The critical role of Hedgehog-responsive mesenchymal progenitors in meniscus development and injury repair
Figures
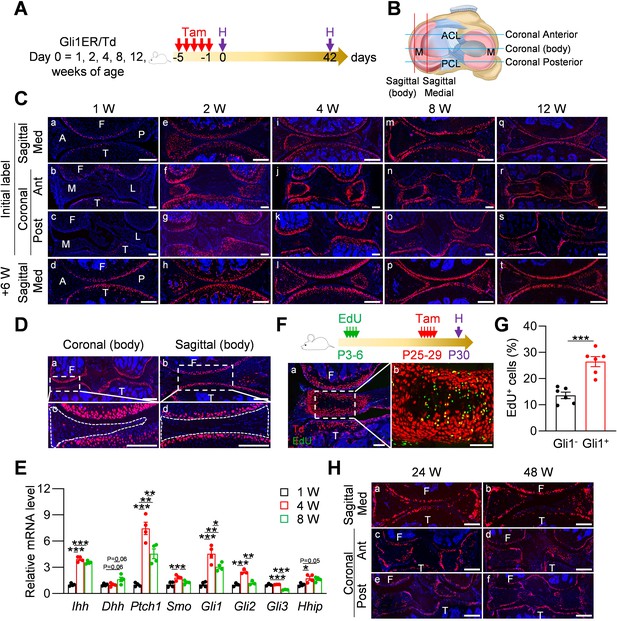
Gli1 labels mesenchymal progenitors in mouse meniscus during development.
(A) Schematic graph of the study protocol. Male Gli1ER/Td mice were treated with Tam at 1, 2, 4, 8, and 12 weeks of age and analyzed at 24 hr (pulse) or 6 weeks (tracing) after the last Tam dosing. (B) Schematic cartoon of meniscus shows sectioning sites. M: meniscus; ACL: anterior cruciate ligament; PCL: posterior cruciate ligament. (C) Representative fluorescence images of meniscus sections at indicated ages and sectioning sites. n = 3 mice/age/sectioning site. Scale bars, 200 μm. F: femur; T: tibia; A: anterior; P: posterior; M: medial meniscus; L: lateral meniscus; Med: medial; Ant: anterior; Post: posterior. Red: Td; Blue: DAPI. (D) Representative fluorescence images of meniscus body at coronal (a) and sagittal (b) planes from 12-week-old Gli1ER/Td mice. Meniscus were harvested at 24 hr after the last Tam injection. n = 3 mice/sectioning site. Scale bars, 200 μm. Boxed areas in a and b are shown at high magnification as c and d, respectively. Dashed lines outline meniscus. (E) qRT-PCR analysis of Hh signaling component genes in mouse meniscus tissues at 1, 4, 8 weeks of age. n = 4 independent experiments. (F) Top panel is a schematic representation of the study protocol. Gli1ER/Td mice were injected with EdU at P3-6 and Tam at P25-29. Joints were harvested 24 hr later. Representative confocal images of coronal sections of mouse knee joints are presented at the bottom panel. Boxed area in a (Scale bars, 200 μm) is shown at high magnification in b (Scale bars, 50 μm). Green: EdU. (G) The percentage of EdU+ cells within Gli1+ or Gli1- meniscus cells was quantified. n = 6 mice/group. (H) Gli1ER/Td mice were treated with Tam at 24 or 48 weeks of age and analyzed 24 hr later. Representative fluorescence images of sagittal (a, b) and coronal (c–f) sections of knee joints are presented. Scale bars, 200 μm. Statistical analysis was performed using unpaired two-tailed t-test and one-way ANOVA with Tukey-Kramer post-hoc test. Data presented as mean ± s.e.m. *p<0.05, **p<0.01, ***p<0.001.
-
Figure 1—source data 1
Raw data for Figure 1G and E.
- https://cdn.elifesciences.org/articles/62917/elife-62917-fig1-data1-v1.xlsx
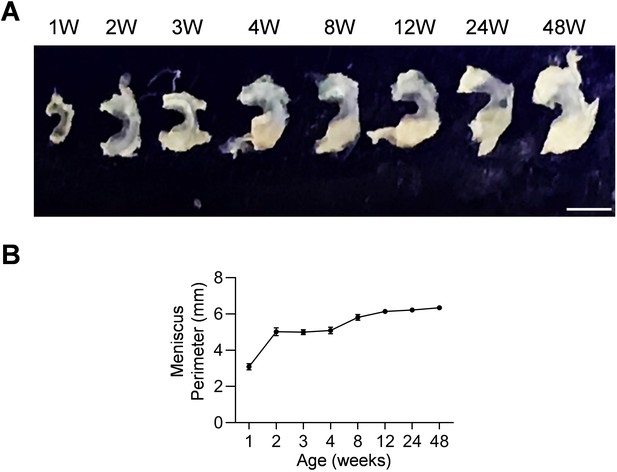
Mouse meniscal morphogenesis during development.
(A) The morphological overview of meniscus at 1, 2, 3, 4, 8, 12, 24, 48 weeks of age. Scale bars, 1 mm. (B) The meniscal perimeter was quantified. n = 3 mice/age.
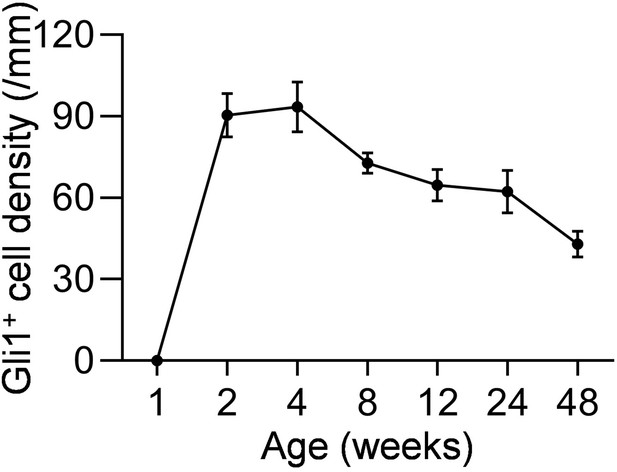
The density of Gli1+ cells along meniscus surface was measured in mice at different ages.
n = 5 mice/age.
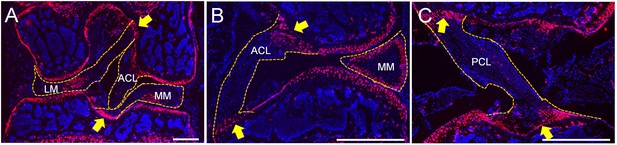
Meniscal enthesis and ligamental enthesis regions in joint are enriched with Gli1+ cells.
Gli1ER/Td mice at 12 weeks of age received Tam injections followed by tissue harvest 24 hr later. Knees were sectioned to show meniscal enthesis regions (A) and ligamental enthesis regions (B, C) within the knee joint. MM: medial meniscus; LM: lateral meniscus; ACL: anterior cruciate ligament; PCL: posterior cruciate ligament. Yellow arrows indicate the enthesis regions.
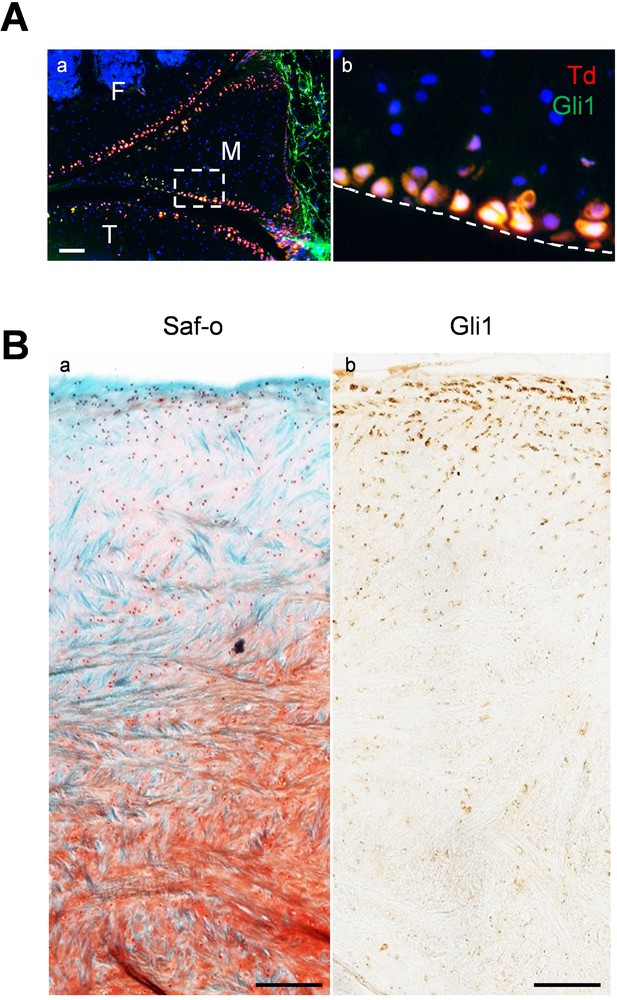
Gli1 labels superficial zone cells of mouse and mini-pig meniscal horns.
(A) Immunofluorescence staining of Gli1 (green) on sagittal sections of 12-week-old Gli1ER/Td mouse knee joints. Boxed area in a is enlarged in b. Dashed line indicates the surface of meniscus. Scale bars, 200 μm. F: femur; T: tibia; M: meniscus. Blue: DAPI, Red: Td; Green: Gli1. (B) Representative safranin O/fast green staining (left) and immunohistochemistry staining of Gli1 (right) in the horn area of mini-pig meniscus. Scale bars, 200 μm.
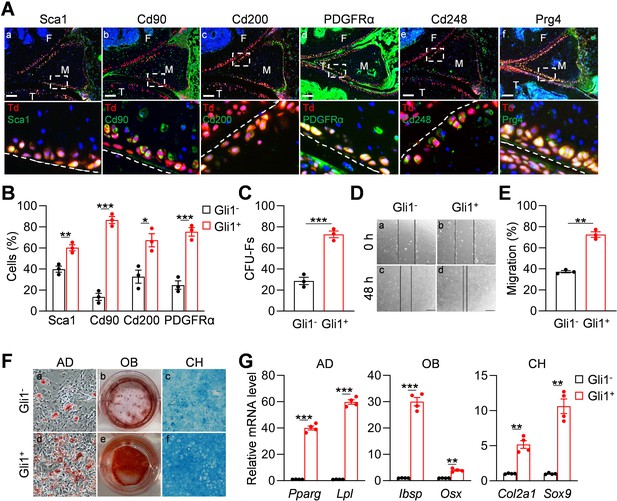
Gli1-labeled meniscus cells possess mesenchymal progenitor properties.
(A) Representative immunofluorescence images of Sca1, Cd90, Cd200, PDGFRα, Cd248, and Prg4 staining in 3-month-old Gli1ER/Td meniscus. Scale bars, 200 μm. Boxed areas are shown at high magnification in corresponding panels at the bottom. Dashed lines indicate meniscus surface. Yellow cells are double positive for progenitor marker and Td. F: femur; T: tibia; M: meniscus. Blue: DAPI. (B) Quantification of the expression level of mesenchymal progenitor markers in Gli1+ and Gli1- cells from meniscus. Digested meniscus cells from 3-month-old Gli1ER/Td mice were subjected to flow cytometry analysis. n = 3 independent experiments. (C) CFU-F assay of digested meniscus cells. Td+colonies and Td- colonies were counted from 1 × 104 seeded cells. n = 3 independent experiments. (D) Representative bright-field images of the scratch-wound closure in Gli1+ or Gli1- meniscus cells at 0 and 48 hr. Scale bars, 200 μm. Solid lines indicate the remaining area not covered by meniscus cells. (E) The relative migration rate was measured by the percentage of scratched area being covered by migrated cells at 48 hr. n = 3 independent experiments. (F) Representative adipogenic (AD), osteogenic (OB), and chondrogenic (CH) differentiation images of Gli1+ and Gli1- cells. Cells were stained by Oil Red, Alizarin red, and Alcian blue, respectively. (G) qRT-PCR analysis of lineage markers in Gli1- or Gli1+meniscal cells after being cultured in adipogenic, osteogenic and chondrogenic differentiation media for 1, 2, and 3 weeks, respectively. n = 4 independent experiments. Statistical analysis was performed using unpaired two-tailed t-test. Data presented as mean ± s.e.m. *p<0.05, **p<0.01, ***p<0.001.
-
Figure 2—source data 1
Raw data for Figure 2B,C,E,G.
- https://cdn.elifesciences.org/articles/62917/elife-62917-fig2-data1-v1.xlsx
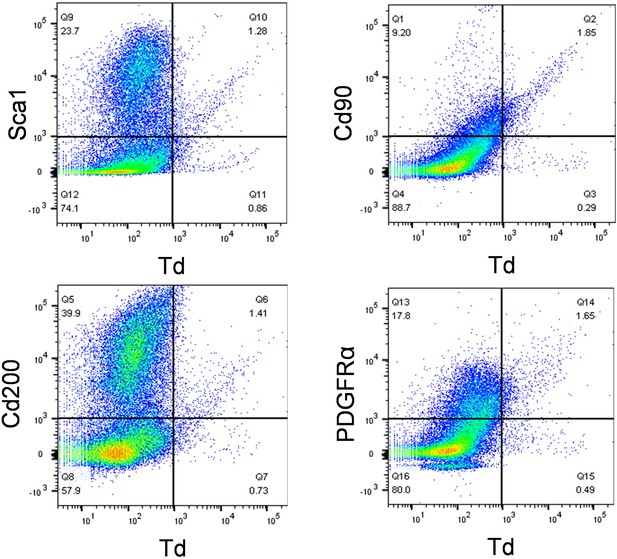
Mesenchymal progenitor markers are enriched in Gli1+ meniscus cells.
Digested meniscus cells from 3-month-old Gli1ER/Td mice were subjected to flow cytometry analysis.
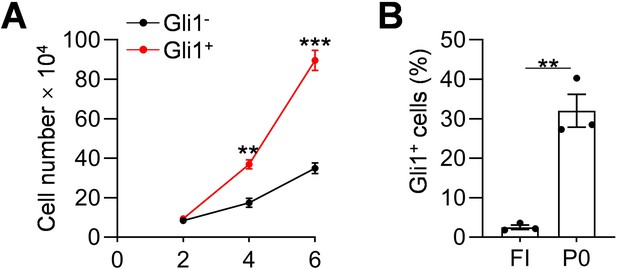
Gli1+ cells proliferate faster than Gli1- cells.
(A) Td(Gli1)+ and Td- cells digested from meniscus cells from Gli1ER/Td mice were grown to confluence and then seeded at 50,000 cells/well on day 0. Cell number/well was counted every other day. n = 3 independent experiments. (B) The percentage of Td+ cells from freshly isolated meniscus cells (FI) and from cells being cultured for 7 days (P0) was quantified by flow cytometry. n = 3 independent experiments. Statistical analysis was performed using unpaired two-tailed t-test. Data presented as mean ± s.e.m. **p<0.01, ***p<0.001.
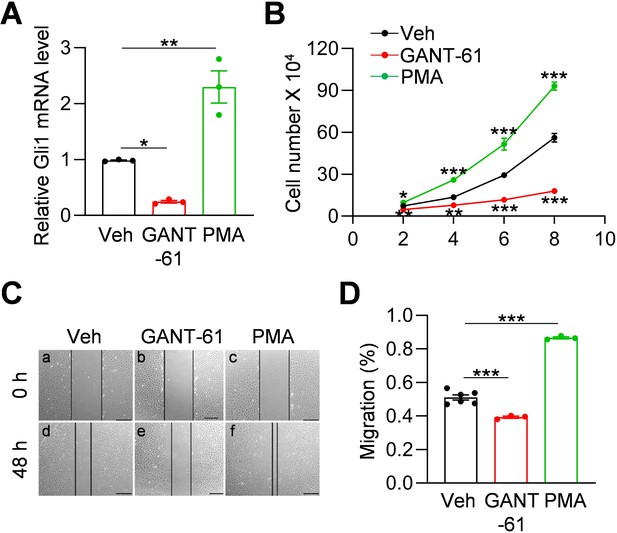
Hh signaling stimulates proliferation and migration of meniscus mesenchymal progenitors.
(A) qRT-PCR analysis of Gli1 mRNA in primary mouse meniscus cells treated with vehicle, GANT-61 (10 μM) or PMA(1 μM) for 48 hr. n = 3 independent experiments. (B) The proliferative ability of primary mouse meniscus cells was up-regulated by PMA and down-regulated by GANT-61 over 8 days of culture. n = 3 independent experiments. (C) Representative bright-field images of the scratch-wound closure in meniscus cells treated with veh, GANT-61 or PMA after 48 hr. Scale bars, 200 μm. Solid lines indicate the remaining area not covered by meniscus cells. (D) The relative migration rate was measured. n = 3–6 independent experiments. Statistical analysis was performed using one-way ANOVA with Dunnett's post-hoc test. Data presented as mean ± s.e.m. *p<0.05, **p<0.01, ***p<0.001.
-
Figure 3—source data 1
Raw data for Figure 3A,B,D.
- https://cdn.elifesciences.org/articles/62917/elife-62917-fig3-data1-v1.xlsx
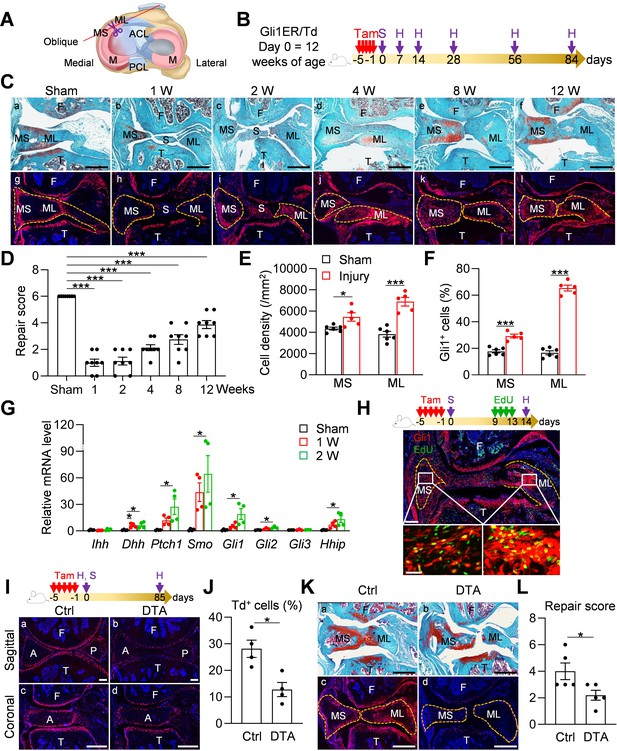
Meniscus injury rapidly expands Gli1-lineage cells.
(A) Schematic cartoon of meniscus shows the sectioning site. M: meniscus; MS: meniscus synovial end; ML: meniscus ligamental end. A pair of scissors indicates the transection site. ACL: anterior cruciate ligament; PCL: posterior cruciate ligament. (B) Male Gli1ER/Td mice received Tam injections (day −5 ~ −1) and meniscus injury (day 0) at 12 weeks of age. Knee joints were harvested at 1, 2, 4, 8, 12 weeks after injury. (C) Representative safranin O/fast green staining (top) and fluorescence images (bottom) of oblique sections of mouse knee joints harvested at indicated time points after injury. Dashed lines outline the meniscus. Scale bars, 200 μm. F: femur; T: tibia; S: synovium; MS: meniscus synovial end; ML: meniscus ligamental end. Red: Td; Blue: DAPI. (D) Repair score was evaluated at indicated time points after meniscus injury. n = 8 mice/group. (E) Cell density in the synovial and ligamental ends of meniscus was quantified at 4 weeks post meniscus injury. n = 5–6 mice/group. (F) The percentage of Td+ cells in the synovial and ligamental ends of meniscus was also quantified. n = 5–6 mice/group. (G) qRT-PCR analysis of Hh signaling component genes in meniscus at 1 and 2 weeks post injury. n = 4 independent experiments. (H) Top panel is a schematic representation of the study protocol. Gli1ER/Td mice at 12 weeks of age were treated with Tam (day −5 to −1), meniscus injury (day 0), and EdU injections (day 9–13). A representative confocal image of knee joint at day 14 is shown below (Scale bars, 250 μm). Boxed areas of synovial and ligamental ends of meniscus (MS and ML, respectively) are shown at high magnification at the bottom (Scale bar, 25 μm). Green: EdU. (I) Top panel is a schematic representation of the study protocol. Gli1ER/Td (Ctrl) or Gli1ER/Td/DTA (DTA) mice were treated with Tam (day −5 to −1) and meniscus injury at 12 weeks of age (day 0). Representative fluorescent images of sagittal and coronal mouse knee joint sections at day 0 without injury are shown below (Scale bars, 200 μm). A: anterior; P: posterior. (J) The percentage of Td+ cells in the anterior horn was quantified. n = 4 mice/group. (K) Representative safranin O/fast green staining (a, b) and fluorescence images (c, d) of oblique sections of mouse knee joints harvested at 12 weeks after injury. Dashed lines outline the meniscus. Scale bars, 200 μm. (L) Repair score was evaluated. n = 5 mice/group. Statistical analysis was performed using one-way ANOVA with Dunnett’s post-hoc test for (D), one-way ANOVA with Tukey-Kramer post-hoc test for (G) and unpaired two-tailed t-test for (E), (F), (J) and (L). Data presented as mean ± s.e.m. *p<0.05, ***p<0.001.
-
Figure 4—source data 1
Raw data for Figure 4D,E,F,G,J,L.
- https://cdn.elifesciences.org/articles/62917/elife-62917-fig4-data1-v1.xlsx
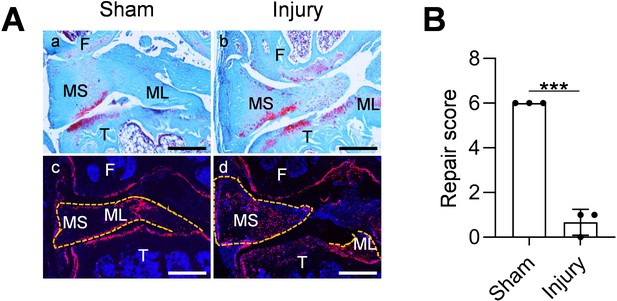
Aging diminishes Gli1-lineage cell expansion and the repair ability of meniscus.
(A) Representative safranin O/fast green staining (top) and fluorescence images (bottom) of aged mouse knee joints at 4 weeks after sham or meniscus injury. Gli1ER/Td mice at 12 months of age received Tam followed by meniscus injury. Dashed lines outline the meniscus. Scale bars, 200 μm. F: femur; T: tibia; MS: meniscus synovial end; ML: meniscus ligamental end; Red: Td; Blue: DAPI. (B) Repair score was quantified. n = 3 mice/group. Statistical analysis was performed using unpaired two-tailed t-test. Data presented as mean ± s.e.m. ***p<0.001.
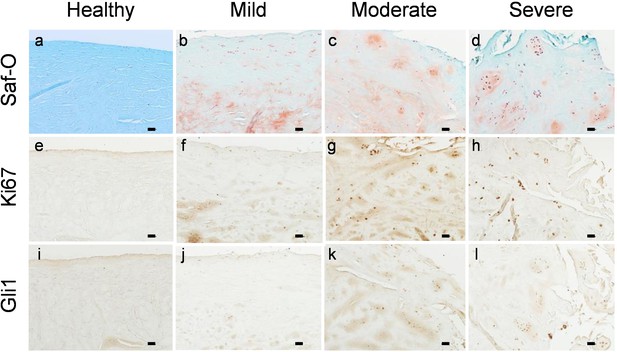
Gli1+ cells appear in proliferative cell clusters of degenerated human meniscus.
Representative safranin O/fast green staining (a–d) and immunohistochemistry staining of Ki67 (e–h) and Gli1 (i–l) in human meniscus tissues at different degenerative stages. n = 3 samples/stage. Scale bars, 200 μm.
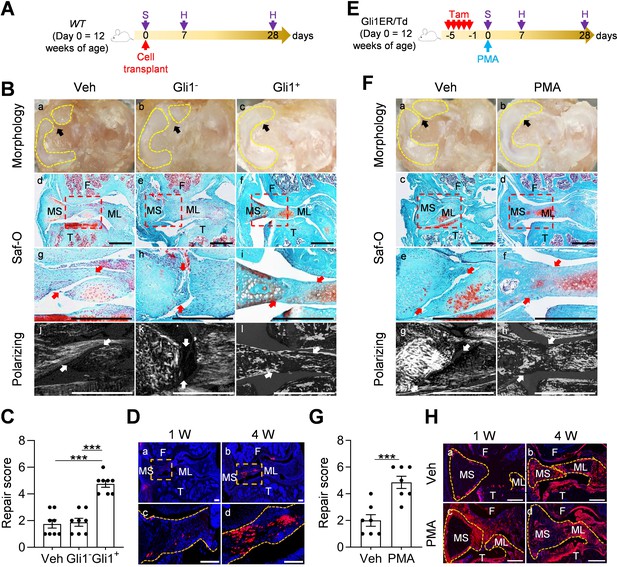
Activation of Hh/Gli1 pathway accelerates mouse meniscus repair.
(A) Schematic representation of the study protocol. WT mice received meniscus injury at 12 weeks of age followed by transplantation of 5000 Gli1+ or Gli1- meniscus cells at the injury site. Knee joints were harvested at 1 and 4 weeks after injury. (B) Representative overview (a–c), safranin O/fast green staining (d–i), and polarizing images (j–l) of mouse knee joints at 4 weeks after injury. Yellow dashed lines outline the overview morphology of injured meniscus. Meniscus is shown attached to tibial plateau. Arrows point to the injury site. Red boxed areas in d-f are shown at high magnification in g-i, respectively. Scale bars, 200 μm. F: femur; T: tibia; MS: meniscus synovial end; ML: meniscus ligamental end. (C) Repair score was evaluated. n = 8 mice/group. (D) Representative confocal images of mouse knee joints at 1 and 4 weeks after injury. Boxed areas in the top panel are shown at a high magnification at the bottom panel. Dashed line outlines meniscus. Scale bars, 200 μm. Blue: DAPI, Red: Td. (E) Schematic representation of the study protocol. Gli1ER/Td mice received Tam injections and meniscus injury at 12 weeks of age (day 0) followed by vehicle and PMA injection. Knee joints were harvested at 1 and 4 weeks after injury. (F) Representative overview (a, b), safranin O/fast green staining (c–f), and polarizing images (g, h) of mouse knee joints at 4 weeks after injury. Yellow dashed lines outline the overview morphology of injured meniscus. Meniscus is shown attached to tibial plateau. Red boxed areas in c and d are shown at high magnification in e and f, respectively. Arrows point to the injury site. Scale bars, 200 μm. (G) Repair score was evaluated. n = 7 mice/group. (H) Representative fluorescence images of vehicle- and PMA-treated mouse meniscus at 1 and 4 weeks after injury. Scale bars, 200 μm. Statistical analysis was performed using one-way ANOVA with Tukey-Kramer post-hoc test for (C) and unpaired two-tailed t-test for (G). Data presented as mean ± s.e.m. ***p<0.001.
-
Figure 5—source data 1
Raw data for Figure 5C and G.
- https://cdn.elifesciences.org/articles/62917/elife-62917-fig5-data1-v1.xlsx
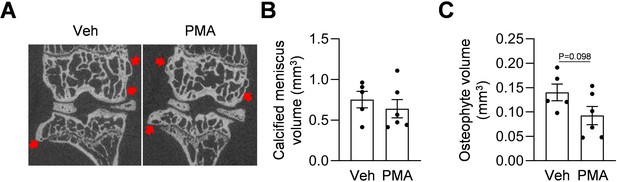
Hh agonist treatment does not promote joint calcification.
(A) Representative microCT images of WT mouse knee joints at 3 months post surgery as well as vehicle (Veh) or PMA treatment. (B) The volume of calcified meniscus was quantified. n = 5–6 mice/group. (C) Osteophyte volume was quantified. n = 5–6 mice/group. Statistical analysis was performed using unpaired two-tailed t-test. Red arrows indicate the osteophytes. Data presented as mean ± s.e.m.
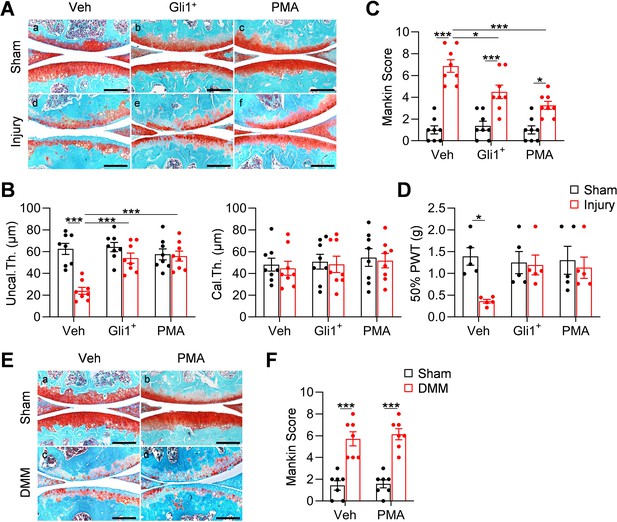
Meniscus repair by enhancing Hh/Gli1 signaling delays OA progression.
(A) Representative safranin O/fast green staining of sagittal sections of vehicle-, Gli1+ cell- and PMA-treated mouse knee joints at 8 weeks after sham or meniscus injury. Scale bars, 200 μm. (B) Average thicknesses of uncalcified zone (Uncal.Th.) and calcified zone (Cal.Th.) of the tibial articular cartilage were quantified. n = 8 mice/group. (C) The OA severity was measured by Mankin score. n = 8 mice/group. (D) Von Frey assay was performed at 8 weeks after injury. PWT: paw withdrawal threshold. n = 5 mice/group. (E) Representative safranin O/fast green staining of sagittal sections of vehicle- and PMA-treated mouse knee joints at 8 weeks after sham or DMM surgery. Scale bars, 200 μm. (F) The OA severity was measured by Mankin score. n = 7 mice/group. Statistical analysis was performed using two-way ANOVA with Tukey-Kramer post-hoc test. Data presented as mean ± s.e.m. *p<0.05, **p<0.01, ***p<0.001.
-
Figure 6—source data 1
Raw data for Figure 6B,C,D and F.
- https://cdn.elifesciences.org/articles/62917/elife-62917-fig6-data1-v1.xlsx
CFU-F assay of total digested meniscus cells.
(A) Crystal-violet staining of CFU-F colonies in a dish. (B) Brightfield and fluorescence images of a representative Gli1+ CFU-F colony in the dish.
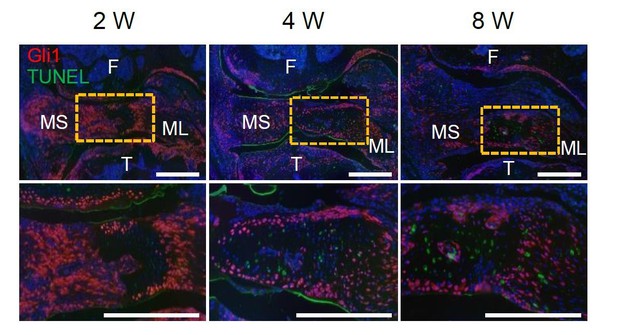
Representative TUNEL fluorescent staining of Gli1ER/Td mouse knee joints harvested at 2, 4, or 8 weeks after meniscus injury.
Mice received Tamoxifen injections right before the injury. Boxed area at the top is shown at a high magnification at the bottom (Scale bars, 200 μm). F: femur; T: tibia; MS: meniscus synovial end; ML: meniscus ligamental end. Red: Td; Green: TUNEL; Blue: DAPI.
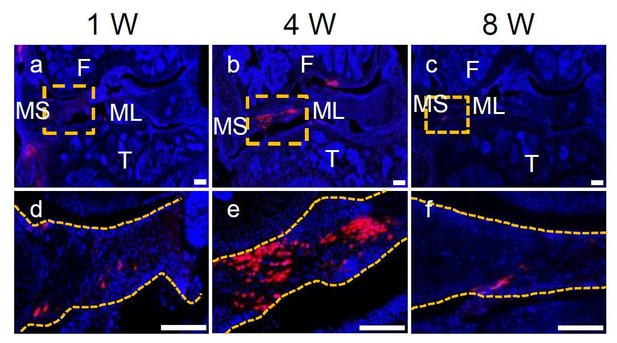
Representative confocal images of WT mouse knee joints at 1, 4, and 8 weeks after meniscus injury and injection of Gli1+ cells derived from Gli1ER/Td meniscus.
Boxed areas in the top panel are shown at high magnification at the bottom. Dashed line outlines meniscus. Scale bars, 200 μm. Blue: DAPI; Red: Td.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, Strain background (Mus musculus) | Gli1CreER | Jackson Laboratory | Stock No: 007913 | |
| Strain, Strain background (Mus musculus) | Rosa26 lsl-tdTomato | Jackson Laboratory | Stock No: 007909 | |
| Strain, Strain background (Mus musculus) | Rosa26 lsl-DTA | Jackson Laboratory | Stock No: 010527 | |
| Sequence-based reagent | Gli1CreER Primer 1 | Jackson Laboratory | PCR Primer | 5’-GCGGTCTGGCAGTAAAAACTATC-3’ |
| Sequence-based reagent | Gli1CreER Primer 2 | Jackson Laboratory | PCR Primer | 5’-GTGAAACAGCATTGCTGTCACTT-3’ |
| Sequence-based reagent | Gli1CreER Primer 3 | Jackson Laboratory | PCR Primer | 5’-CACGTGGGCTCCAGCATT-3’ |
| Sequence-based reagent | Gli1CreER Primer 4 | Jackson Laboratory | PCR Primer | 5’-TCACCAGTCATTTCTGCCTTTG-3’ |
| Sequence-based reagent | Rosa26 lsl-tdTomato Primer 1 | Jackson Laboratory | PCR Primer | 5’-AAGGGAGCTGCAGTGGAGTA-3’ |
| Sequence-based reagent | Rosa26 lsl-tdTomato Primer 2 | Jackson Laboratory | PCR Primer | 5’-CCGAAAATCTGTGGGAAGTC-3’ |
| Sequence-based reagent | Rosa26 lsl-tdTomato Primer 3 | Jackson Laboratory | PCR Primer | 5’-GGCATTAAAGCAGCGTATCC-3’ |
| Sequence-based reagent | Rosa26 lsl-tdTomato Primer 4 | Jackson Laboratory | PCR Primer | 5’-CTGTTCCTGTACGGCATGG-3’ |
| Sequence-based reagent | Rosa26 lsl-DTA Primer1 | Jackson Laboratory | PCR Primer | 5’-CGACCTGCAGGTCCTCG-3’ |
| Sequence-based reagent | Rosa26 lsl-DTA Primer 2 | Jackson Laboratory | PCR Primer | 5’-CCAAAGTCGCTCTGAGTTGTTATC-3’ |
| Sequence-based reagent | Rosa26 lsl-DTA Primer 3 | Jackson Laboratory | PCR Primer | 5’-GAGCGGGAGAAATGGATATG-3’ |
| Sequence-based reagent | Rosa26 lsl-DTA Primer 4 | Jackson Laboratory | PCR Primer | 5’-CTCGAGTTTGTCCAATTATGTCAC-3’ |
| Antibody | Rabbit polyclonal Anti-Gli1 | NOVUS biologicals | NB600-600 | IF (1:100) |
| Antibody | Rabbit polyclonal Anti-Ki67 | Abcam | ab15580 | IF (1:50) |
| Antibody | Rat monoclonal Anti-Sca1 | Santa Cruz Bio | sc-52601 | IF (1:200) |
| Antibody | Rat monoclonal Anti-Cd200 | Santa Cruz Bio | sc-53100 | IF (1:100) |
| Antibody | Mouse monoclonal Anti-Cd90 | Santa Cruz Bio | sc-53456 | IF (1:100) |
| Antibody | Mouse monoclonal Anti-PDGFRα | Santa Cruz Bio | sc-398206 | IF (1:200) |
| Antibody | Mouse monoclonal Anti-Cd248 | Santa Cruz Bio | sc-377221 | IF (1:200) |
| Antibody | Rabbit polyclonal anti-Prg4 | Abcam | ab28484 | IF (1:50) |
| Antibody | Rat monoclonal Anti-Sca1 | BioLegend | 108131 | Flow analysis (1:100) |
| Antibody | Mouse monoclonal Anti-Cd90 | BioLegend | 202526 | Flow analysis (1:100) |
| Antibody | Rat monoclonal Anti-Cd200 | BioLegend | 123809 | Flow analysis (1:100) |
| Antibody | Rat monoclonal Anti-PDGFRα | BioLegend | 135907 | Flow analysis (1:100) |
| Software, algorithm | Graphpad 8.0 | Statistical Analysis | Graph preparation, statistical analysis |
Additional files
-
Supplementary file 1
Mouse real-time PCR primer sequences.
- https://cdn.elifesciences.org/articles/62917/elife-62917-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62917/elife-62917-transrepform-v1.pdf





