Multiple pathways of toxicity induced by C9orf72 dipeptide repeat aggregates and G4C2 RNA in a cellular model
Figures

Cytoplasmic and nuclear poly-GA aggregates are amyloid-like but differ in solubility.
(A) Schematic representation of the poly-GA encoding constructs used. Two different classes of poly-GA encoding constructs were used to study dipeptide repeat protein (DPR)-mediated toxicity in the presence or absence of RNA repeat regions: synthetic constructs that do not contain G4C2 motifs and are translated into GA65 (left), and constructs containing 73 G4C2 repeats that encode GA73 (right). Both classes contained ATG start codons and were fused in frame to GFP. N-terminal nuclear import (NLS) or export sequences (NES) were present when indicated. (B) The indicated constructs were transfected into HEK293 cells. Antibodies against nuclear pore complexes (NPC, red) were used to detect the nuclear membrane, poly-GA was visualized by GFP fluorescence (green), and nuclei were counterstained with DAPI. (C) NES-GA65-GFP aggregates co-localize with p62. The indicated constructs were transfected into HEK293 cells. p62 (red) was detected by immunofluorescence, and poly-GA was visualized by GFP fluorescence (green). White dashed lines delineate the nucleus based on DAPI staining. (D) Cytoplasmic and nuclear aggregates can be stained with an amyloid-specific dye. The indicated constructs were transfected into HEK293 cells and stained with AmyTracker (AmyT, red). Poly-GA DPRs were visualized by GFP fluorescence (green). White dashed lines delineate the nucleus based on DAPI staining. (E) Cytoplasmic GA aggregates are SDS insoluble. The indicated constructs were transfected into HEK293 cells. GFP was expressed as a soluble control protein. Cells were lysed and analyzed for SDS-insoluble poly-GA aggregates by filter retardation assay. GFP antibody was used for detection. Scale bars represent 10 µm.
Cytoplasmic and nuclear poly-GA aggregates differ in their staining properties.
(A) Sequences of the GA-expressing constructs used in this study. Sequences (1) and (2) are synthetic degenerated sequences that do not contain G4C2. A third construct only contains G4C2 repeats and is translated into GA73. All three constructs contain an ATG start codon in frame with the GA coding sequence. (B) Cytoplasmic and nuclear aggregates can be stained with the OC antibody against amyloid fibrils. GA65-GFP was expressed in HEK293 cells, and immunofluorescence analysis was performed against amyloid fibrils (OC, red). Poly-GA was visualized by GFP fluorescence (green). White dashed lines delineate the nucleus based on DAPI staining. Scale bar represents 10 µm.
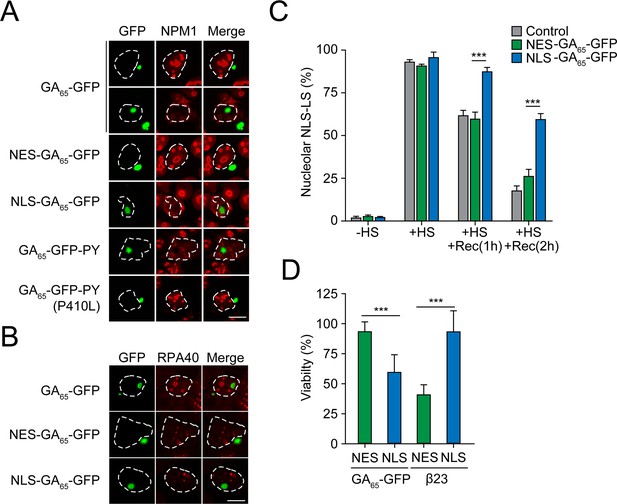
Nuclear poly-GA aggregates compromise nucleolar integrity and impair cell viability.
(A) Nuclear poly-GA aggregates alter nucleophosmin (NPM1) localization. The indicated constructs were transfected into HEK293 cells. Cells were fixed and stained with anti-NPM1 antibodies (red). Poly-GA was visualized by GFP fluorescence (green). (B) Nuclear poly-GA aggregates are not nucleolar. The indicated constructs were transfected into HEK293 cells, followed by staining with antibodies against DNA-directed RNA polymerases I and III subunit RPAC1 (RPA40) (red). Poly-GA was visualized by GFP fluorescence (green). (C) Nuclear poly-GA aggregates disrupt nucleolar protein quality control. HEK293 cells were co-transfected with NLS-firefly luciferase fused to mScarlet (NLS-LS) and the indicated poly-GA constructs or GFP as a control. Cells were maintained at 37°C (–HS) or subjected to heat stress 43°C (+HS) for 2 hr or heat stress and recovery (+HS + Rec) for 1 hr and 2 hr. Cells with nucleolar NLS-LS were counted, and the results plotted as percentage of transfected cells. Data are shown as mean + SD (n = 3). p-Value of two-sided t-test is displayed (***p≤0.001). Representative immunofluorescence images are shown in Figure 2—figure supplement 1A. (D) Nuclear poly-GA is toxic. HEK293 cells were transfected with the indicated constructs, and MTT cell viability assays were performed 4 days after transfection. Data were normalized to cells transfected with empty vector. Data are shown as means + SD (n ≥ 3); p-values of two-sided t-test are shown (***p≤0.001). White dashed lines delineate the nucleus based on DAPI staining. Scale bars represent 10 µm.
-
Figure 2—source data 1
Numerical values for graph in Figure 2C.
- https://cdn.elifesciences.org/articles/62718/elife-62718-fig2-data1-v1.docx
-
Figure 2—source data 2
Numerical values for graph in Figure 2D.
- https://cdn.elifesciences.org/articles/62718/elife-62718-fig2-data2-v1.docx
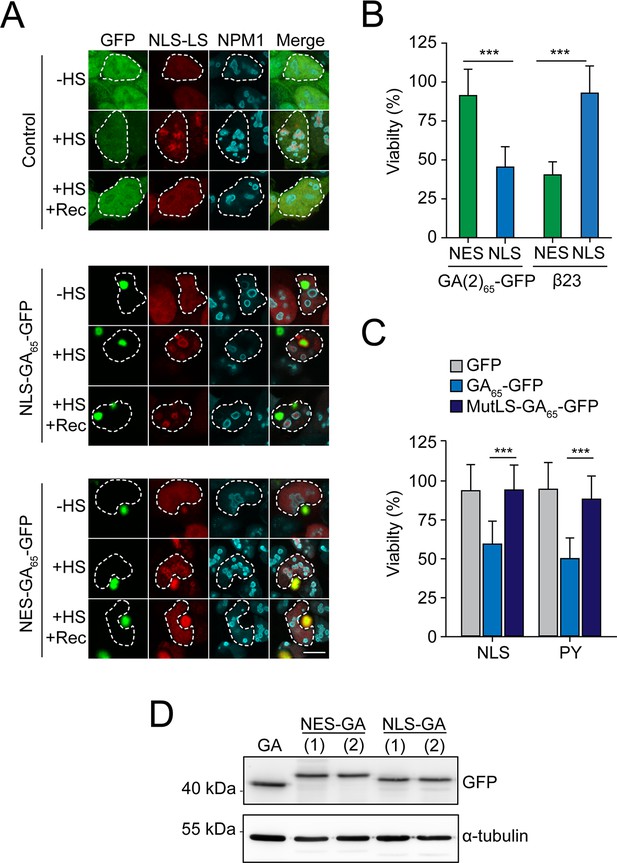
Nuclear poly-GA aggregates impair nucleolar protein quality control and cell viability.
(A) Nuclear poly-GA aggregates disrupt nucleolar protein quality control. HEK293 cells were co-transfected with NLS-LS and the indicated poly-GA (NLS-GA65-GFP or NES-GA65-GFP) constructs or GFP as control. Cells were maintained at 37°C (–HS) or subjected to heat stress at 43°C for 2 hr (+HS) and recovery (+HS + Rec) for 2 hr. Samples were immunostained for NPM1 (cyan), and NLS-LS and poly-GA proteins were visualized by mScarlet and GFP fluorescence, respectively. Merged panels are shown, and a white dashed line delineates the nucleus based on DAPI staining. Scale bars represent 10 µm. Representative images of three independent experiments are shown. See Figure 2C for quantification. (B) NLS-GA65-GFP produced from an alternative degenerated and G4C2-free DNA sequence also decreases cellular viability. HEK293 cells were transfected with the indicated constructs, and MTT cell viability assays performed 4 days after transfection. Data were normalized to cells transfected with empty vector. Data are means + SD (n ≥ 3), and two-sided t-test is shown (***p≤0.001). β23 data are from Figure 2D and are shown as control. (C) Disabling nuclear targeting sequence functionality of GA65-GFP rescues cellular viability. HEK293 cells were transfected with constructs encoding two different nuclear localization sequences (NLS or PY) fused to GFP, GA65-GFP, or GA65-GFP with an inactivating mutation in the nuclear localization sequence (MutLS-GA65-GFP), followed by MTT cell viability assay 4 days after transfection. Data were normalized to cells transfected with empty vector. Data are shown as means + SD (n ≥ 3). ***p≤0.001 by two-sided t-test. Part of this data is also shown in Figure 2D. (D) HEK293 cells were transfected with the indicated constructs (GA65-GFP; NES-GA65-GFP; NES-GA65(2)-GFP; NLS-GA65-GFP; and NLS-GA65(2)-GFP). Expression levels were determined by immunoblotting with anti-GFP antibodies, α-tubulin served as loading control.
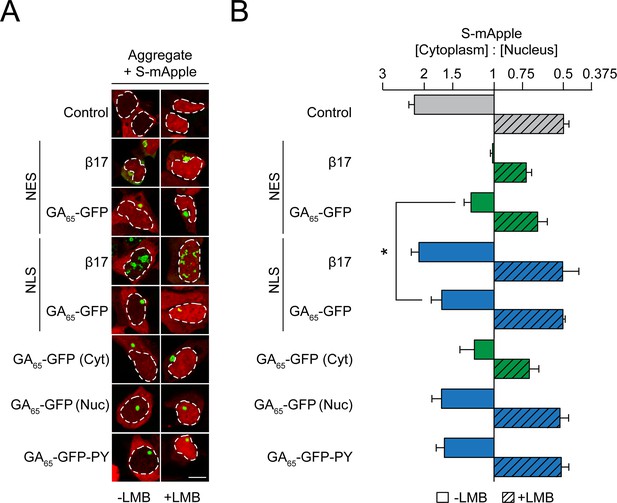
Cytoplasmic poly-GA aggregates impair nucleocytoplasmic protein transport.
(A) Cytoplasmic poly-GA aggregates alter nuclear transport of a shuttling reporter protein. HEK293 cells were co-transfected with S-mApple (red) and either empty vector (Control), NES-β17, NES-GA65-GFP, NLS-β17, NLS-GA65-GFP, GA65-GFP, or GA65-GFP-PY (green). Leptomycin B (LMB; 10 ng/ml) was added for 15 min when indicated. White dashed lines delineate nuclei based on DAPI staining. Scale bar represents 10 µm. (B) Quantification of S-mApple distribution from data in (A). The x-axis shows the enrichment of S-mApple concentration in the cytoplasm relative to the nucleus. Cells transfected with GA65-GFP were further analyzed and divided into cells with cytoplasmic (Cyt) or nuclear (Nuc) aggregates. Data are means + SD, n = 3 independent experiments, >70 cells were analyzed per condition. *p≤0.05 from two-sided t-test.
-
Figure 3—source data 1
Numerical values for graph in Figure 3B.
- https://cdn.elifesciences.org/articles/62718/elife-62718-fig3-data1-v1.docx
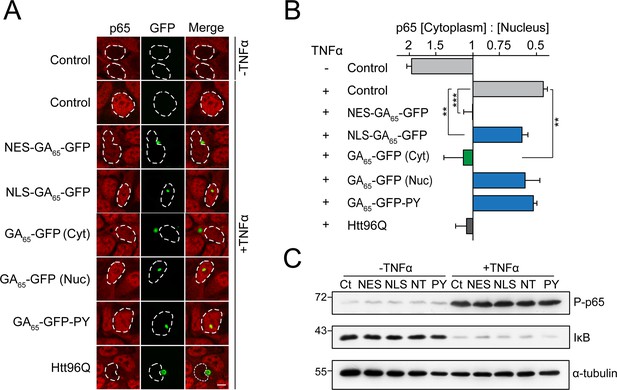
Cytoplasmic poly-GA aggregates inhibit nuclear import of p65.
(A) Cytoplasmic poly-GA aggregates inhibit p65 nuclear translocation. HEK293 cells were transfected with empty vector (Control), NES-GA65-GFP, NLS-GA65-GFP, GA65-GFP, GA65-GFP-PY, or Htt96Q-GFP (Htt96Q) (green) and analyzed for NF-κB p65 localization (red) with and without TNFα treatment (30 min). White dashed lines delineate nuclei based on DAPI staining. Scale bar represents 10 µm. (B) Quantification of NF-κB p65 distribution from data in (A). The x-axis shows the enrichment of p65 in the cytoplasm relative to the nucleus. Data are means + SD (n = 3), >100 cells were analyzed per condition. **p≤0.01, ***p≤0.001 from two-sided t-test. (C) Expression of poly-GA does not alter the degradation of IκB and phosphorylation of p65. HEK293 cells were transfected with the indicated constructs (Ct: Control; NES: NES-GA65-GFP; NLS: NLS-GA65-GFP; NT: GA65-GFP; PY: GA65-GFP-PY) and treated as described in (A). Levels of IκB and phosphorylated NF-κB p65 (P–p65) were analyzed by immunoblotting. α-tubulin served as loading control.
-
Figure 4—source data 1
Numerical values for graph in Figure 4B.
- https://cdn.elifesciences.org/articles/62718/elife-62718-fig4-data1-v1.docx
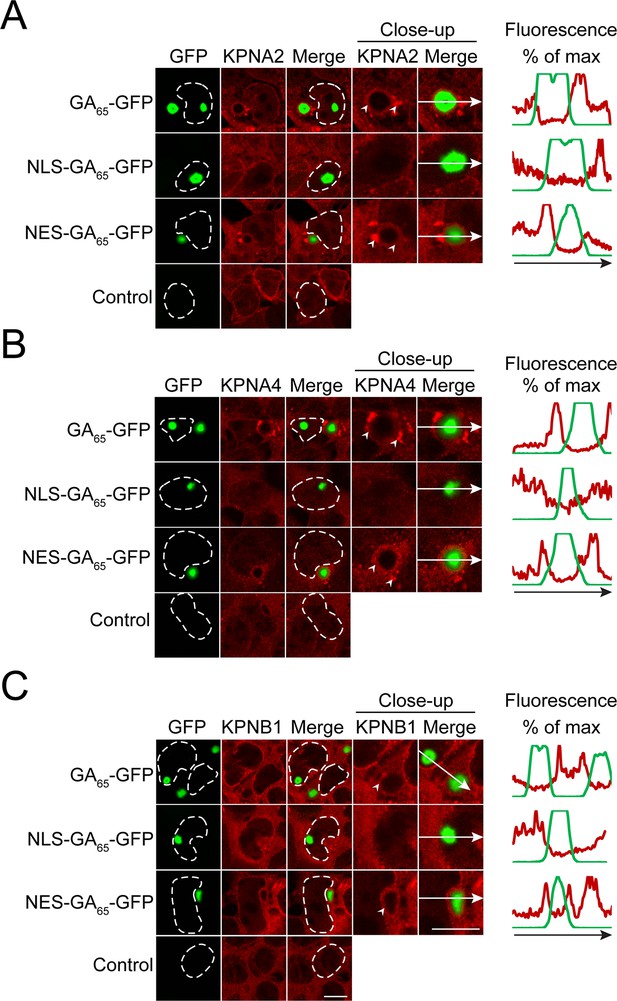
Importins form aggregates in cells containing cytoplasmic poly-GA aggregates and are partially sequestered.
HEK293 cells were left untreated (Control) or were transfected with GA65-GFP, NES-GA65-GFP or NLS-GA65-GFP (green). Cells were then analyzed by immunofluorescence with antibodies against importin α-1 (KPNA2; A), importin α-3 (KPNA4; B), and importin β-1 (KPNB1; C) (red). Close-up views of inclusions are shown, and arrowheads indicate importin aggregates and/or their sequestration at the periphery of the inclusion. The co-localization profile along the white arrow is shown, plotted as percentage of maximal fluorescence on the y-axis. The white dashed lines delineate nuclei based on DAPI staining. Scale bars are 10 µm.
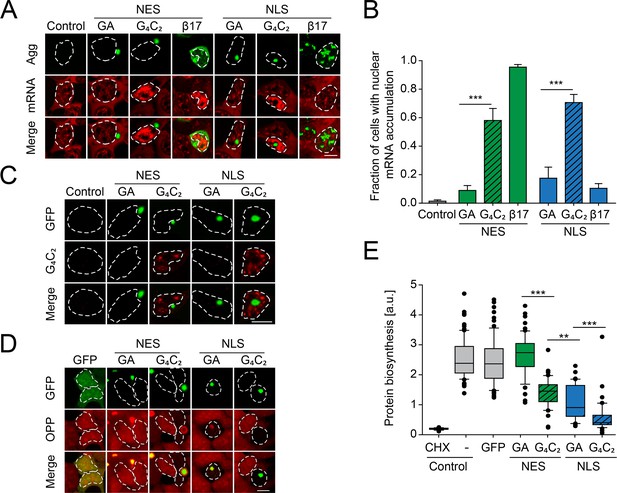
Protein biosynthesis defects correlate with retention of mRNA in the nucleus induced by G4C2 mRNA as well as presence of nuclear poly-GA aggregates.
(A) G4C2-containing constructs induce strong nuclear mRNA accumulation. HEK293 cells were transfected with the indicated constructs: empty vector (Control); NES-GA65-GFP (NES-GA); NES-G4C2-GFP (NES-G4C2); NES-β17; NLS-GA65-GFP (NLS-GA); NLS-G4C2-GFP (NLS-G4C2); NLS-β17. PolyA-RNA was detected by fluorescence in situ hybridization using a poly-dT probe (red); protein aggregates (Agg) green. (B) Quantification of data in (A). The graph shows the fraction of cells with nuclear mRNA accumulation. Data are means + SD (n = 3), ***p≤0.001 from two-sided t-test. (C) G4C2-containing constructs induce the formation of G4C2 RNA foci. HEK293 cells were transfected with the indicated constructs: empty vector (Control); NES-GA65-GFP (NES-GA); NES-G4C2-GFP (NES-G4C2); NLS-GA65-GFP (NLS-GA); NLS-G4C2-GFP (NLS-G4C2). Cells were analyzed for GFP fluorescence (green) and C4G2 fluorescence by in situ hybridization (red). (D) Decreased protein biosynthesis in the presence of nuclear poly-GA and G4C2 mRNA. Newly synthesized proteins were labeled with O-propargyl-puromycin (OPP; red) in HEK293 cells transfected with the indicated constructs (green). The white dashed lines delineate the nucleus based on DAPI staining, and the scale bar represents 10 µm. (E) Quantification of data in (D). Analysis of control cells transfected with empty vector and treated with the translation inhibitor cycloheximide (CHX) when indicated is included as control. Boxplot of a representative experiment is shown. Center lines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the 10th and 90th percentiles, outliers are plotted as circles. Welch's t-test was used to assess statistical significance (**p≤0.01; ***p≤0.001).
-
Figure 5—source data 1
Numerical values for graph in Figure 5B.
- https://cdn.elifesciences.org/articles/62718/elife-62718-fig5-data1-v1.docx
-
Figure 5—source data 2
Numerical values for repeats for Figure 5E.
- https://cdn.elifesciences.org/articles/62718/elife-62718-fig5-data2-v1.docx
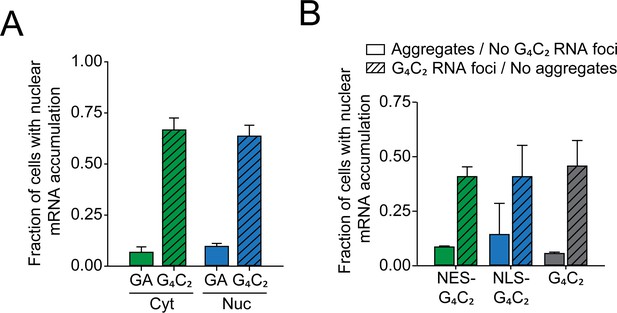
Expression of G4C2-containing constructs induces mRNA retention in the nucleus.
(A) Presence of G4C2 rather than aggregate localization causes nuclear mRNA retention. HEK293 cells were transfected with either GA65-GFP (GA) or G4C2-GFP (G4C2) and analyzed for nuclear mRNA accumulation as in Figure 5A, B. Cells containing GA-GFP aggregates in the cytoplasm (Cyt) or nucleus (Nuc) were analyzed separately. Data are shown as means + SD (n = 3). (B) mRNA retention correlates with the presence of G4C2 positive nuclear foci. HEK293 cells were transfected with G4C2-GFP (G4C2), NES-G4C2-GFP (NES-G4C2), or NLS-G4C2-GFP (NLS-G4C2) and analyzed for mRNA nuclear accumulation as in Figure 5C. Fluorescence in situ hybridization was performed as in Figure 5C to detect G4C2 foci. Cells were categorized according to the presence of G4C2-positive foci as well as aggregates. Data are shown as means + SD (n = 3).
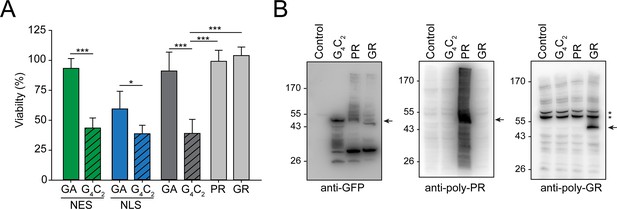
Production of G4C2 mRNA strongly decreases cellular viability.
(A) G4C2 mRNA induces strong toxicity. HEK293 cells were transfected with the indicated constructs: G4C2-GFP (G4C2), NES-G4C2-GFP (NES-G4C2), NLS-G4C2-GFP (NLS-G4C2), GA65-GFP (GA), NES-GA65-GFP (NES-GA), NLS-GA65-GFP (NLS-GA), PR73-GFP (PR), or GR73-GFP (GR). MTT cell viability assays were performed 4 days after transfection. Data were normalized to cells transfected with empty vector. Data are means + SD (n ≥ 3). Part of this data is also shown in Figure 2D. *p≤0.05; ***p≤0.001 from two-sided t-test. (B) (G4C2)73-GFP does not produce detectable amounts of arginine containing dipeptide repeats (R-DPRs). HEK293 cells were transfected with the indicated constructs: empty vector (control), (G4C2)73-GFP (G4C2), PR73-GFP (PR), and GR73-GFP (GR). Immunoblot analysis was then performed against GFP (left), poly-PR (center), and poly-GR (right). A representative result of three biological repeats is shown. The arrows indicate the main band of the respective DPRs, and * indicate non-specific bands recognized by the anti-GR antibody.
-
Figure 6—source data 1
Numerical values for graph in Figure 6A.
- https://cdn.elifesciences.org/articles/62718/elife-62718-fig6-data1-v1.docx
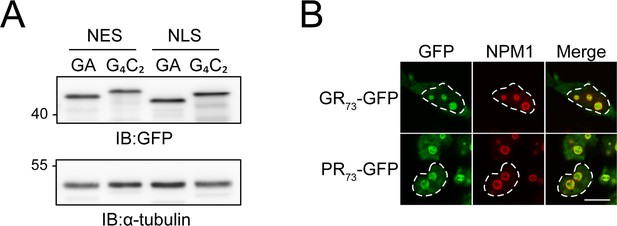
Expression levels of GA-DPRs and localization of R-DPRs.
(A) Toxicity induced by G4C2 mRNA is not due to a higher poly-GA level. HEK293 cells were transfected with the indicated constructs: NES-GA65-GFP (NES-GA); NES-G4C2-GFP (NES-G4C2); NLS-GA65-GFP (NLS-GA); and NLS-G4C2-GFP (NLS-G4C2). Poly-GA-GFP protein levels were analyzed by immunoblotting against GFP. α-tubulin served as loading control. A representative result of three independent experiments is shown. (B) Arginine-containing dipeptide repeats (R-DPRs) accumulate in the nucleolus. HEK293 cells were transfected with the indicated constructs (GR73-GFP (GR); PR73-GFP (PR)). Cells were then stained with anti-nucleophosmin (NPM1, red). Expressed constructs are shown in green. The white dashed lines delineate the nucleus based on DAPI staining, and the scale bar represents 10 µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens, female) | HEK293 | ATCC | Cat. #: ATCC-CRL-1573 RRID:CVCL_0045 | Lot/Batch No: 63777489 |
| Recombinant DNA reagent | pcDNA3.1 GA65-GFP | This paper | Described in Results part 1 and Materials and methods section | |
| Recombinant DNA reagent | pcDNA3.1 NES-GA65-GFP | This paper | Described in Results part 1 and Materials and methods section | |
| Recombinant DNA reagent | pcDNA3.1 NLS-GA65-GFP | This paper | Described in Results part 1 and Materials and methods section | |
| Recombinant DNA reagent | pcDNA3.1 NLS-GA65-GFP (K7T, K15T) | This paper | Described in Results part 1 and Materials and methods section | |
| Recombinant DNA reagent | pcDNA3.1 GA65-GFP-PY | This paper | Described in Results part 1 and Materials and methods section | |
| Recombinant DNA reagent | pcDNA3.1 GA65-GFP-PY(P410L) | This paper | Described in Results part 1 and Materials and methods section | |
| Recombinant DNA reagent | pcDNA3.1 GFP | This paper | Described in Results part 1 and Materials and methods section | |
| Recombinant DNA reagent | pcDNA3.1 PR73-GFP | This paper | Described in Results part 3 and Materials and methods section | |
| Recombinant DNA reagent | pcDNA3.1 GR73-GFP | This paper | Described in Results part 3 and Materials and methods section | |
| Recombinant DNA reagent | pcDNA3.1 (G4C2)73-GFP | This paper | Described in Results part 3 and Materials and methods section | |
| Recombinant DNA reagent | pcDNA3.1 NLS-(G4C2)73-GFP | This paper | Described in Results part 3 and Materials and methods section | |
| Recombinant DNA reagent | pcDNA3.1 NES-(G4C2)73-GFP | This paper | Described in Results part 3 and Materials and methods section | |
| Recombinant DNA reagent | Plasmid expressing NLS-LS | PMID:31296649 | Prof. F. Ulrich Hartl (Max Planck Institute for Biochemistry) | |
| Recombinant DNA reagent | Plasmid expressing S-mApple | This paper | Described in Results part 2 and Materials and methods section | |
| Recombinant DNA reagent | Plasmid expressing c-myc-NES-β17 | PMID:26634439 | Prof. F. Ulrich Hartl (Max Planck Institute for Biochemistry) | |
| Recombinant DNA reagent | Plasmid expressing c-myc-NLS-β17 | PMID:26634439 | Prof. F. Ulrich Hartl (Max Planck Institute for Biochemistry) | |
| Recombinant DNA reagent | Plasmid expressing Htt96Q | PMID:26634439 | Prof. F. Ulrich Hartl (Max Planck Institute for Biochemistry) | |
| Antibody | Mouse monoclonal anti-nuclear pore complex proteins | Abcam | Cat. #: ab24609 RRID:AB_448181 | IF (1:1000) |
| Antibody | Mouse monoclonal anti-NPM1 | Invitrogen | Cat. #: 32-5200 RRID:AB_2533084 | IF (1:1000) |
| Antibody | Rabbit monoclonal anti-NF-κB p65 (D14E12) | Cell Signaling Technology | Cat. #: 8242 RRID:AB_10859369 | IF (1:1000) |
| Antibody | Mouse monoclonal anti-p62 | Abcam | Cat. #: ab203430 RRID:AB_2728795 | IF (1:1000) |
| Antibody | Mouse monoclonal anti-c-Myc-Cy3 (9E10) | Sigma | Cat. #: C6594 RRID:AB_258958 | IF (1:1000) |
| Antibody | Rabbit polyclonal anti-amyloid fibrils OC | Millipore | Cat. #: AB2286 RRID:AB_1977024 | IF (1:500) |
| Antibody | Mouse biclonal anti-GFP | Roche | Cat. #: 11814460001 RRID:AB_390913 | WB (1:1000) |
| Antibody | Mouse monoclonal anti-α-tubulin | Sigma | Cat. #: T6199 RRID:AB_477583 | WB (1:1000) |
| Antibody | Mouse monoclonal anti-RPA40 | Santa Cruz | Cat. #: sc-374443 RRID:AB_10991310 | IF (1:1000) |
| Antibody | Mouse monoclonal anti-IκBα (L35A5) | Cell Signaling Technology | Cat. #: 4814 RRID:AB_390781 | WB (1:/1000) |
| Antibody | Rabbit monoclonal anti-phospho-NF-κB p65 (Ser533) (93H1) | Cell Signaling Technology | Cat. #: 3033 RRID:AB_331284 | WB (1:1000) |
| Antibody | Rabbit polyclonal anti-KPNA2 | Abcam | Cat. #: ab70160 RRID:AB_2133673 | IF (1:1000) |
| Antibody | Rabbit polyclonal anti-KPNA4 | Abcam | Cat. #: ab84735 RRID:AB_1860702 | IF (1:1000) |
| Antibody | Mouse monoclonal anti-KPNB1 | Abcam | Cat. #: ab2811 RRID:AB_2133989 | IF (1:1000) |
| Antibody | Mouse monoclonal anti-GAPDH | Millipore | Cat. #: MAB374 RRID:AB_2107445 | WB (1:2000) |
| Antibody | Rabbit polyclonal anti-poly-PR | Proteintech | Cat. #: 23979-1-AP RRID:AB_2879388 | WB (1:1000) |
| Antibody | Mouse monoclonal anti-poly-GR (5A2) | Millipore | Cat. #: MABN778 RRID:AB_2728664 | WB (1:1000) |
| Antibody | Goat Secondary anti-mouse IgG-Alexa 488 | Cell Signaling Technology | Cat. #: 4408 RRID:AB_10694704 | IF (1:1000) |
| Antibody | Goat Secondary anti-mouse IgG-Alexa 555 | Cell Signaling Technology | Cat. #: 4409 RRID:AB_1904022 | IF (1:1000) |
| Antibody | Goat Secondary anti-rabbit IgG-Alexa 555 | Cell Signaling Technology | Cat. #: 4413 RRID:AB_10694110 | IF (1:1000) |
| Antibody | Secondary chicken anti-mouse IgG-Alexa 488 | Invitrogen | Cat. #: 21200 RRID:AB_2535786 | IF (1:1000) |
| Antibody | Secondary goat anti-mouse IgG-Alexa 633 | Invitrogen | Cat. #: A21053 RRID:AB_2535720 | IF (1:1000) |
| Antibody | Secondary goat anti-mouse IgG-peroxidase | Sigma-Aldrich | Cat. #: A4416 RRID:AB_258167 | WB (1:10,000) |
| Antibody | Secondary goat anti-rabbit IgG-peroxidase | Sigma-Aldrich | Cat. #: A9169 RRID:AB_258434 | WB (1:10,000) |
| Sequence-based reagent | T30 | PMID:26634439 | FISH probe | Cy5-conjugated |
| Sequence-based reagent | (C4G2)5 | This paper | FISH probe | Cy3-conjugated |
| Chemical compound, drug | UltraPure SSC buffer | Thermo Fisher Scientific | Cat. #: 15557044 | For FISH |
| Chemical compound, drug | Formamide | Sigma-Aldrich | Cat. #: 47671 | For FISH |
| Chemical compound, drug | Dextran sulphate | Sigma-Aldrich | Cat. #: D6001 | For FISH |
| Chemical compound, drug | AmyTracker 680 (AmyT) | Ebba Biotech AB | Cat. #: AmyTracker 680 | Amyloid dye |
| Chemical compound, drug | DAPI | Invitrogen | Cat. #: D1306 RRID:AB_2629482 | Nuclear stain |
| Chemical compound, drug | Thiazolyl blue tetrazolium bromide | Sigma Aldrich | Cat. #: M2128 | For cell viability |
| Chemical compound, drug | N,N-dimethylformamide | Sigma-Aldrich | Cat. #: D4551 | For cell viability |
| Chemical compound, drug | SDS | Sigma-Aldrich | Cat. #: L3771 | For cell viability |
| Chemical compound, drug | Acetic acid | Sigma-Aldrich | Cat. #: A6283 | For cell viability |
| Chemical compound, drug | Leptomycin B | Sigma Aldrich | Cat. #: L2913 | See Materials and methods section |
| Commercial assay or kit | CellTiter-Glo 2.0 | Promega | Cat. #: G9241 | For cell viability |
| Commercial assay or kit | Lipofectamine 2000 transfection reagent | Thermo Fischer Scientific | Cat. #: 11668019 | |
| Commercial assay or kit | Click-iT Plus OPP Alexa Fluor 594 Protein Synthesis Assay Kit | Thermo Fischer Scientific | Cat. #: C10457 | |
| Peptide, recombinant protein | Human recombinant TNFα | Jena Biosciences | Cat. #: PR-430 |





