Mapping immune variation and var gene switching in naive hosts infected with Plasmodium falciparum
Figures
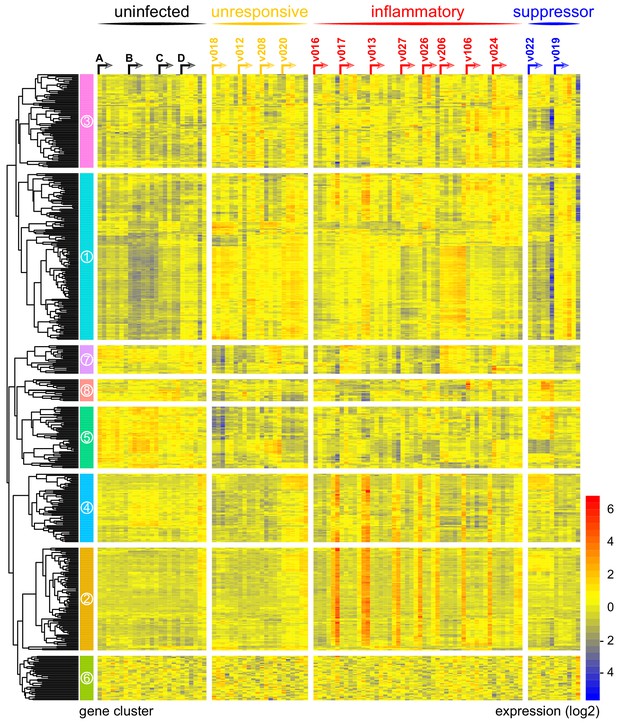
Immune variation in falciparum malaria.
Log2 expression values of 517 protein-coding genes in whole blood during infection. Genes (rows) are ordered by hierarchical clustering whereas whole blood samples (columns) are ordered by volunteer and time-point (pre-infection to diagnosis, left to right). Arrows start from the pre-infection sample and volunteers are grouped by host response. Uninfected controls demonstrate minimal within-host variation in expression of these genes. Median sample number per volunteer = 6.
-
Figure 1—source data 1
An identical immune challenge leads to diverse outcomes in falciparum malaria.
Log2 fold-change of the 100 protein-coding genes with highest variance in whole blood during infection. Data are presented as deviation from median (within each volunteer). Genes (rows) are ordered by hierarchical clustering, whereas whole blood samples (columns) are ordered by time-point (pre-infection to diagnosis, left to right). Each volunteer was analysed independently and therefore every top 100 gene list is unique.
- https://cdn.elifesciences.org/articles/62800/elife-62800-fig1-data1-v1.pdf
-
Figure 1—source data 2
Log2 transformed expression values of the 517-gene superset in whole blood during infection.
- https://cdn.elifesciences.org/articles/62800/elife-62800-fig1-data2-v1.xlsx
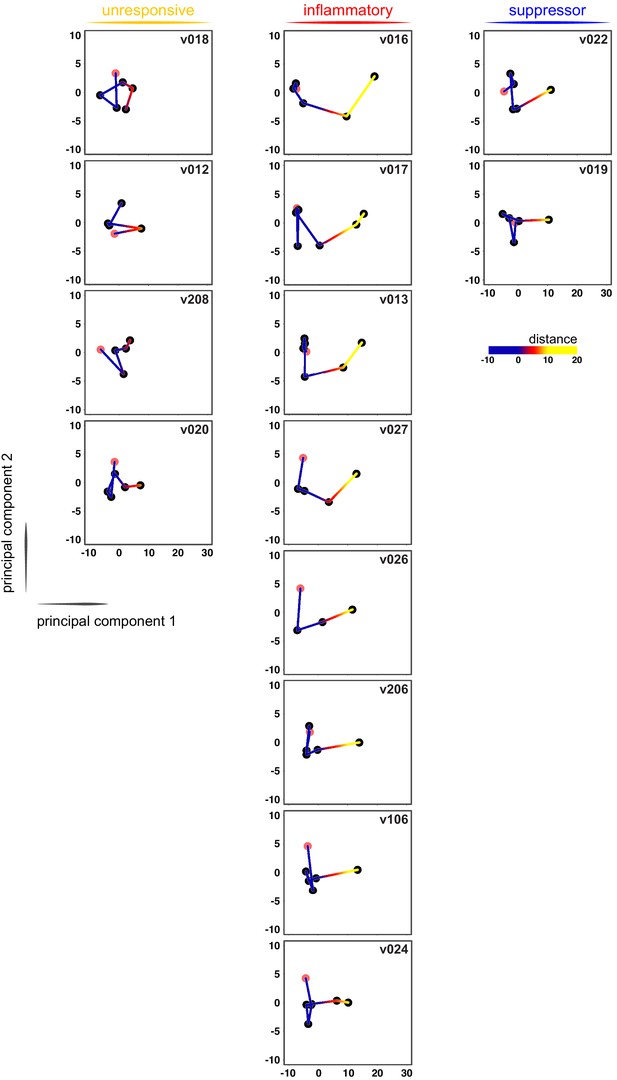
Immune quiescence is a common early outcome of infection.
Principal component analysis of the 517-gene superset in whole blood during infection. Each dot represents one time-point (pink is pre-infection) and every volunteer is centred around their own average position through time (mean x-y coordinates set to zero). Each volunteer was analysed independently and distance travelled (relative to average position) measures the magnitude of their immune response.
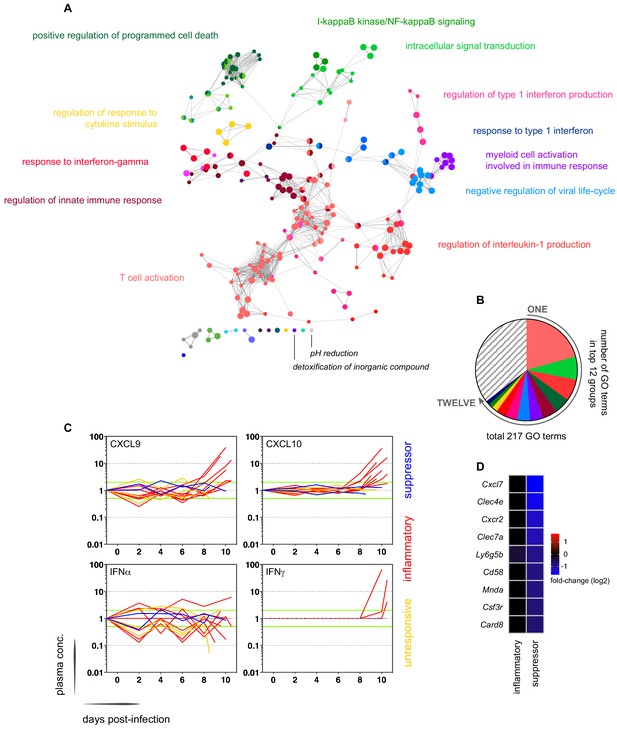
Interferon-stimulated inflammation is the dominant response to blood-stage infection.
(A) Gene ontology network of 2028 genes differentially expressed at diagnosis in the inflammatory group. Each node represents a significantly enriched GO term (adj p<0.05) and node size is determined by significance (bigger nodes have lower p values). Nodes are interconnected according to their relatedness (kappa score >0.4) and grouped if they are connected and share >40% genes. Each functional group is then given a unique colour and the leading GO term in the top 12 groups is highlighted. Two GO terms of interest, which are not part of any functional group, are also shown in italics. (B) The proportion of GO terms in each of the top 12 functional groups; collectively, these account for two thirds of all significantly enriched GO terms in inflammatory volunteers. (C) Plasma concentration of interferon alpha and gamma and interferon-stimulated chemokines (CXCL9 and CXCL10) during infection. One line represents one volunteer (no data for v020) and lines are colour-coded by host response. For each volunteer, all data points are normalised to their own baseline (day −1); horizontal green lines represent a twofold increase or decrease compared to baseline. (D) Log2 fold-change of nine immune genes involved in myeloid cell differentiation and activation in whole blood at diagnosis. Data are presented relative to pre-infection samples and all genes are significantly downregulated in the two suppressor volunteers (adj p<0.05).
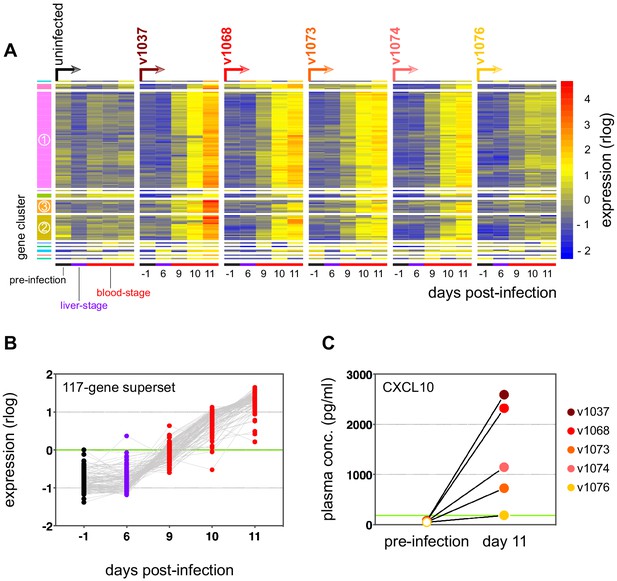
Sporozoites do not trigger a systemic transcriptional response in human malaria.
(A) Rlog expression values of the 117-gene superset in whole blood after mosquito challenge. Genes (rows) are ordered by hierarchical clustering, whereas whole blood samples (columns) are ordered by volunteer and time-point. An uninfected control volunteer displayed minimal within-host variation in expression of these genes. Sample number per volunteer = 5. (B) Rlog expression values of the 117-gene superset shown as the mean of all infected volunteers; each dot represents a single gene. (A and B) Day 6 was chosen as the end point of liver-stage infection as there were no detectable circulating parasites at this time; 24 hr later every volunteer was parasitaemic. (C) Plasma concentration of CXCL10 after mosquito challenge; one dot represents one volunteer and the green line shows the mean concentration in the uninfected control.
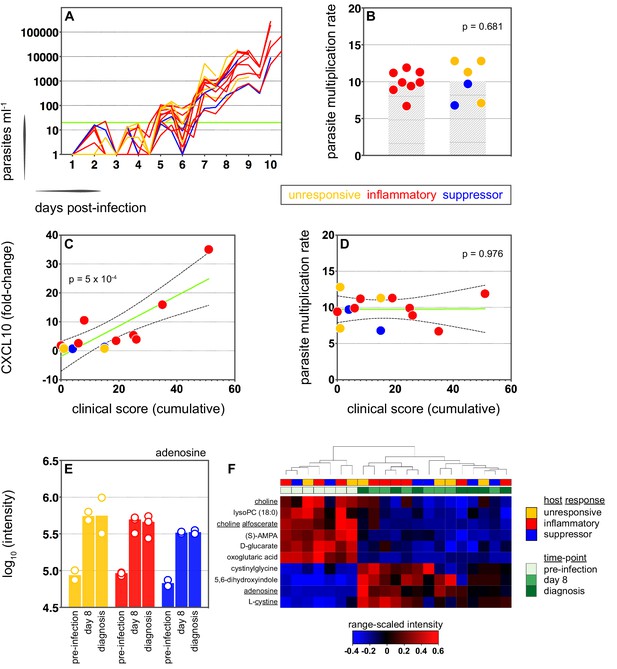
Systemic inflammation coincides with the onset of clinical symptoms.
(A) Parasite growth curves colour-coded by host response; each line represents one volunteer. Blood samples were collected every 12 hr for qPCR analysis of circulating parasite density and the horizontal green line represents the lower limit of quantification (20 parasites ml−1). (B) Parasite multiplication rates colour-coded by host response; each dot represents one volunteer and shaded areas show the mean value. A Mann Whitney test was used to ask whether the parasite multiplication rate observed in the inflammatory group was different to all other volunteers (p value is shown). (C and D) Linear regression of CXCL10 (C) or parasite multiplication rate (D) plotted against clinical score (the sum of adverse events during infection). CXCL10 fold-change measures plasma concentration at diagnosis over baseline (day −1). One dot represents one volunteer (no data for v020) and dots are colour-coded by host response. The green line represents the best-fit model (p value of the slope is shown) and dashed lines are the 95% confidence intervals. (E) Log10 transformed intensity values of adenosine in plasma during infection. An authentic standard was run in tandem with all samples to validate adenosine detection. (F) Range-scaled intensity values of 10 plasma metabolites that were differentially abundant during infection. Metabolites (rows) and samples (columns) are ordered by hierarchical clustering. Note that an authentic standard was used to validate detection of all underlined metabolites and the full name for oxoglutaric acid is 4-hydroxy-2-oxoglutaric acid. (E and F) Only plasma samples from the most inflammatory and symptomatic volunteers (v016, v017, and v013), the suppressor volunteers (v022 and v019) and two unresponsive volunteers (v018 and v208) were analysed for metabolite abundance.
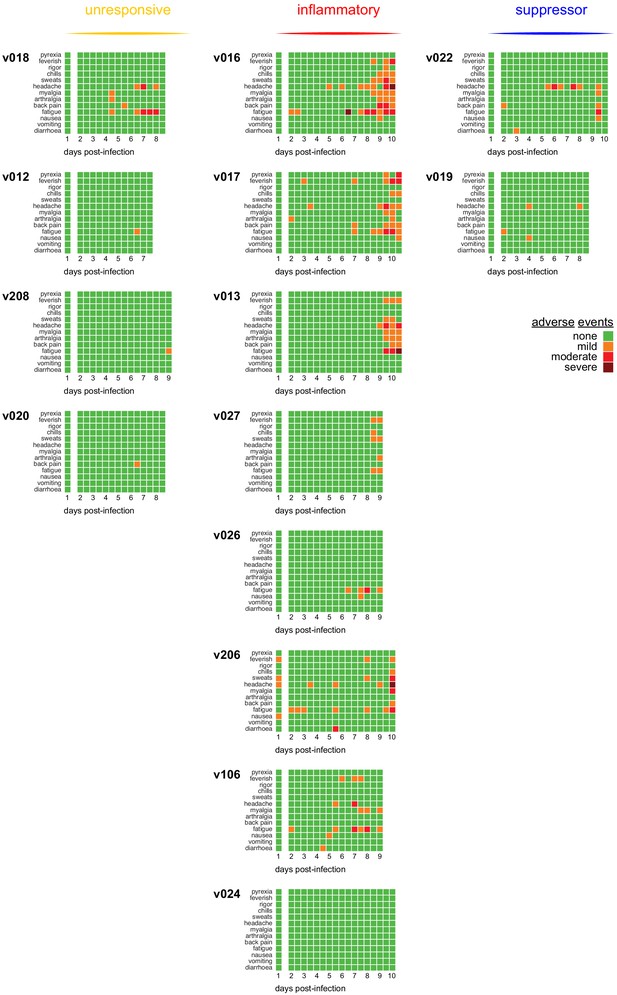
Adverse events after blood challenge.
Data on symptoms were collected every 12 hr from day 2 post-infection either on diary cards (self-reporting) or during clinic visits. Adverse events were graded as mild (transient or mild discomfort – no medical intervention required); moderate (mild to moderate limitation in activity – no or minimal medical intervention required); or severe (marked limitation in activity – may require medical intervention).
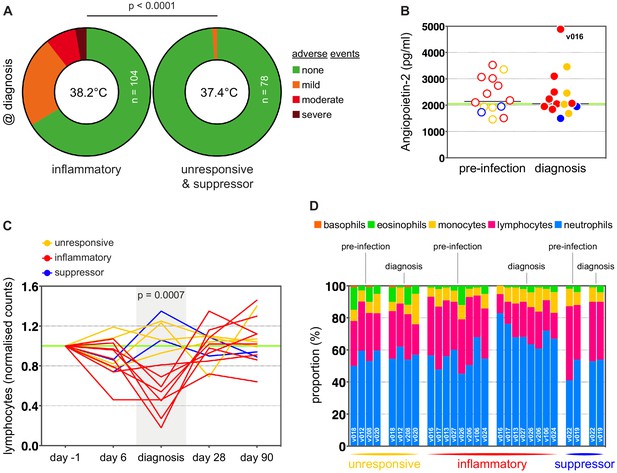
Inflammation is linked to hallmark symptoms of malaria.
(A) Proportion of mild, moderate and severe adverse events at diagnosis. For each volunteer, adverse events were recorded for 13 clinical categories and these were summed to give a clinical score at diagnosis (mild = 1, moderate = 2 and severe = 3). A Fisher’s exact test was then used to assess the significance of the difference in clinical score between the inflammatory group and unresponsive/suppressor hosts (p value is shown and n = total number of data-points recorded). The maximum core body temperature measured in any volunteer within a group is shown in each doughnut plot. (B) Plasma concentration of Angiopoietin-2 comparing pre-infection (open circle) and diagnosis (filled circle) samples. Each dot represents one volunteer and dots are colour-coded by host response. The green line represents the mean reference value measured in healthy human donors (heparin plasma samples). (C) Lymphocyte counts in whole blood during infection; each line represents one volunteer and lines are colour-coded by host response. Data are shown as fold-change over baseline (day −1). A Mann Whitney test was used to assess the significance of differences in cell counts between inflammatory volunteers and all other hosts at diagnosis (p value is shown). (D) Five-part differential whole blood counts comparing pre-infection and diagnosis samples. Volunteer numbers are inset into stacked bars and samples are ordered by host response followed by time-point.
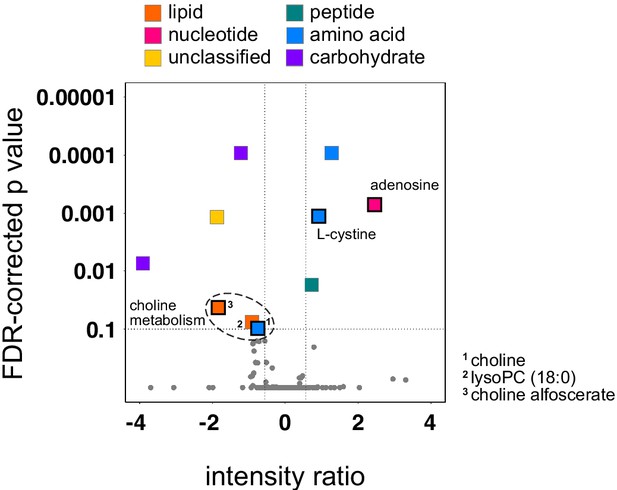
A conserved early metabolic signature of falciparum malaria.
Volcano plot showing the 10 plasma metabolites that were differentially abundant during infection. We grouped unresponsive (v018 and v208), inflammatory (v016, v017 and v013), and suppressor (v022 and v019) hosts and grouped two post-infection time-points (day 8 and diagnosis) to identify a conserved and persistent signature, respectively. These samples (n = 14) were tested against pre-infection plasma (n = 7) using an FDR-corrected p value < 0.1. A positive intensity ratio indicates metabolites are more abundant during infection, whereas a negative ratio indicates metabolite depletion. Vertical black dotted lines represent a 1.5-fold change in abundance. Squares with a solid black border show that an authentic standard was run in tandem with the samples to validate metabolite detection.
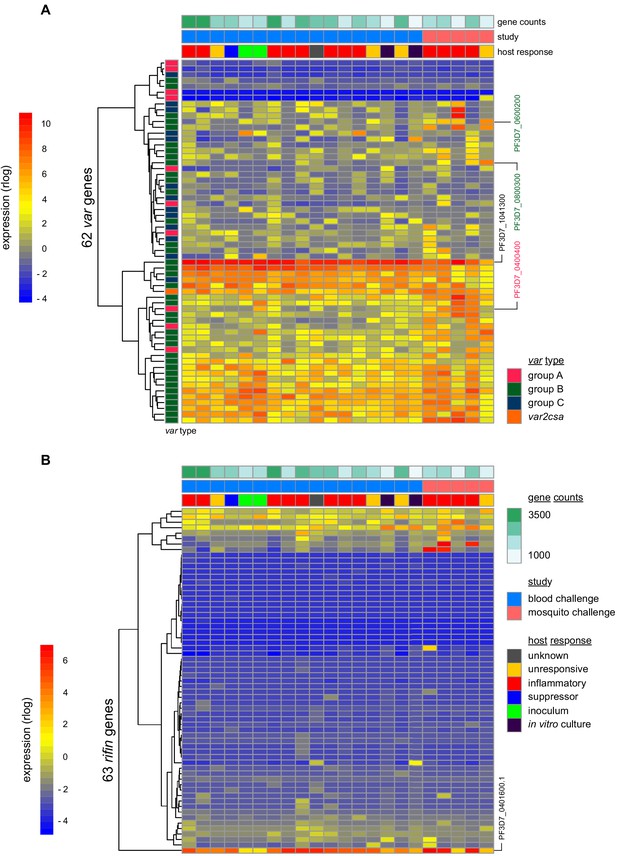
Parasite variants associated with severe disease do not rapidly expand in naive hosts.
(A and B) Rlog expression values of var (A) and rifin (B) genes in the inoculum and diagnosis parasite samples after blood challenge (blue study); parasites obtained from volunteers infected by mosquito bite are also shown (pink study). Genes (rows) are ordered by hierarchical clustering and colour-coded by var type; parasite samples are colour-coded by host response; and in vitro cultured ring-stage parasites are shown for comparison. Two volunteers did not have parasite sequencing data (unresponsive volunteer v208 and suppressor volunteer v022) and the two inoculum samples are technical replicates of one biological sample. The var (PF3D7_1041300) and rifin (PF3D7_0401600) genes dominantly expressed across all samples are labelled. Var genes associated with severe disease (group A variant PF3D7_0400400 and DC8-like variants PF3D7_0600200 and PF3D7_0800300) are also labelled. Gene counts show the number of parasite genes that have at least three uniquely mapping reads; this provides a measure of genome coverage in every sample.
-
Figure 4—source data 1
Rlog expression values of var and rifin genes in parasites isolated from whole blood after controlled human malaria infection.
- https://cdn.elifesciences.org/articles/62800/elife-62800-fig4-data1-v1.xlsx
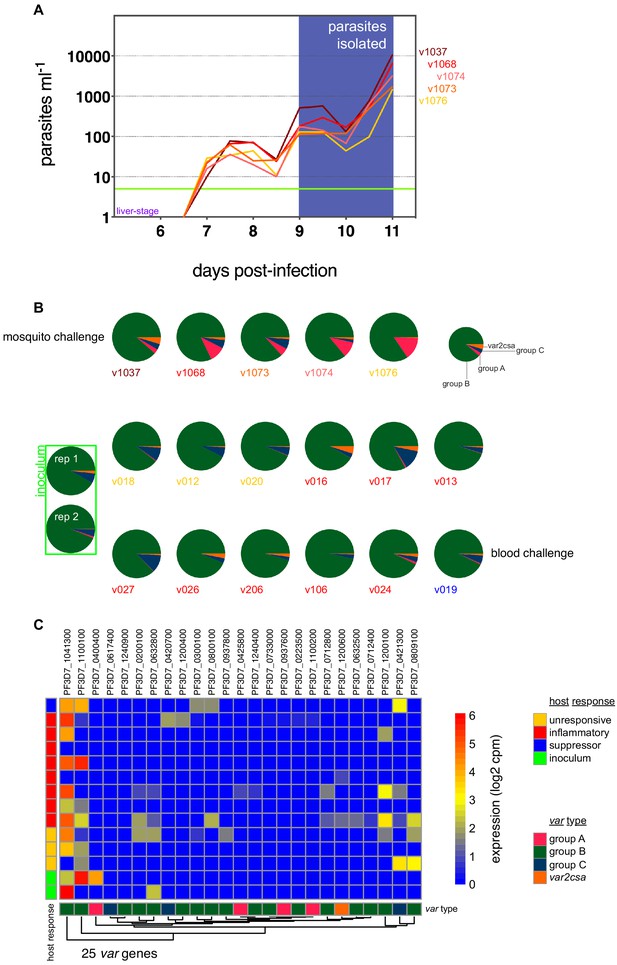
Group B var genes dominate the PfEMP1 landscape in naive hosts.
(A) Parasite growth curves in five volunteers infected by mosquito bite; each line represents one volunteer and the horizontal green line represents the lower limit of detection (five parasites ml−1). On days 9, 10, and 11 post-infection parasites were isolated from whole blood and immediately processed for ex vivo RNA-sequencing – for every volunteer, read counts were pooled from all three time-points to generate a comprehensive parasite transcriptome 2–3 cycles after liver egress. (B) Proportion of read counts that map to groups A, B, C, or E (var2csa) var genes after mosquito challenge (top row) or blood challenge (bottom two rows); parasite samples are colour-coded by host response. Note that the two inoculum samples are a technical replicate of a single biological sample. (C) Log2(cpm) expression values of var gene intron-spanning reads in the inoculum and diagnosis samples after blood challenge. Genes (columns) are ordered by hierarchical clustering and colour-coded by var type; parasite samples by host response.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Plasmodium falciparum) | clone 3D7 | Cheng et al., 1997 | PMID:9347970 | |
| Sequence-based reagent | TaqMan probe | Applied Biosystems | 5′ FAM AACAATTGGAGGGCAAG NFQ-MGB 3′ | |
| Sequence-based reagent | ISPCR primer | biomers.net | 5’- AAG CAGTGGTATCAACGCAG AGT −3’ | |
| Sequence-based reagent | LNA-modified TSO | exiqon.com | 5’- AAGCAGTGGTATCAACGCAGAGTACATrGrG+G −3’ | |
| Sequence-based reagent | Oligo-dT30VN | biomers.net | 5’- AAGC AGTGGTATCAACGCAGAGT ACT30VN −3’ | |
| Commercial assay or kit | Tempus spin RNA isolation | Applied Biosystems | cat. no. 4378926 | |
| Commercial assay or kit | RNA clean and concentrator | Zymo Research | cat. no. R1016 | |
| Commercial assay or kit | Globin-Zero Gold | Illumina | cat. no. GZG1224 | |
| Commercial assay or kit | RNA 6000 pico chip | Agilent | cat. no. 5067–1513 | |
| Commercial assay or kit | High sensitivity DNA kit bioanalyzer | Agilent | cat. no. 5067–4626 | |
| Commercial assay or kit | TruSeq stranded mRNA library | Illumina | cat. no. RS-122–2101 | |
| Commercial assay or kit | NEBNext Ultra DNA library prep kit | New England Biolabs | cat. no. E7370 | |
| Software, algorithm | Kallisto v0.42.3 | Bray et al., 2016 | RRID:SCR_016582 | PMID:27043002 |
| Software, algorithm | EdgeR | Robinson et al., 2010 | RRID:SCR_012802 | PMID:19910308 |
| Software, algorithm | ClueGO v2.5.4 | Bindea et al., 2009 | RRID:SCR_005748 | PMID:19237447 |
| Software, algorithm | IDEOM | Creek et al., 2012 | PMID:22308147 | |
| Software, algorithm | mzCloud | RRID:SCR_014669 | ||
| Software, algorithm | DESeq2 | Love et al., 2014 | RRID:SCR_015687 | PMID:25516281 |
| Other | Saponin from quillaja bark | Sigma Aldrich | cat. no. S7900 | test each batch |
| Other | Leucoflex LXT filters | Macopharma | macopharma.com/transfusion | |
| Other | KAPA HiFi HotStart ReadyMix | Biosystems | cat. no. KK2601 | |
| Other | AMPure XP beads | Beckman Coulter | cat. no. A63881 |
Additional files
-
Supplementary file 1
Demographics of volunteers infected by blood or mosquito challenge; includes genetic and non-genetic variables known to influence human immune variation in vitro.
Classification of volunteers by host response after blood challenge is described in the results and methods.
- https://cdn.elifesciences.org/articles/62800/elife-62800-supp1-v1.xlsx
-
Supplementary file 2
Three metrics that measure the magnitude of the human immune response to blood challenge.
[1] The mean variance of the top 50 most variable protein-coding genes in each volunteer. [2] The Euclidean distance travelled during principal component analysis of the 517-gene superset. [3] The number of differentially expressed genes at diagnosis in each group. Note that uninfected control volunteers set a threshold for baseline variation in gene expression through time; a Mann Whitney test was then used to support the observation that unresponsive volunteers were comparable to uninfected controls. Classification of volunteers by host response after blood challenge is described in the results and methods.
- https://cdn.elifesciences.org/articles/62800/elife-62800-supp2-v1.xlsx
-
Supplementary file 3
List of the 2028 differentially expressed genes identified in inflammatory volunteers at diagnosis (compared to pre-infection samples).
These data underpin the gene ontology network in Figure 2a.
- https://cdn.elifesciences.org/articles/62800/elife-62800-supp3-v1.xlsx
-
Supplementary file 4
List of the 217 significantly enriched GO terms identified in inflammatory volunteers at diagnosis.
GO terms are arranged into their functional groups and groups are ordered by size. The leading GO term in the top 12 functional groups is highlighted in red. On sheet 2, we highlight the top 20 GO terms as ordered by p value.
- https://cdn.elifesciences.org/articles/62800/elife-62800-supp4-v1.xlsx
-
Supplementary file 5
List of the 77 differentially expressed genes identified in suppressor volunteers at diagnosis (compared to pre-infection samples).
These data underpin the heatmap in Figure 2d.
- https://cdn.elifesciences.org/articles/62800/elife-62800-supp5-v1.xlsx
-
Supplementary file 6
List of the 893 genes that were differentially expressed between inflammatory and suppressor hosts at diagnosis.
There were no differentially expressed genes between these volunteers prior to infection (adj p<0.05).
- https://cdn.elifesciences.org/articles/62800/elife-62800-supp6-v1.xlsx
-
Supplementary file 7
Rlog expression values of the 117-gene superset in whole blood after mosquito challenge.
These data underpin the heatmap in Figure 2—figure supplement 1a. On sheet two we highlight the volunteers in which EdgeR identified these genes as differentially expressed. And on sheet three we look at the 21 genes that were most upregulated in inflammatory volunteers after blood challenge.
- https://cdn.elifesciences.org/articles/62800/elife-62800-supp7-v1.xlsx
-
Supplementary file 8
List of the 44 significantly enriched GO terms identified in the 117-gene superset.
GO terms are arranged into their functional groups and groups are ordered by size. On sheet two we highlight the top 20 GO terms as ordered by p value.
- https://cdn.elifesciences.org/articles/62800/elife-62800-supp8-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62800/elife-62800-transrepform-v1.docx





