Developmental hourglass and heterochronic shifts in fin and limb development
Figures
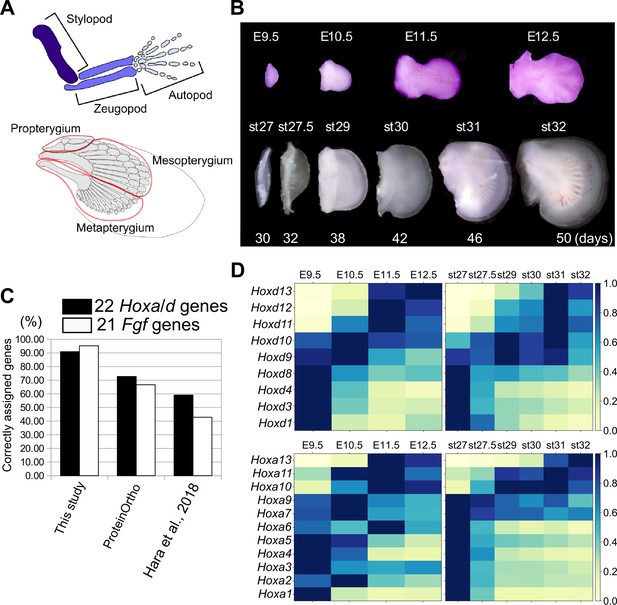
Transcriptome analysis and orthology assignment.
(A) The skeletal patterns of a mouse limb (top) and a bamboo shark pectoral fin (bottom). Anterior is to the top; distal is to the right. (B) Mouse forelimb buds and bamboo shark pectoral fin buds that were analyzed by RNA-seq. (C) Comparison of the accuracy of three orthology assignment methods. Vertical axis, the percentages of correctly assigned Hoxa and Hoxd paralogs (black bars) and Fgf paralogs (white bars). (D) Heat map visualization of the transcription profile of Hoxa and Hoxd genes in mouse limb buds (left) and bamboo shark fin buds (right) with scaled TPMs.
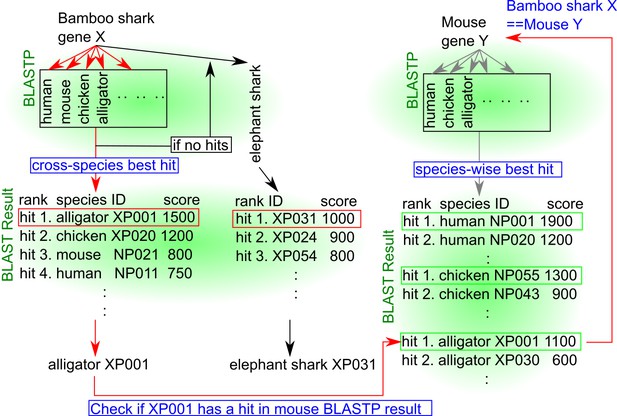
Schematic representation of the orthology assignment algorithm.
Red arrows, the main flow of the algorithm. Black arrows, orthology assignment for cartilaginous fish-specific genes. Gray arrows, parallel retrieving of orthologs of mouse genes from other animals. Red rectangles, best hits across other animal genes or in elephantfish genes. Green rectangles, best hits among each animal genome. Note that this schematic explains how the orthology of abstract genes ‘bamboo shark gene X’ and ‘mouse gene Y’ are assigned. First, using BLASTP, putative orthologs of bamboo shark genes are retrieved from other animal genomes, such as human, mouse, alligator, and elephantfish. Then, all BLASTP results except those from elephantfish are concatenated to find a best scored gene across species (cross-species best hit). In this schematic, the alligator gene XP001 is the best hit. In parallel, putative orthologs of mouse genes are also retrieved from the same set of animal genomes. If there is a mouse gene Y that has a best hit with alligator XP001, this mouse gene Y and bamboo shark gene X are considered to be an orthologous pair.
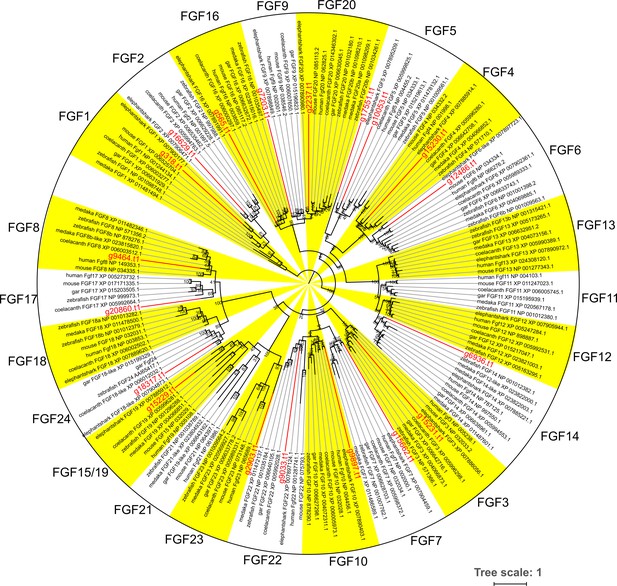
Molecular phylogenetic tree for Fgf family.
The tree was inferred with the maximum-likelihood method. The support values at nodes indicate bootstrap probabilities. Genes highlighted in red are bamboo shark genes (can be converted into the original gene ID by replacing ‘g’ with ‘Chipu’ and fill the digit with 0 to be seven figure number in total).
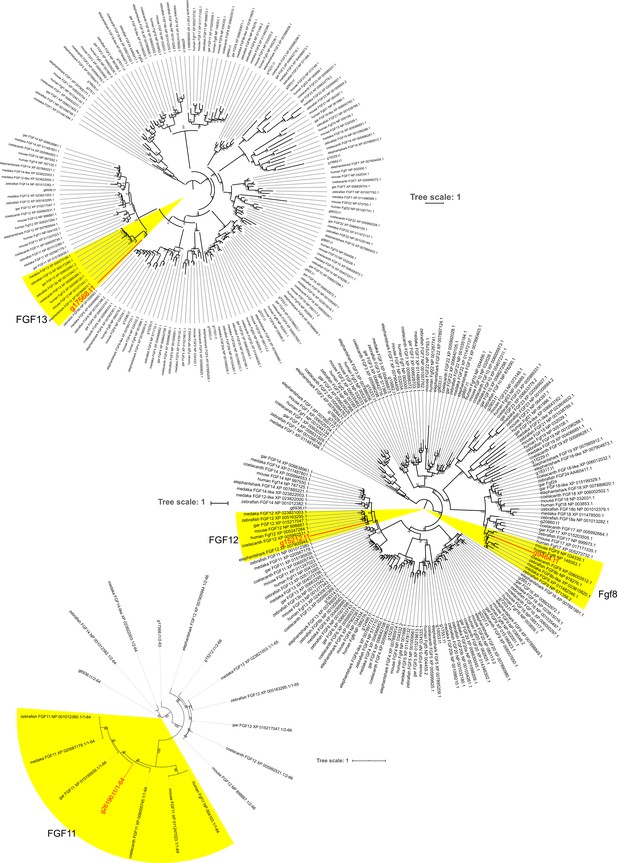
Additional molecular phylogenetic trees for Fgf8, Fgf11, Fgf12, and Fgf13.
These trees are shown because alignment sequences used in Figure 1—figure supplement 2 are truncated or absent in these genes. The tree was inferred with the maximum-likelihood method. The support values at nodes indicate bootstrap probabilities. Genes highlighted in red are bamboo shark genes.
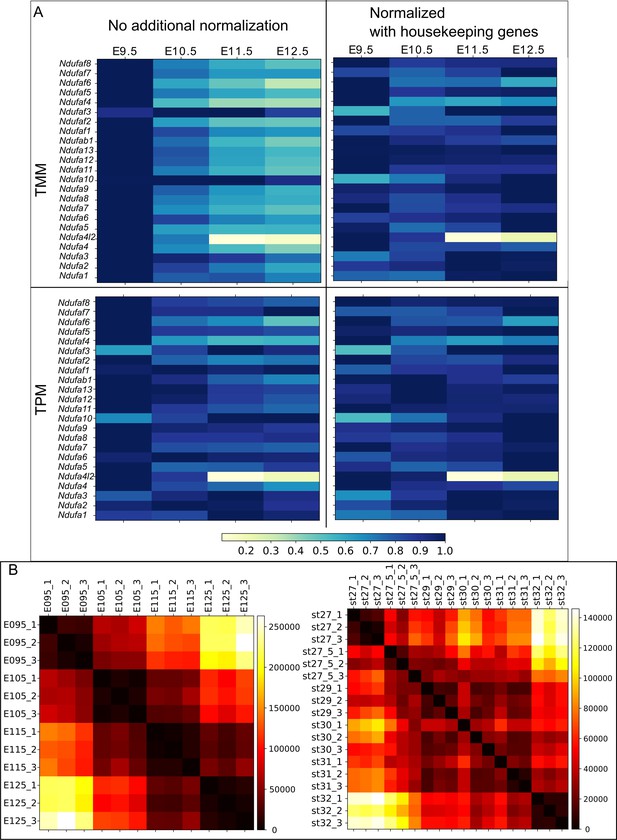
Comparison between the TPM and TMM.
(A) Visualization of the effect of normalization by showing a housekeeping gene family, Ndufa. Left panels show TMM (trimmed mean of M) and TPM (transcripts per million) calculated by RSEM. Right panels show these values with additional normalization using several other housekeeping genes (Atp5j, Atp5h, Atp5g3, Psmc3, Psmc5, Psmd7, Mrpl54, Mrpl46, Polr2e, Polr1b, Mrpl2). Housekeeping genes are selected from a previously published list (https://www.tau.ac.il/~elieis/HKG/HK_genes.txt; Eisenberg and Levanon, 2013). All expression values are standardized by setting the maximum expression value of each gene as ‘1’. Note that because housekeeping genes do not change their expression amount over time, these expression values should be close to ‘1’ (i.e. all colors should be dark blue) with some exceptions. However, the intact TMM (top, left panel) is apparently biased, in that the majority of Ndufa genes show their strongest expression at E9.5, with sharp decreases at other stages. This bias can be corrected by normalization with other housekeeping genes (top right panel). In contrast, the intact TPM (bottom, left panel) has a weaker bias than TMM. Additional normalization (bottom, right panel) has less of an effect. Therefore, this study used the intact TPM. (B) Euclidean distances of transcriptome data between mouse samples (left) and between bamboo shark samples (right). Whereas the close relation of the replicates of mouse samples can be seen from this heat map, the replicates of bamboo shark samples show less similarity. This noisy data may be attributed to the fact that there is no established strain of the bamboo shark and/or that bamboo shark embryos were staged by morphology but not physical time. However, the average of replicates seems to mitigate the noise of the bamboo shark samples, because Hox gene expression showed a smooth temporal collinearity as seen in Figure 1D.
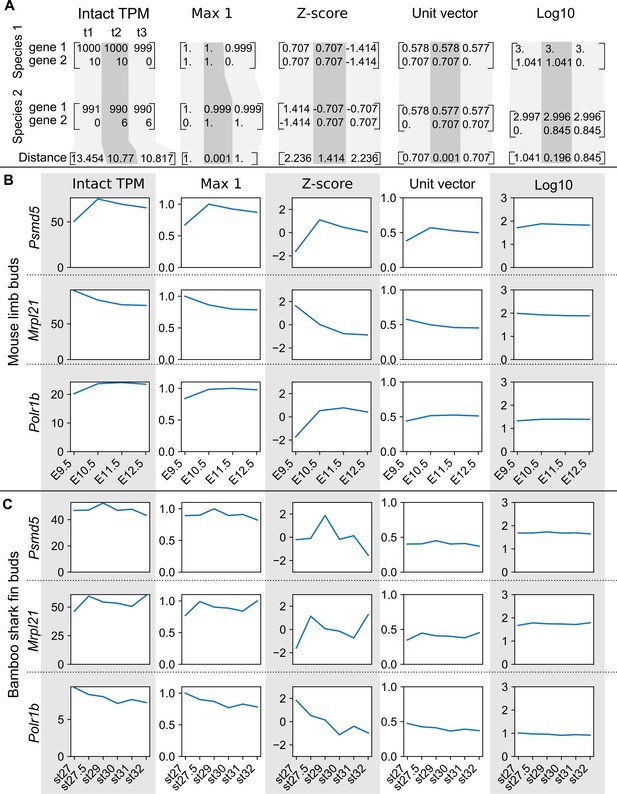
The effect of scaling methods to housekeeping genes.
(A) A simple example for comparing expression distances between two species. Species 1 and 2 are imaginery simple species that have two genes (genes 1 and 2) and three developmental time points (t1, t2, and t3). Distances in the bottom are the Euclidean distance between two species at each stage. (B, C) Housekeeping gene expressions with intact TPM values and different scaling methods in mouse limb buds (B) and bambooshark fin buds (C). Intact TPM, TPM values without any scaling methods; Max 1, TPM values were scaled by setting the highest TPM in each gene of each species to ‘1’; Z-score, the mean expression value was subtracted from each expression value and each result was then divided by the standard deviation; Unit vector, expression values were divided by the norm; Log10, log10 transformation. These housekeeping genes are listed in both a human housekeeping gene list (https://www.tau.ac.il/~elieis/HKG/HK_genes.txt) and the BUSCO data set (thus these genes are likely conserved in most vertebrates). Note that although the expression values of the housekeeping genes were almost constant during development, Z-score scaling amplifies subtle differences between stages. In addition, intact TPM values were not readily comparable between limb buds and bamboo shark fin buds (e.g. the maximum value of POLR1B in mouse limb buds was roughly twice as high as that of bamboo shark fin buds). Error bars are not displayed.
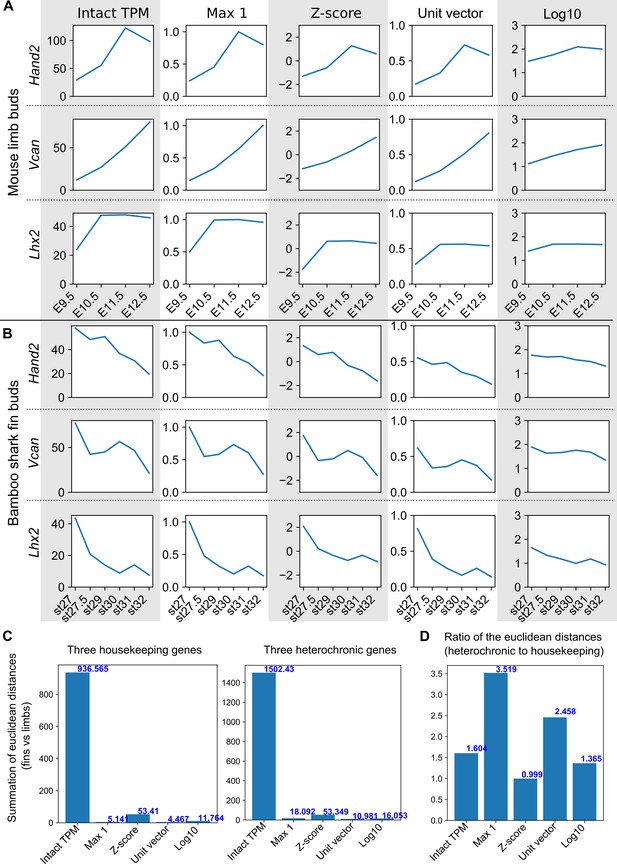
The effect of scaling methods to heterochronic genes.
(A, B) Heterochronic gene expressions with intact TPM values and different scaling methods in mouse limb buds (A) and bambooshark fin buds (B). See the legend of Figure 1—figure supplement 5 for scaling methods. Error bars are omitted. (C) The total Euclidean distance with respect to gene expression for the three housekeeping and the three heterochronic genes between mouse limb buds and bamboo shark fin buds. Using the housekeeping genes shown in (A, B) and the different scaling methods, the graph shows the summation of Euclidean distances between all combinations of mouse limb and bamboo shark fin stages. (D) The ratios of the Euclidean distances for housekeeping genes to those for heterochronic genes as shown in C.
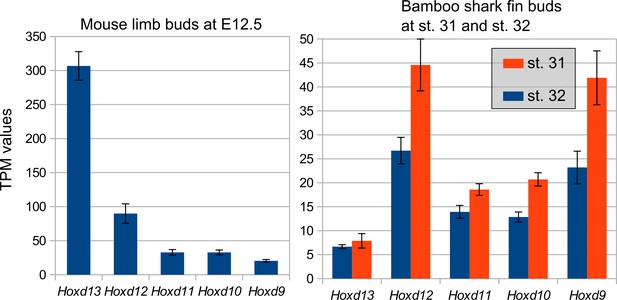
Examination of quantitative collinearity of Hoxd genes.
TPM values of 5ʹ Hoxd genes in mouse limb buds at E12.5 (left) and bamboo shark fin buds at st. 31 (right, orange bars) and st. 32 (right, blue bars). Error bars, SEM. Note that the genomic locus of Hoxd genes was positively correlated with their expression amount in the mouse limb bud, whereas no such correlation was found in the bamboo shark fin bud.
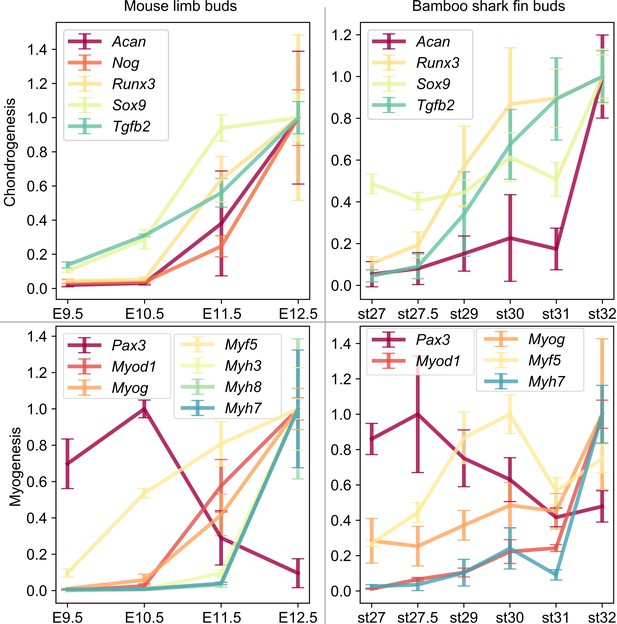
Expression profile of genes related to cellular differentiation.
(A, B) Scaled TPM values of indicated genes related to chondrogenesis (A) and myogenesis (B). Error bars, SEM.
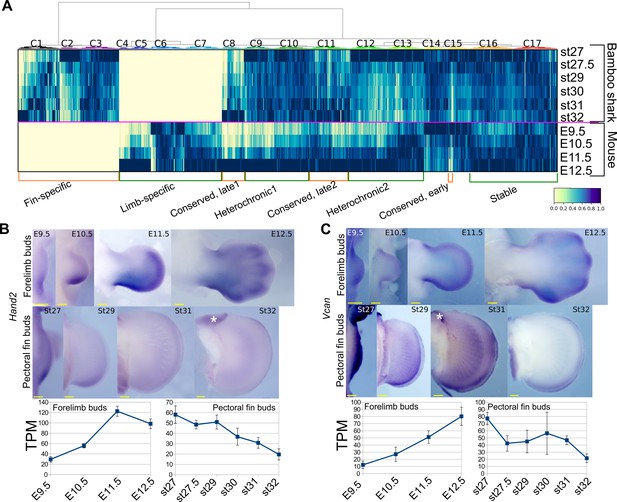
Detection of heterochronic gene expression between mouse limb buds and bamboo shark fin buds.
(A) Clustering analysis of gene expression dynamics. Each column represents an ortholog pair between the bamboo shark and the mouse. Each row indicates scaled gene expression at a time point indicated to the right of the heat map. Values are scaled TPMs. (B, C) Whole-mount in situ hybridization of Hand2 (B) and Vcan (C) as examples of the heterochronic genes detected in (A). Asterisks, background signals; scale bars, 200 μm. Error bars: SEM.
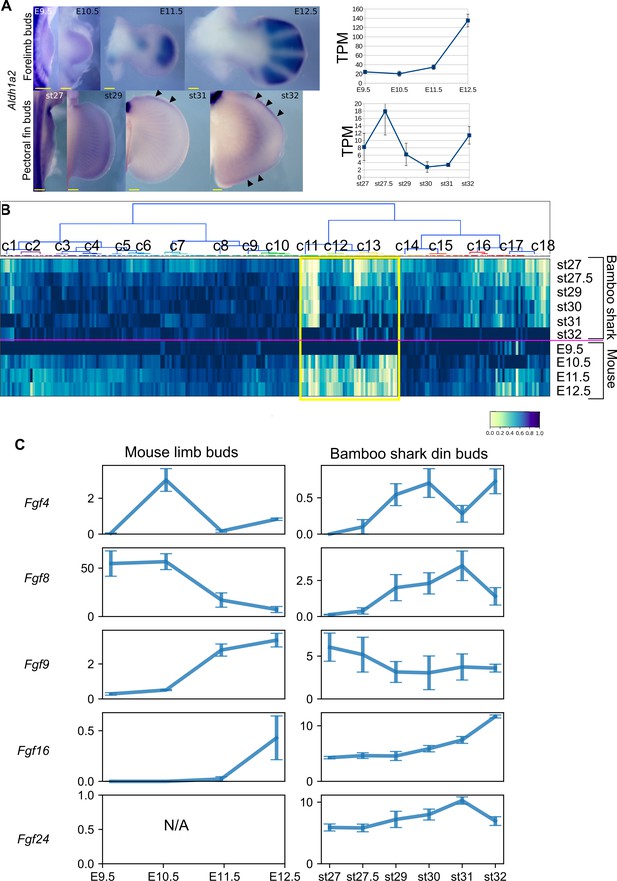
Other heterochronic genes.
(A) Left panels, whole mount in situ hybridization of Aldh1a2 (one of the genes from the cluster Heterochronic1) in bamboo shark fin buds and mouse limb buds. Right panels, TPM values of Aldh1a2. Arrowheads indicate the late-stage expression of Aldh1a2 in bamboo shark fin buds. Scale bars, 200 μm. Error bars, SEM. (B) Heatmap of genes that exhibit an inverse relation to Heterochronic2 genes in Figure 2A. Yellow empty box, genes that exhibit relatively sharp upregulation in bamboo shark fin buds and downregulation in mouse limb buds over time. (C) Comparison of Fgf gene expression. Vertical axis, TPM values; error bars, SEM; N/A, not applicable because of the absence of Fgf24 in the mouse genome.
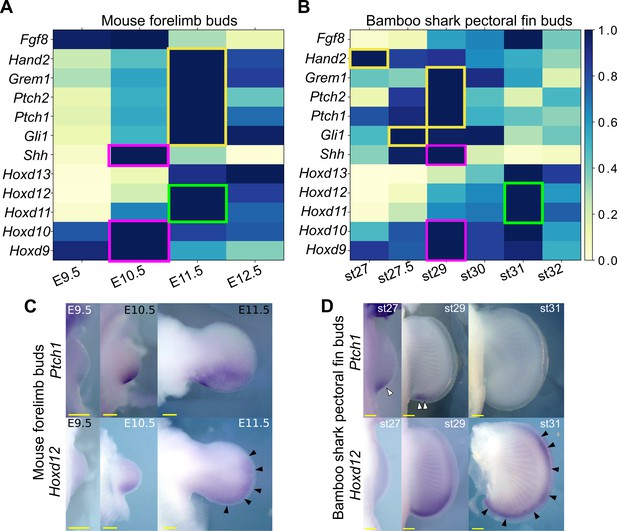
Shh pathway in mouse limb buds and bamboo shark fin buds.
(A, B) Scaled expression of Shh and related genes in mouse limb buds (A) and bamboo shark fin buds (B), respectively. The rectangles indicate the expression peaks of Shh, Hoxd9, and Hoxd10 (magenta), Shh target genes (yellow) and Hoxd11 and Hoxd12 (green). (C, D) Whole-mount in situ hybridization of Ptch1 and Hoxd12 in mouse limb buds (C) and bamboo shark fin buds (D); scale bars, 200 μm. White arrowheads in (D) indicate restricted expression of Ptch1 in bamboo shark fin buds. Black arrowheads in (C and D) indicate anteriorly extended expression of Hoxd12.
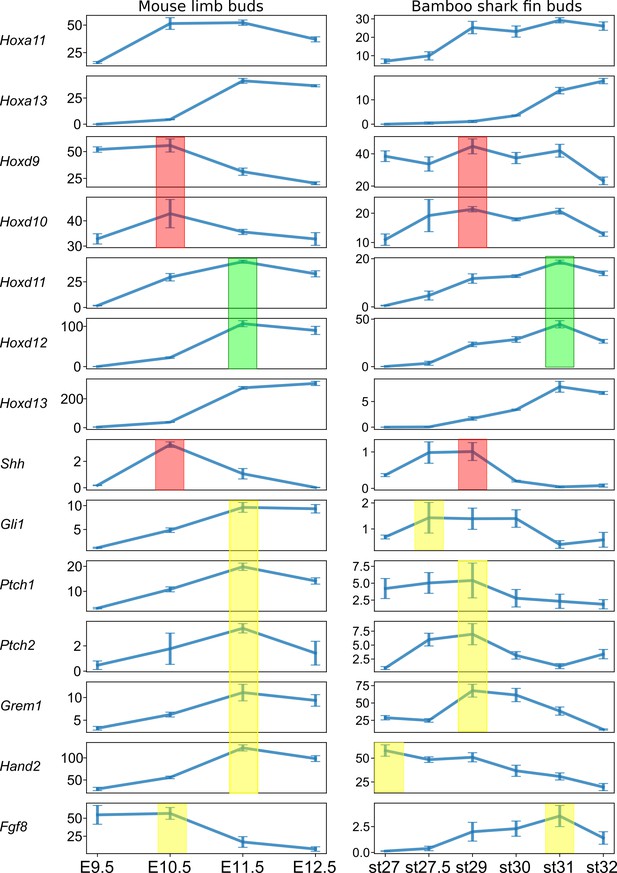
The temporal dynamics of the Shh pathway based on intact TPM values.
Red filled rectangles, the expression peak of Shh, Hoxd9, and Hoxd10; yellow filled rectangles, the expression peak of Shh target genes; green filled rectangles, the expression peak of Hoxd11 and Hoxd12.
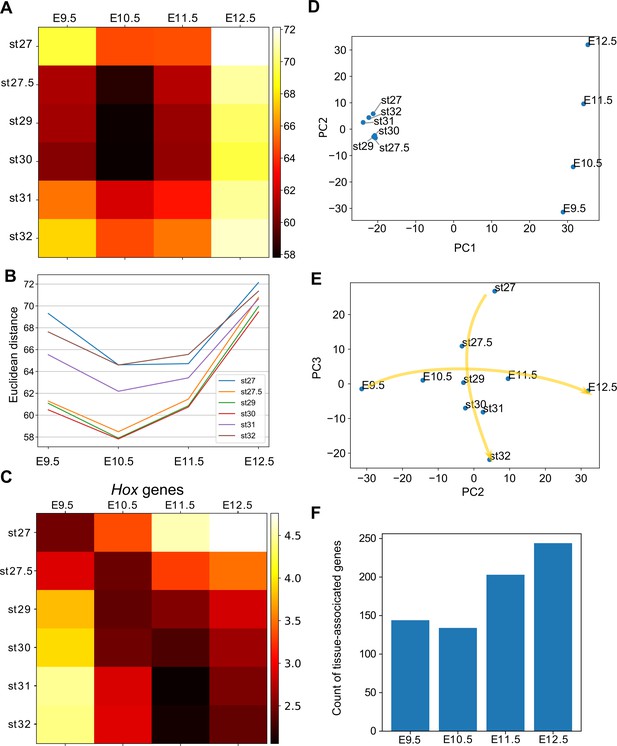
Hourglass-shaped conservation of the transcriptome profile between fins and limbs.
(A) Euclidean distances of the transcriptome profiles. Every combination of time points of bamboo shark fin buds and mouse limb buds is shown. The darker colors indicate a greater similarity between gene expression profiles. (B) A line plot of the Euclidean distances shown in (A). The x axis indicates the mouse limb stages, and the y axis is the Euclidean distance. (C) The same as (A) except that only Hoxd genes are included. (D, E) Scatter plots of the first and second principal components (D) and of the second and third components (e). Arrows in (E) indicate the time-order of transcriptome data. (F) Count of tissue-associated genes expressed in mouse forelimb buds. Genes with 0.65 ≤ entropy were counted.
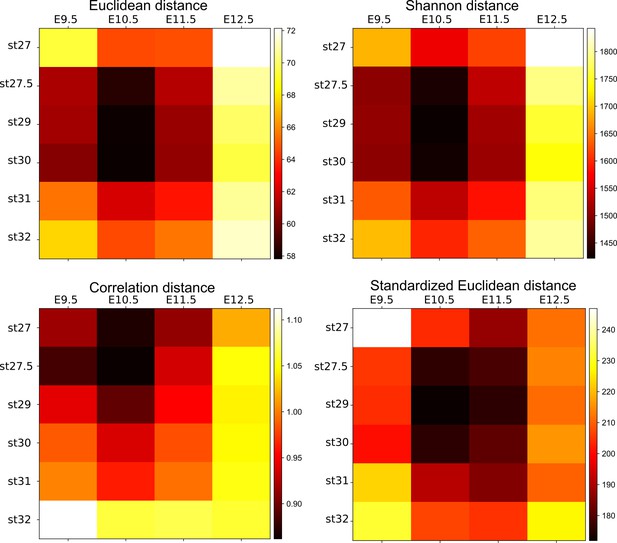
Confirmation analyses of the transcriptome comparison.
Cross-species comparisons of transcriptome data between the two species with indicated distance methods. Note that these methods consistently show the closest distance around E10.5 and st. 27.5–30.
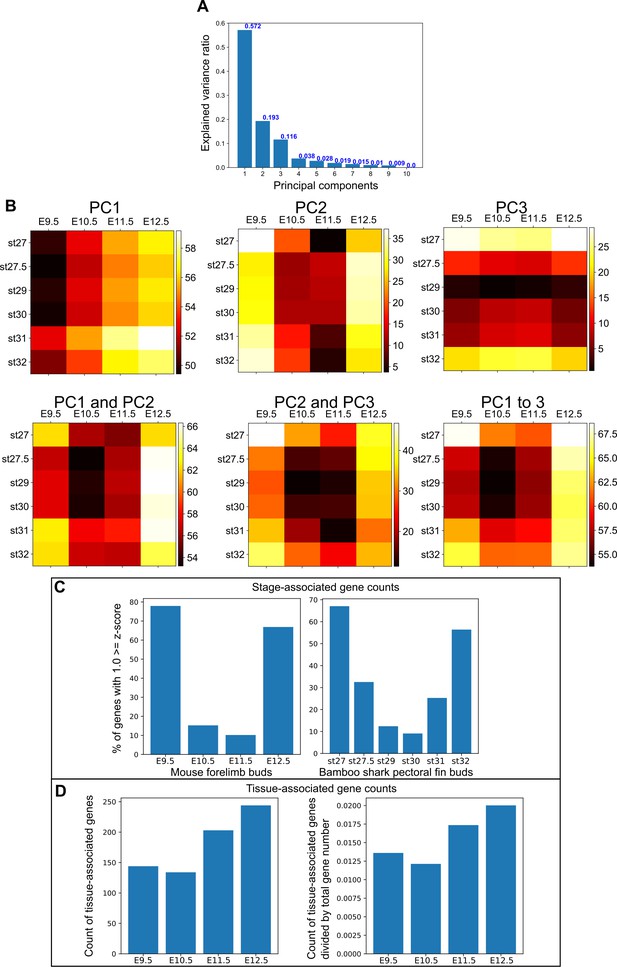
Additional PCA data and counts for stage- and tissue-associated genes.
(A) The ratio of explained variable for each of the principal components from Figure 4D and E. (B) Euclidean distance measures using the indicated principal components. Note that individual principal components do not reproduce the hourglass-shaped conservation shown in Figure 4A, but PC1, PC2, and PC3 are sufficient for the most part to reproduce Figure 4A. (C) Percentage of stage-associated genes with [z ≤ 1.0] for mouse limb buds (left) and bamboo shark fin buds (right). Note that both species showed a low percentage of stage-associated genes during the middle stages of development. (D) Number (left) and fraction (right) of tissue-associated genes expressed in mouse limb buds. Tissue specificity was evaluated by entropy using RNA-seq data from 71 mouse tissues. A gene with entropy ≥0.65 was considered a tissue-specific gene. In the right panel, gene counts were normalized based on the number of total expressed genes. Note that the number of tissue-associated genes was lowest at E10.5.
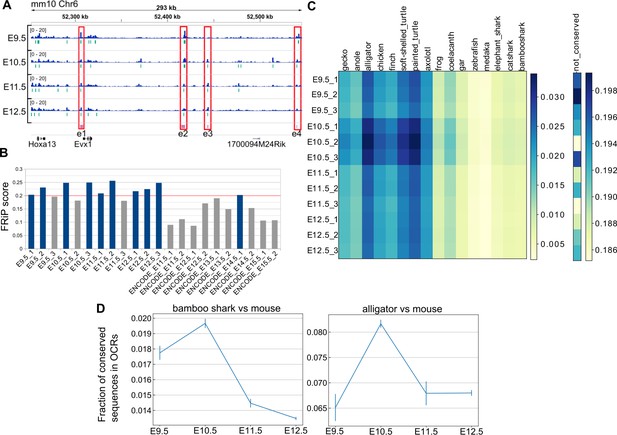
Hourglass-shaped conservation of OCRs in mouse limb development.
(A) ATAC-seq signals in the enhancer regions of the HoxA cluster. e1 to e4, known limb enhancers. Green vertical lines below the signals, peak regions determined by MACS2. (B) Comparison of a quality index, FRiP, for ATAC-seq data. Blue bars are samples with a FRiP score >0.2. The number in the end of the label name indicates the replicate number. (C) Conservation analysis of sequences in ATAC-seq peaks with BLASTN. The values to the right of each graph indicate the fraction of conserved sequences in the total peak regions. The common name of each genome sequence is indicated above the graph. The not-conserved heatmap indicates the fraction of sequences that were not aligned to any genome sequences and thus serve as a negative control. (D) Temporal changes of sequence conservation frequency in ATAC-seq peaks with LAST. Error bars: SEM.
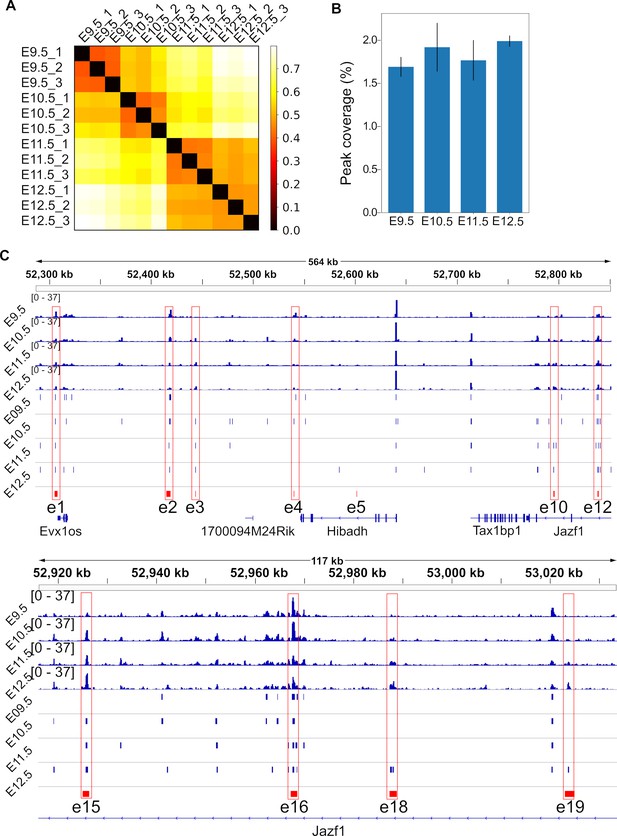
ATAC-seq quality control.
(A) Correlation distance between samples. The numbers in the end of the sample names indicate the replicates of indicated stages. Darker color means more similar gene expressions. (B) Percentage of peak regions in the genome sequence. (C) ATAC-seq signals in BPM (blue signals), peak regions (blue rectangles) and the known limb enhancers of HoxA cluster (red rectangles, e1–e19). Note that only e5 is not covered by ATAC-seq data.
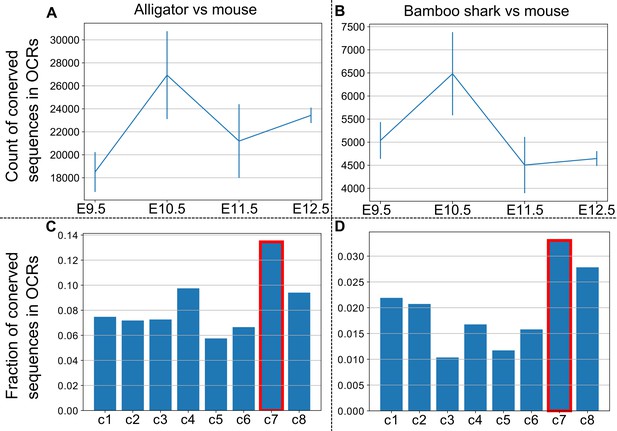
Conservation measures of OCRs.
(A, B) The absolute count of OCRs that overlap with sequences conserved between the mouse and the alligator (A) and the bamboo shark (B). Error bars, SEM. (C, D) The fraction of conserved OCRs sorted by the identified clusters in Figure 6A. Sequence conservation was estimated by pairwise alignment using LAST (A–D).
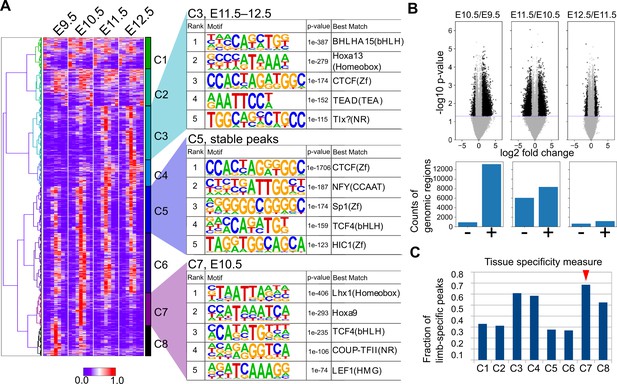
Temporal dynamics of OCRs during mouse limb development.
(A) The heatmap (left) shows whole-genome clustering of ATAC-seq peaks. Each row indicates a particular genome region with a length of 1400 bp. Columns indicate developmental stages. C1−C8 are cluster numbers. The motifs (right) show the rank of enriched motifs in the sequences of each cluster. (B) Top, volcano plots of ATAC-seq signals between indicated stages (p-values, two-sided Student’s t-test). Bottom, the counts of differential signals (black dots in the top panel). + and − are genomic regions with increased or decreased signals, respectively. (C) The fraction of limb-specific OCRs for each cluster.
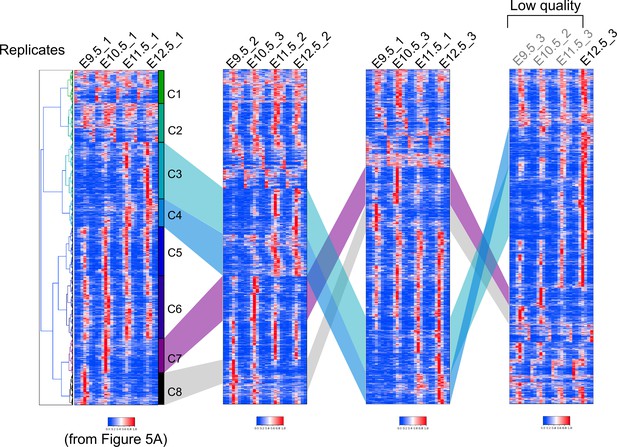
Clustering analyses of ATAC-seq peaks with different replicates.
DDifferent replicates were used for the same analysis as shown in Figure 6A. The number after the stage name indicates the replication number. Note that the clustering analyses with different replicates identified clusters similar to those in Figure 6A (compare the left-most panel with the second and third panels from the left). Including replicates with a low-quality score resulted in a relatively small fraction of early stage-specific peaks and a large fraction of late stage-specific peaks (right-most panel).

Analysis of enrichment for known motifs in ATAC-seq peaks.
The top five motifs from clusters C5 and C6 determined while using all other peaks as the background sequence.
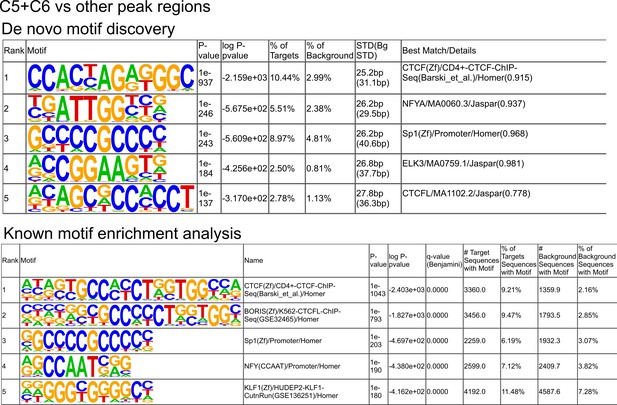
De novo motif discoveries and known motif enrichment analysis of ATAC-seq peaks with an alternative background.
The top five motifs from clusters C5 and C6 determined while using all other peaks as the background sequence.
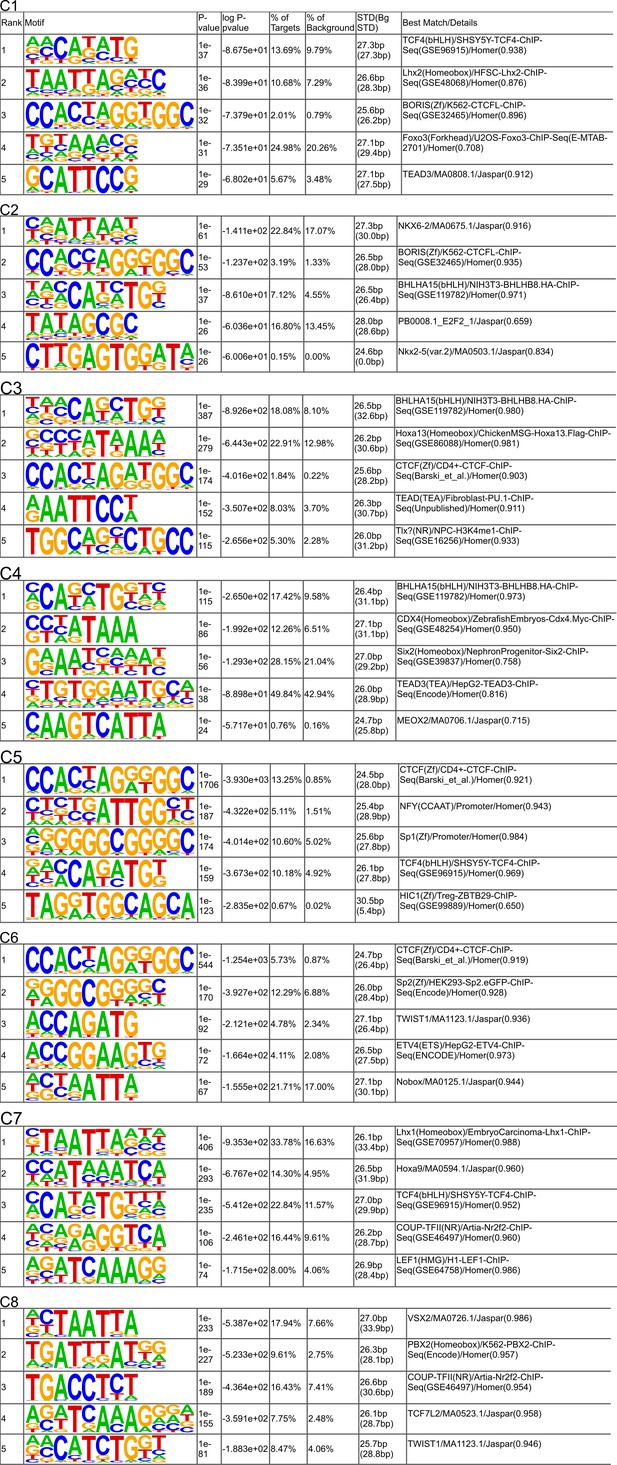
De novo motif discoveries of ATAC-seq peaks.
The top five motifs from each cluster. See Supplementary data for the full list of motifs. C1–C8 correspond to the clusters in Figure 6A.
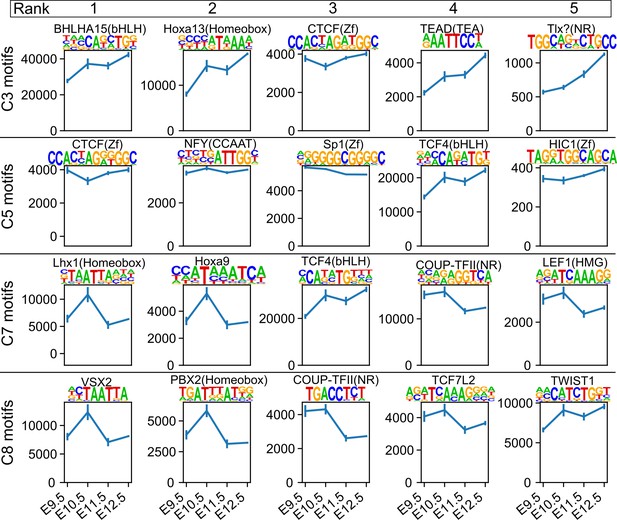
Counts of accessible motifs at each stage.
The average number of top-ranked motifs identified by de novo motif discovery in Figure 6—figure supplement 4 are plotted against mouse embryonic stages. The average numbers were calculated using all three replicates of ATAC-seq peaks at each stage. Rows indicate clusters identified in Figure 6A; columns indicate motif rank. Error bars, SEM. C1–C8 correspond to the clusters in Figure 6A. Note that the number of CTCF motifs (top-ranked in C5) was relatively stable over time, which is consistent with the clustering analysis shown in Figure 6A. In addition, the number of motifs enriched for C3, such as BHLHA15, HOX13, TEAD, and Tlx?, increased over time. In contrast, COUP-TFII and TCF7L2 motifs decreased over time. Interestingly, LHX and HOX9 motifs were transiently increased at E10.5.
Tables
Assembly statistics of bamboo shark transcriptome.
| Characteristic | Bamboo shark transcriptome | Bamboo shark gene model (Hara et al., 2018) |
|---|---|---|
| Total number of sequences | 222015 | 34038 |
| Total sequence length (bp) | 195541367 | 36633751 |
| Average length (bp) | 880 | 1076 |
| Maximum length (bp) | 18451 | 108594 |
| N count | 0 | 10208 |
| L50 | 24765 | 5666 |
| N50 length (bp) | 2075 | 1749 |
| Protein coding | 63898 | 34038 |
| Orthology detected | 41633 | 18180 |
| Unique orthologs | 14139 | 14907 |
| Unique orthologs without gene symbols | 1821 | 1780 |
| Unique orthologs only in elephantfish | 826 | 552 |
| Sequences with no orthology | 20892 | 15254 |
| Orthologs with mouse genes | 12326 | 13005 |
PCA loadings.
| Loading axis: PC2 | ||
|---|---|---|
| Gene symbol | Cluster name | Loading |
| TRHDE | C8 | 0.31 |
| PAX9 | C11 | 0.3 |
| COL9A2 | C8 | 0.3 |
| RTN4R | C8 | 0.3 |
| APC2 | C9 | 0.3 |
| CNMD | C8 | 0.29 |
| HOXD13 | C8 | 0.29 |
| FAM69C | C8 | 0.29 |
| WFIKKN2 | C8 | 0.29 |
| HOXA13 | C8 | 0.29 |
| LRRN3 | C12 | 0.29 |
| HPSE2 | C9 | 0.29 |
| SERPINB1A | C11 | 0.29 |
| CDKN2B | C8 | 0.28 |
| LTBP1 | C8 | 0.28 |
| CDH19 | C8 | 0.28 |
| PDZD2 | C8 | 0.28 |
| NLGN3 | C9 | 0.28 |
| MATN1 | C8 | 0.28 |
| MYOD1 | C8 | 0.28 |
| TSPAN11 | C12 | 0.28 |
| SERINC2 | C9 | 0.28 |
| FYB | C8 | 0.28 |
| KIF1A | C8 | 0.28 |
| COL9A3 | C8 | 0.28 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Hand2 | ENSEMBL | ENSMUST00000040104.4 | N/A |
| Gene (Mus musculus) | Vcan | ENSEMBL | ENSMUST00000109546.8 | N/A |
| Gene (Mus musculus) | Aldh1a2 | ENSEMBL | ENSMUST00000034723.5 | N/A |
| Gene (Mus musculus) | Ptch1 | ENSEMBL | ENSMUST00000192155.5 | N/A |
| Gene (Mus musculus) | Hoxd12 | ENSEMBL | ENSMUST00000001878.5 | N/A |
| Gene (Chiloscyllium punctatum) | Hand2 | ENSEMBL | Chipun0004250/g4250.t1/ TRINITY_DN85524_c0_g1_i1 | N/A |
| Gene (Chiloscyllium punctatum) | Vcan | This paper | Chipun0003941/g3941.t1/ TRINITY_DN95522_c0_g1_i8 | N/A |
| Gene (Chiloscyllium punctatum) | Hoxd12 | This paper | Chipun0005654/g5654.t1/TRINITY_DN85970_c0_TRINITYg1_i1 | N/A |
| Gene (Chiloscyllium punctatum) | Ptch1 | This paper | Chipun0003320/g3320.t1/TRINITY_DN92499_c0_g1_i3 | N/A |
| Gene (Chiloscyllium punctatum) | Aldh1a2 | This paper | Chipun0010503/g10503.t1/TRINITY_DN81423_c0_g1_i1 | N/A |
| Strain, strain background (Mus musculus) | C52BL/6 | Laboratory for Animal Resources and Genetic Engineering RIKEN, | N/A | N/A |
| Antibody | Anti-Digoxigenin-AP, Fab fragments (Sheep) | Millipore Sigma | Cat# 11093274910 | polyclonal (1:4000) |
| Sequence-based reagent | Mus musculus Hand2 forward primer | This paper | PCR primers | ACCAAACTCTCCAAGATCAAGACACTG |
| Sequence-based reagent | Mus musculus Hand2 reverse primer | This paper | PCR primers | TTGAATACTTACAATGTTTACACCTTC |
| Sequence-based reagent | Mus musculus Vcan forward primer | This paper | PCR primers | TGCAAAGATGGTTTCATTCAGCGACAC |
| Sequence-based reagent | Mus musculus Vcan reverse primer | This paper | PCR primers | ACACGTGCAGAGACCTGCAAGATGCTG |
| Sequence-based reagent | Mus musculus Aldh1a2 forward primer | This paper | PCR primers | ACCGTGTTCTCCAACGTCACTGATGAC |
| Sequence-based reagent | Mus musculus Aldh1a2 reverse primer | This paper | PCR primers | TCTGTCAGTAACAGTATGGAGAGCTTG |
| Sequence-based reagent | Mus musculus Hoxd12 forward primer | This paper | PCR primers | CTCAACTTGAACATGGCAGTGCAAGTG |
| Sequence-based reagent | Mus musculus Hoxd12 reverse primer | This paper | PCR primers | AGCTCTAGCTAGGCTCCTGTTTCATGC |
| Sequence-based reagent | Mus musculus Ptch1 forward primer | This paper | PCR primers | GGGAAGGCAGTTCATTGTTACTGTAACTG |
| Sequence-based reagent | Mus musculus Ptch1 reverse primer | This paper | PCR primers | TGTAATACGACTCACTATAGGTCAGAAGCTGCCACACACAGGCATGAAGC |
| Sequence-based reagent | Chiloscyllium punctatum Hand2 forward primer | This paper | PCR primers | ACCAGCTACATTGCCTACCTCATGGAC |
| Sequence-based reagent | Chiloscyllium punctatum Hand2 reverse primer | This paper | PCR primers | CACTTGTTGAACGGAAGTGCACAAGTC |
| Sequence-based reagent | Chiloscyllium punctatum Vcan forward primer | This paper | PCR primers | AGCTTGGGAAGATGCAGAGAAGGAATG |
| Sequence-based reagent | Chiloscyllium punctatum Vcan reverse primer | This paper | PCR primers | AGAGCAGCTTCACAATGCAGTCTCTGG |
| Sequence-based reagent | Chiloscyllium punctatum Aldh1a2 forward primer | This paper | PCR primers | TTGAACTTGTACTAAGTGGTATCGCTG |
| Sequence-based reagent | Chiloscyllium punctatum Aldh1a2 reverse primer | This paper | PCR primers | AGGATGTGAACATTAGGCTGACCTCAC |
| Sequence-based reagent | Chiloscyllium punctatum Hoxd12 forward primer | This paper | PCR primers | GCCAGTATGCAACAGATCCTCTGATGG |
| Sequence-based reagent | Chiloscyllium punctatum Hoxd12 reverse primer | This paper | PCR primers | CTAATGACCTGTTGTACTTACATTCTC |
| Sequence-based reagent | Chiloscyllium punctatum Ptch1 forward primer | This paper | PCR primers | TTCAGCCAGATTGCAGATTACATCAACC |
| Sequence-based reagent | Chiloscyllium punctatum Ptch1 reverse primer | This paper | PCR primers | TTCTCTGTGTTTCACATTCAACGTCCTG |
| Commercial assay or kit | Nextera DNA Sample Preparation Kit | Illumina | Cat# FC-121–1031 | |
| Commercial assay or kit | TruSeq Stranded mRNA LT Sample Prep Kit | Illumina | Cat# RS-122–2101 | |
| Software, algorithm | Trinity | https://github.com/trinityrnaseq/trinityrnaseq | RRID:SCR_013048 | N/A |
| Software, algorithm | Bowtie2 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | RRID:SCR_016368 | N/A |
| Software, algorithm | BWA | http://bio-bwa.sourceforge.net/ | RRID:SCR_010910 | N/A |
| Software, algorithm | MACS2 | https://github.com/macs3-project/MACS | RRID:SCR_013291 | N/A |
| Software, algorithm | HOMER | http://homer.ucsd.edu/homer/motif/ | RRID:SCR_010881 | N/A |
| Software, algorithm | RSEM | https://github.com/deweylab/RSEM | RRID:SCR_013027 | N/A |
| Software, algorithm | scikit-learn | https://scikit-learn.org/stable/ | RRID:SCR_002577 | N/A |
Additional files
-
Supplementary file 1
Summary of short-read sequencing data.
- https://cdn.elifesciences.org/articles/62865/elife-62865-supp1-v2.xlsx
-
Supplementary file 2
Orthology asignment for the transcriptome of the brown-banded bamboo shark.
Columns 1–4: transcriptome assembly ID, NCBI gene ID, gene symbol, blast score.
- https://cdn.elifesciences.org/articles/62865/elife-62865-supp2-v2.tsv
-
Supplementary file 3
Orthology asignment for the gene model of the brown-banded bamboo shark.
Columns 1–4: gene model ID, NCBI gene ID, gene symbol, blast score.
- https://cdn.elifesciences.org/articles/62865/elife-62865-supp3-v2.tsv
-
Supplementary file 4
Quality control of orthology assignment.
Source data to create Figure 1C.
- https://cdn.elifesciences.org/articles/62865/elife-62865-supp4-v2.xlsx
-
Supplementary file 5
The mean and SEM of TPM values of mouse limb RNA-seq data.
Source data for Figure 1D and other plots related to gene expression amount.
- https://cdn.elifesciences.org/articles/62865/elife-62865-supp5-v2.txt
-
Supplementary file 6
The mean and SEM of TPM values of bamboo shark fin RNA-seq data.
Source data for Figure 1D and other plots related to gene expression amount.
- https://cdn.elifesciences.org/articles/62865/elife-62865-supp6-v2.txt
-
Supplementary file 7
Clustered gene expression table with phenotype annotation.
The details of Figure 2A.
- https://cdn.elifesciences.org/articles/62865/elife-62865-supp7-v2.xlsx
-
Supplementary file 8
The list of genes downregulated over time in mouse limb buds being upregulated in bamboo shark fin buds over time (related to Figure 2—figure supplement 1B).
- https://cdn.elifesciences.org/articles/62865/elife-62865-supp8-v2.txt
-
Supplementary file 9
PCA loadings of Figure 4D and E.
- https://cdn.elifesciences.org/articles/62865/elife-62865-supp9-v2.txt
-
Supplementary file 10
- https://cdn.elifesciences.org/articles/62865/elife-62865-supp10-v2.xlsx
-
Supplementary file 11
GO analysis of ATAC-seq peaks.
c1 to c8 correspond to the clusters in Figure 5A.
- https://cdn.elifesciences.org/articles/62865/elife-62865-supp11-v2.tsv
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62865/elife-62865-transrepform-v2.pdf





