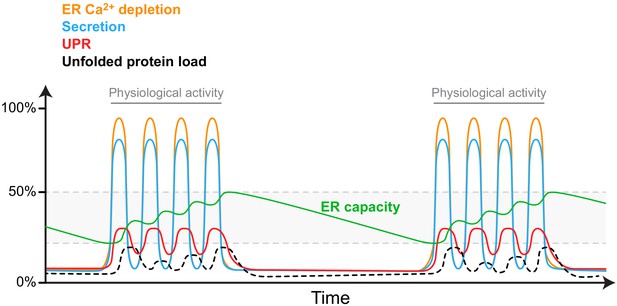
A speculative model of the fitness-promoting features of the observed selective Ca2+ sensitivity of metazoan ER-localised Hsp70s.
Upper panel: Bursts of physiologically entrained cellular activity (e.g. secretion; blue trace) triggered by bursts of signal-regulated ER Ca2+ depletion (orange trace) are coupled by BiP’s special features to UPR activity (red trace). Whilst UPR activity is transient in such circumstances, the stability of the proteins encoded by many UPR target genes converts fleeting UPR signals to maintenance of a sustained increase in ER capacity (green trace). ER capacity is thus matched to organellar activity by a process that responds to a physiological stimulus without or with minimal perturbation to ER protein folding homeostasis (unfolded protein load; black dotted line).
Tables
Data collection and refinement statistics.
| BiP(NBD) | Hsp70(NBD) | Hsc70(NBD) | Apo BiP oligomer | ADP BiP oligomer | |
|---|---|---|---|---|---|
| Data collection | |||||
| Synchrotron stations (DLS) | I03 | I04 | I04 | I24 | I24 |
| Space group | P1 | P212121 | P1211 | P1211 | P1211 |
| a,b,c; Å | 48.36, 51.86, 93.08 | 46.17, 63.71, 144.50 | 73.68, 78.04, 75.38 | 50.33, 51.29, 119.72 | 50.18, 51.25, 188.80 |
| α, β, γ; ⁰ | 78.07, 86.72, 62.28 | 90.00, 90.00, 90.00 | 90.00, 101.26, 90.00 | 90.00, 100.17, 90.00 | 90.00, 99.77, 90.00 |
| Resolution, Å* | 29.14-1.88 (1.92-1.88) | 29.53-1.52 (1.54-1.52) | 73.73-1.85 (1.89-1.85) | 59.92-1.77 (1.81-1.77) | 29.27-1.94 (1.99-1.94) |
| Rmerge* | 0.063 (0.989) | 0.065 (0.703) | 0.120 (1.005) | 0.054 (0.844) | 0.047 (0.533) |
| <I/σ(I)>* | 18.2 (1.8) | 14.0 (2.3) | 8.9 (1.4) | 9.9 (1.1) | 13.7 (2.0) |
| CC1/2* | 0.999 (0.669) | 0.998 (0.814) | 0.998 (0.633) | 0.975 (0.531) | 0.999 (0.717) |
| No. of unique reflections* | 62200 (3792) | 66932 (3125) | 71566 (4400) | 58872 (3347) | 43705 (2937) |
| Completeness, %* | 97.4 (90.4) | 99.7 (95.1) | 100.0 (100.0) | 99.9 (99.8) | 99.1 (94.5) |
| Redundancy* | 8.9 (7.5) | 6.7 (6.3) | 6.7 (5.7) | 3.2 (3.2) | 3.3 (3.2) |
| Refinement | |||||
| Rwork/Rfree | 0.209/0.225 | 0.196/0.212 | 0.218/0.247 | 0.185/0.217 | 0.184/0.226 |
| No. of atoms (non H) | 6137 | 3417 | 6399 | 4432 | 4327 |
| Average B-factors | 32.5 | 18.6 | 22.9 | 32.8 | 34.4 |
| RMS bond lengths Å | 0.002 | 0.002 | 0.002 | 0.006 | 0.006 |
| RMS bond angles,⁰ | 1.163 | 1.163 | 1.172 | 1.341 | 1.338 |
| Ramachandran favoured region, % | 99.34 | 99.17 | 98.28 | 98.66 | 98.27 |
| Ramachandran outliers, % | 0 | 0 | 0 | 0 | 0 |
| MolProbity score† | 0.78 (100) | 0.83 (100) | 0.80 (100) | 0.96 (100) | 0.99 (100) |
| PDB code | 6ZYH | 6ZYI | 6ZYJ | 7A4U | 7A4V |
-
*Values in parentheses are for highest-resolution shell.†100thpercentile is the best among structures of comparable resolutions. 0thpercentile is the worst.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | M15 | Qiagen | Recombinant protein expression | |
| Strain, strain background (Escherichia coli) | BL21 T7 Express lysY/Iq cells | New England Biolabs | Cat. #: C3013 | Recombinant protein expression |
| Cell line (Cricetulus griseus) | CHO-K1 | ATCC | RRID:CVCL_0214 | |
| Cell line (Cricetulus griseus) | CHO-K1 CHOP::GFP XBP1::Turquoise (clone S21) | PMID:27812215 | CHO-K1 S21 | CHO CHOP::GFP, XBP1s::Turquoise dual UPR reporter cell line; a derivative of RRID:CVCL_0214 |
| Cell line (Homo sapiens) | HEK293T | ATCC | RRID:CVCL_0063 | |
| Cell line (Cricetulus griseus) | CHO-K1 TCRα (clone 24) | This study | Derivative of CHO-K1 cell line | |
| Cell line (Cricetulus griseus) | CHO-K1 TCRα-N/Q (clone 12) | This study | Derivative of CHO-K1 cell line | |
| Antibody | Mouse monoclonal anti-FLAG M1; ANTI-FLAG M1 | Sigma | Cat. #: F3040 | Flow cytometry (1:500), WB (1:1000) |
| Antibody | Goat polyclonal anti-mouse IgG; Alexa Fluor 488 AffiniPure Goat Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | Cat. #: 115-545-146 | Flow cytometry (1:750) |
| Antibody | Chicken polyclonal anti-BiP IgY | PMID:23589496 | WB (1:2000) | |
| Antibody | Donkey polyclonal anti-chicken IgY; IRDye 800CW Donkey anti-Chicken Secondary Antibody | Li-Cor | Cat. #: 926–32218 | WB (1:2000) |
| Antibody | Goat polyclonal anti-mouse IgG (H+L); IRDye 800CW Goat anti-Mouse IgG Secondary Antibody | Li-Cor | Cat. #: 926–32210 | WB (1:2000) |
| Peptide, recombinant protein | P15; ALLLSAPRRGAGKK | GenScript (custom synthesis) | Biotinylated on the C-terminal lysine | |
| Peptide, recombinant protein | NR; NRLLLTG | GenScript (custom synthesis) | Carrying a fluorescein moiety at the N-terminus | |
| Chemical compound, drug | Tunicamycin; Tm | Melford | Cat. #: T2250 | |
| Chemical compound, drug | Thapsigargin; Tg | Calbiochem | Cat. #: 586005 | |
| Chemical compound, drug | A23187 | Sigma | Cat. #: C7522 | |
| Chemical compound, drug | 2-Deoxy-D-glucose; 2DG | ACROS Organics | Cat. #: D6134 | |
| Chemical compound, drug | PERK inhibitor; PERKi | GSK | Cat. #: GSK2606414 | |
| Chemical compound, drug | Brefeldin A; BFA | LC Laboratories | Cat. #: B-8500 | |
| Chemical compound, drug | Puromycin | Cayman Chemical | Cat. #: 13884 | |
| Chemical compound, drug | TransIT-293 Transfection Reagent | Mirus | Cat. #: MIR 2704 | |
| Chemical compound, drug | Hexokinase Type F-300 | Sigma | Cat. #: H4502 | |
| Chemical compound, drug | MABA-ADP; 8-[(4-Amino)butyl]-amino-ADP-MANT | Jena Bioscience | Cat. #: NU-893-MNT | |
| Chemical compound, drug | MABA-ATP; 8-[(4-Amino)butyl]-amino-ATP-MANT | Jena Bioscience | Cat. #: NU-806-MNT | |
| Chemical compound, drug | SYPRO orange protein gel stain | Invitrogen | Cat. #: S6651 | |
| Software, algorithm | Prism | GraphPad | ||
| Software, algorithm | XIA2 | DOI: 10.1107/S0021889809045701 | ||
| Software, algorithm | Mosflm | PMID:21460445 | ||
| Software, algorithm | XDS | PMID:20124692 | ||
| Software, algorithm | Pointless | PMID:21460446 | ||
| Software, algorithm | Aimless | PMID:21460441 | ||
| Software, algorithm | Phaser | PMID:19461840 | ||
| Software, algorithm | COOT | PMID:20383002 | ||
| Software, algorithm | refmac5 | PMID:11134934 | ||
| Software, algorithm | phenix.refine | PMID:31588918 | ||
| Software, algorithm | UCSF Chimera | PMID:15264254 | ||
| Software, algorithm | PyMOL | PyMOL | Version 1.3 educational | |
| Software, algorithm | CheckMyMetal | PMID:28291757 | ||
| Software, algorithm | phenix.polder | PMID:28177311 |
Additional files
-
Supplementary file 1
Source data for the flow cytometry experiment shown in Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/62601/elife-62601-supp1-v2.xlsx
-
Supplementary file 2
Source data and calculated rates for MABA-ADP release experiment shown in Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/62601/elife-62601-supp2-v2.xlsx
-
Supplementary file 3
Source data and calculated rates for the MABA-ADP release experiment shown in Figure 6—figure supplement 2A.
- https://cdn.elifesciences.org/articles/62601/elife-62601-supp3-v2.xlsx
-
Supplementary file 4
Source data and calculated melting temperatures for the DSF experiment shown in Figure 6—figure supplement 2B.
- https://cdn.elifesciences.org/articles/62601/elife-62601-supp4-v2.xlsx
-
Supplementary file 5
Source data and calculated rates for the MABA-ADP release experiment shown in Figure 6—figure supplement 3B.
- https://cdn.elifesciences.org/articles/62601/elife-62601-supp5-v2.xlsx
-
Supplementary file 6
Source data for the SEC traces shown in Figure 7—figure supplement 2.
- https://cdn.elifesciences.org/articles/62601/elife-62601-supp6-v2.xlsx
-
Supplementary file 7
Table of plasmids.
- https://cdn.elifesciences.org/articles/62601/elife-62601-supp7-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62601/elife-62601-transrepform-v2.docx





