Targeted molecular profiling of rare olfactory sensory neurons identifies fate, wiring, and functional determinants
Figures
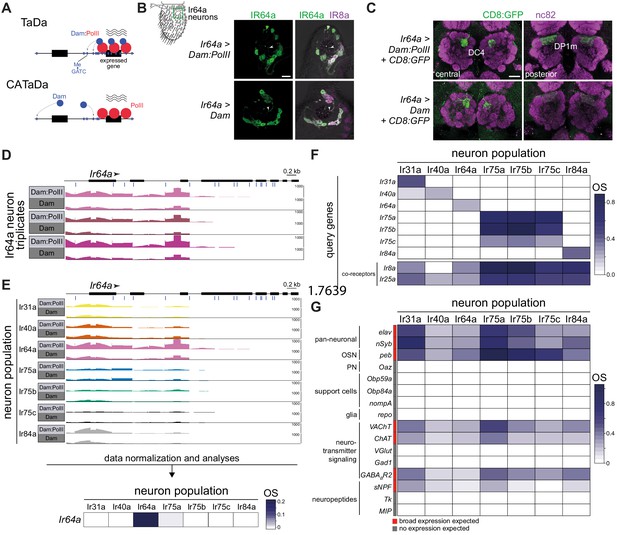
Targeted DamID of OSN populations.
(A) Schematic of the principles of Targeted DamID (TaDa) and Chromatin accessibility TaDa (CATaDa). TaDa reports on the enrichment of GATC methylation (Me; blue lines) by a Dam:PolII fusion (top) relative to Dam alone (bottom), thereby identifying PolII-transcribed genes. In CATaDa, analysis of methylation patterns of free Dam provides information on chromatin accessibility. (B) Immunofluorescence for IR64a and IR8a on antennal sections of animals expressing in Ir64a neurons either Dam:PolII (Ir64a-Gal4/+;UAS-LT3-Dam:RpII15/+) or Dam alone (Ir64a-Gal4/+;UAS-LT3-Dam/+). The approximate field of view shown is indicated by the green box on the antennal schematic on the left. The merged fluorescent channels are overlaid on a bright-field background to reveal anatomical landmarks. The arrowheads point to IR64a that is localized in the neuron sensory endings within the sensillar hairs. Scale bar = 10 µm. (C) Immunofluorescence for GFP and the neuropil marker nc82 on whole-mount brains of animals expressing in Ir64a neurons a CD8:GFP reporter together with Dam:PolII (Ir64a-Gal4/+;UAS-LT3-Dam:RpII15/UAS-mCD8:GFP) or free Dam (Ir64a-Gal4/+;UAS-LT3-Dam/UAS-mCD8:GFP). Two focal planes are shown to reveal the two glomeruli (DC4 and DP1m) innervated by Ir64a neurons. Background fluorescence in the GFP channel was slightly higher across the brain in the Dam-alone samples. Scale bar = 20 µm. (D) Illustration of raw sequence read data underlying the TaDa analyses. Bar heights indicate the read coverage of methylated GATC fragments mapping to the Ir64a gene (transcribed left-to-right, as indicated by the arrowhead; blue lines under the gene model indicate the position of GATC motifs). The precise transcription start site of Ir64a is unknown. For each of three replicate experiments, the top row is the Dam:PolII sample, which has higher read counts compared to the Dam-alone sample below. The y-axes (range 0–1000) indicate the read depth. (E) Top: comparisons among the seven Ir neuron datasets of the raw sequence reads mapping to the Ir64a gene for the Dam:PolII and Dam samples (a single representative replicate is shown for each). Genotypes are of the form shown in (B), except for Ir75a and Ir75c neuron populations where third chromosome Ir-Gal4 driver transgenes were used. Only the Ir64a population displays a higher read count for Dam:PolII samples compared to Dam-alone samples. The y-axes (range 0–1000) indicates the read depth. Bottom: heatmap representation of the occupancy scores (OSs) following data normalization and statistical analyses (based on data across the full gene body (see Materials and methods); color scale on the right) for Ir64a calculated across triplicates for each of the seven neuron populations. (F) OS heatmap for the seven target Ir genes, as well as the main Ir co-receptor genes, in the seven Ir neuron populations. OSs and FDR values associated with each Ir gene within the corresponding neuron population are shown in Supplementary file 5. (G) OS heatmap of diverse positive- and negative-control genes in the seven Ir neuron populations (i.e. with known or expected expression/lack of expression). Abbreviations are defined in the main text. See also Supplementary file 1 and DamID_files.zip (https://gitlab.com/roman.arguello/ir_tada/-/tree/main/DamID_analyses) for data.
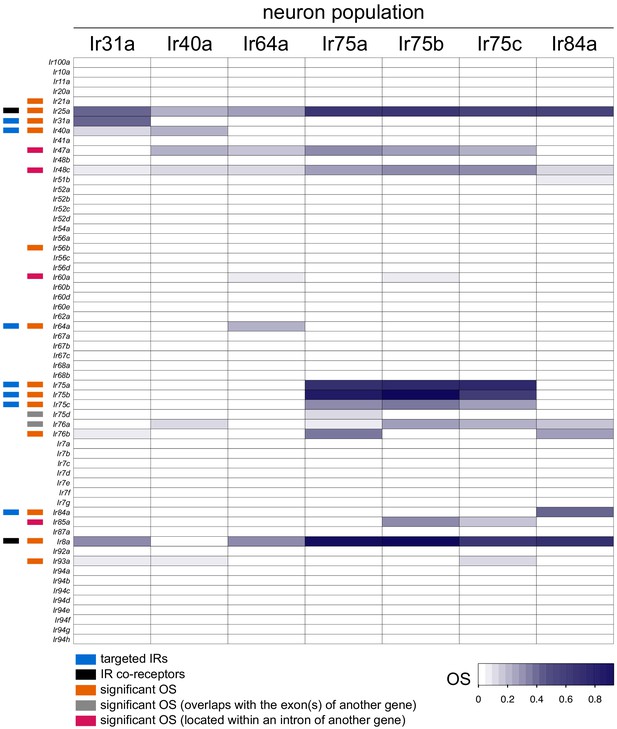
Ir gene Occupancy Scores across Ir populations.
Heatmap of OSs for Ir genes across the seven Ir neuron populations. Information on individual genes displaying significant OSs in one or more populations is shown with colored bars to the left (key is at the bottom). See also Supplementary files 4–5.
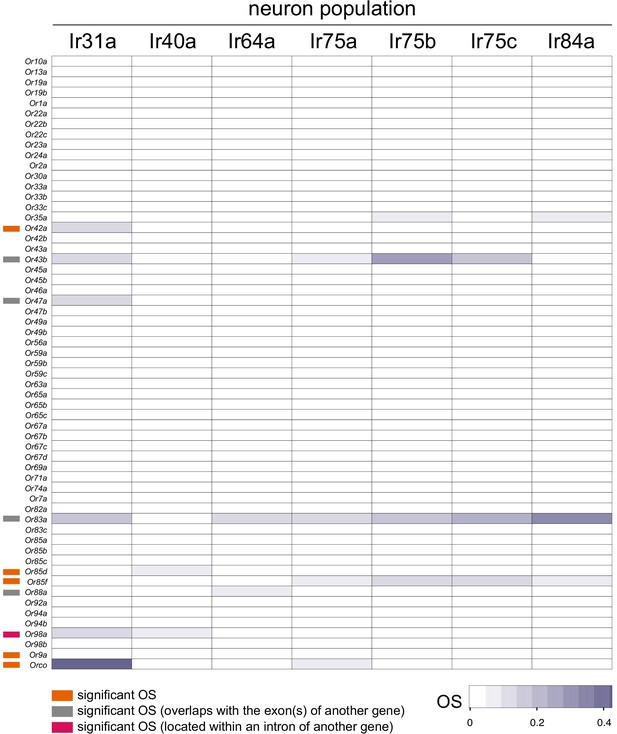
Or gene Occupancy Scores across Ir populations.
Heatmap of OSs for Or genes across the seven Ir neuron populations. Information on individual genes displaying significant OSs in one or more populations is shown with colored bars to the left (key is at the bottom). See also Supplementary files 4–5.
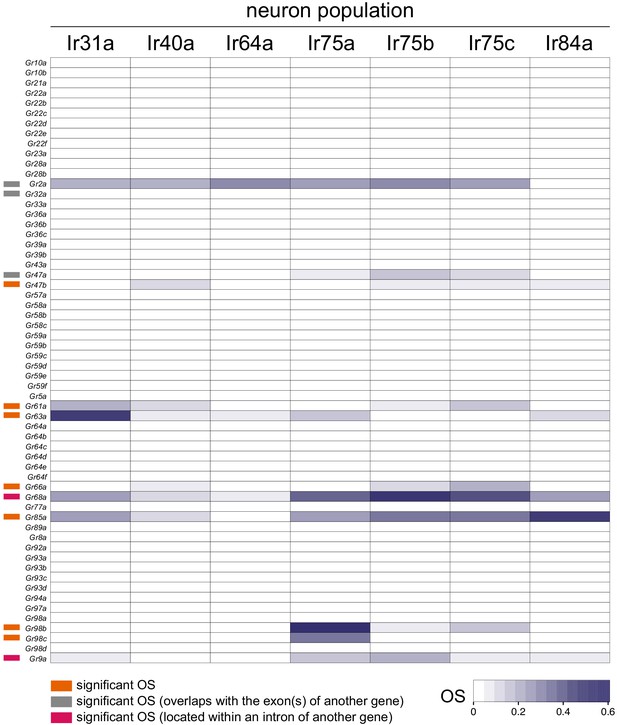
Gr gene Occupancy Scores across Ir populations.
Heatmap of OSs for Gr genes across the seven Ir neuron populations. Information on individual genes displaying significant OSs in one or more populations is shown with colored bars to the left (key is at the bottom). See also Supplementary files 4–5.
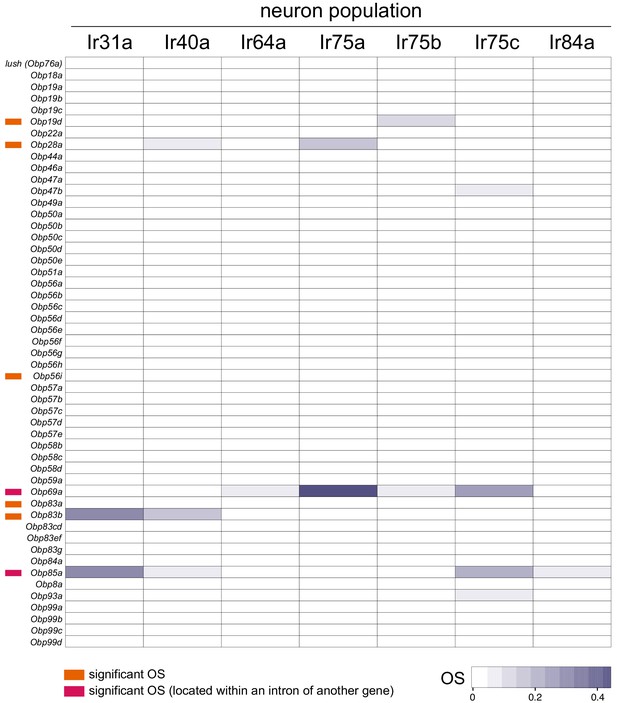
Obp gene Occupancy Scores across Ir populations.
Heatmap of OSs for Obp genes across the seven Ir neuron populations. Information on individual genes displaying significant OSs in one or more populations is shown with colored bars to the left (key is at the bottom). See also Supplementary files 4–5.
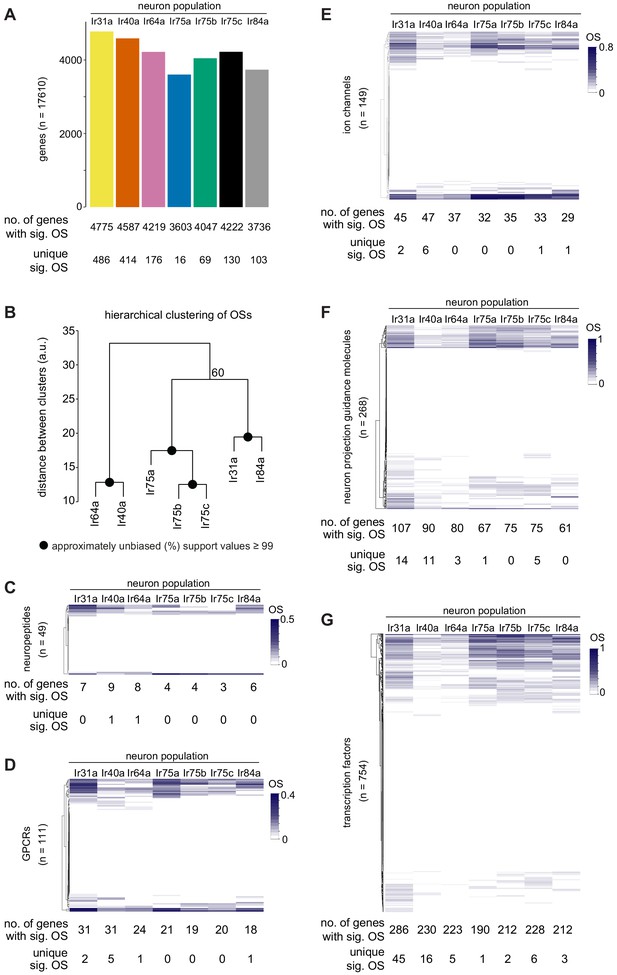
Global analyses of TaDa datasets.
(A) Bar plot summarizing the number of genes in the genome that have significantly positive OSs within each Ir neuron population, and the number of significantly occupied genes that are unique to a given neuron population. See also Supplementary files 1–3. (B) Hierarchical clustering (arbitrary units, a.u.) of the seven Ir neuron populations based upon the OSs of all genes. All nodes had approximately unbiased support values of 100% except for the one denoted with 60%. (C–G) Heatmaps displaying the hierarchical clustering of OSs for the genes encoding (C) neuropeptides, (D) GPCRs, (E) ion channels, (F) neuron projection guidance molecules, and (G) transcription factors. For each population, the total number of genes with a significant OS, and the number of those that are unique to a given population, are shown below (see also Supplementary files 6–20) which list, for each category: (i) all analyzed genes, (ii) significantly occupied genes, and (iii) population-specific occupied genes.
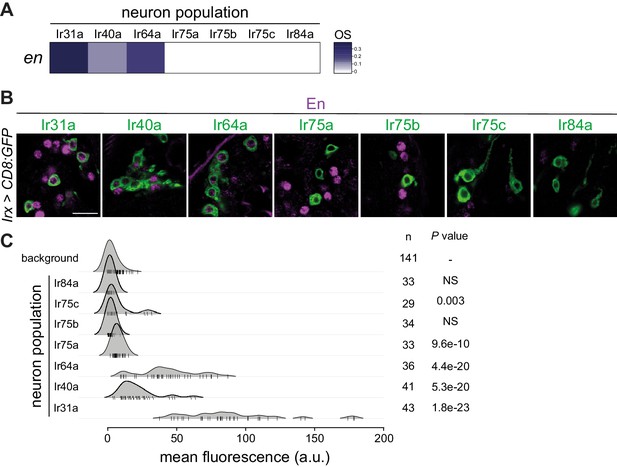
Heterogeneous expression of Engrailed in Ir neuron populations.
(A) OS heatmap of en in the seven Ir neuron populations. (B) Immunofluorescence for En (magenta) and GFP (green) on antennal sections of animals in which the indicated Ir neuron populations are labeled with a CD8:GFP reporter. Genotypes are of the form: Irxx-Gal4/+;UAS-mCD8:GFP/+ or, for Ir75a and Ir75c neurons, UAS-mCD8:GFP/+;Irxx-Gal4/+. Scale bar = 10 µm. (C) Density plots for En immunofluorescence signals (arbitrary units, a.u.) quantified from Ir neuron nuclei. Most signals from Ir75a, Ir75b, Ir75c, and Ir84a neuron nuclei fall within the range of background signals; Ir31a, Ir40a, and Ir64a neuron nuclei have consistently higher levels of immunofluorescence. n = sample size; p- value based on one-sided Wilcoxon tests, with Bonferroni correction for multiple comparisons.
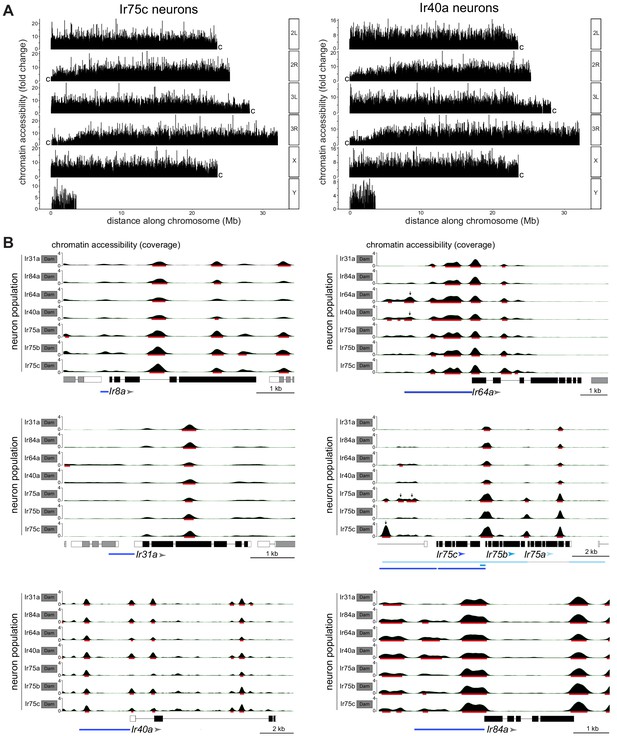
CATaDa reveals global similarity in chromatin accessibility at Ir genes across Ir neuron populations.
(A) Comparison of chromatin accessibility peak locations (q < 0.05) over the D. melanogaster genome for two example neuron populations, Ir75c and Ir40a. The y-axis represents the fold enrichment for the peak summit against random Poisson distribution of the small local region (1000 bp). 2L/3L and 2R/3R refer to left and right arms of chromosomes 2/3; ‘c’ at the end of each arm indicates the centromeric (heterochromatic) end of the arm (the Y chromosome is mainly heterochromatic). (B) Plots of Ir gene exons are shown in black, and UTRs in white; exons of flanking genes are shown in gray. The arrowheads indicate the direction of Ir transcription. The minimal defined regulatory sequences to recapitulate Ir expression are indicated with blue bars (color-coordinated with the arrowheads for Ir75a, Ir75b, and Ir75c) (Prieto-Godino et al., 2017; Silbering et al., 2011). The y-axis scale shows the CATaDa accessibility at the genomic region with the RPGC normalization method. The red bars are the significant peaks with q < 0.05. In the Ir75a/Ir75b/Ir75c genomic region, the arrows mark significant peaks that are unique to the corresponding Ir neuron populations and which are contained within known regulatory regions of these Ir genes. In the Ir64a genomic region, the arrows mark significant peaks observed in both Ir64a and Ir40a neurons. Note that the smooth boundaries around peaks of Dam-accessible regions are visual artifacts of the analysis; they do not reflect the resolution of CATaDa, which is discrete at the GATC motifs.
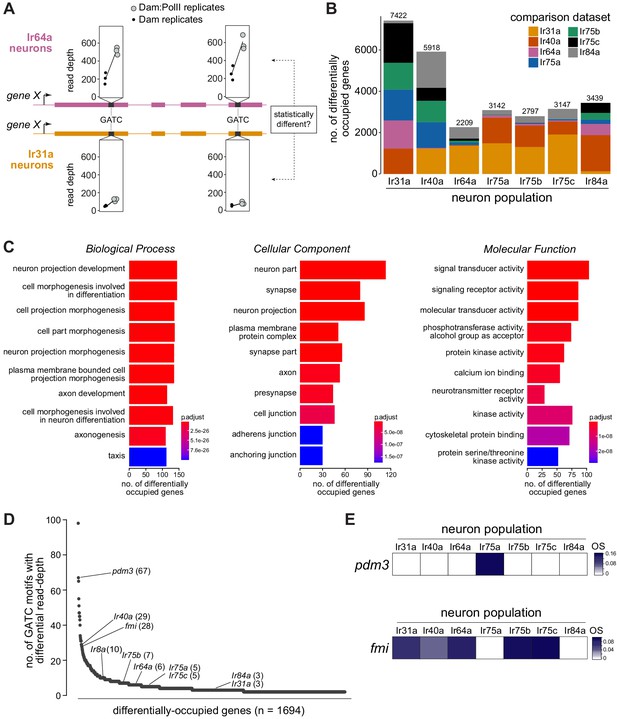
Differentially occupied genes across Ir populations.
(A) Schematic illustrating the identification of a differentially occupied gene between two Ir populations by using variation in read depth at GATC motifs (see Materials and methods). In this example, occupancy of ‘gene X’ is compared between Ir64a neurons and Ir31a neurons. At GATC motifs (black bars), a test is applied to determine if significant variation exists in the relative read depths (in triplicate Dam:PolII versus Dam-alone experiments) between these two neuron populations. Fictive data for two GATC motifs are depicted: for both, the relative read depth in the Dam:PolII experiments (compared to the Dam-alone experiments) is greater in the Ir64a neurons. (B) Stacked bar plots of the numbers of genes in the genome that are differentially occupied among Ir neuron populations based on pairwise tests (see Materials and methods). The colors indicate the proportions of genes emerging from comparisons with the six other neuron populations. The figures above the bars are the total numbers of genes from all pairwise comparisons; note that a given gene may be counted multiple times. (C) Gene Ontology (GO) analysis of the 1694 differentially occupied genes, showing the top ten over-represented GO terms. The x-axis represents the number of genes annotated to a particular GO term in the input subset. The value indicated by the colored bar is the probability of observing at least the same number of genes associated to that GO term compared to what would have occurred by chance. (D) Ranking of the 1694 differentially occupied genes ordered by the number of GATC motifs contributing to their between-neuron differences (uncorrected for gene length). (E) OS heatmaps (calculated as in Figures 1–2) of pdm3 and fmi in the seven Ir neuron populations.
-
Figure 4—source data 1
Candidate genes for pairwise differential occupancy based on the Wald Test with DESeq2.
21 Bed-formatted files (representing all possible pairwise comparisons between the seven datasets) containing statistics and annotations.
- https://cdn.elifesciences.org/articles/63036/elife-63036-fig4-data1-v2.zip
-
Figure 4—source data 2
Candidate genes for differential occupancy based on the Likelihood Ratio Test within DESeq2.
One file contains the list of genes, the other contains the list of GATC fragments and the associated annotations and statistics.
- https://cdn.elifesciences.org/articles/63036/elife-63036-fig4-data2-v2.zip
-
Figure 4—source data 3
Number of GATC motifs within candidate differentially expressed genes contributing to significant differences.
Values correspond to those plotted in Figure 4D.
- https://cdn.elifesciences.org/articles/63036/elife-63036-fig4-data3-v2.txt
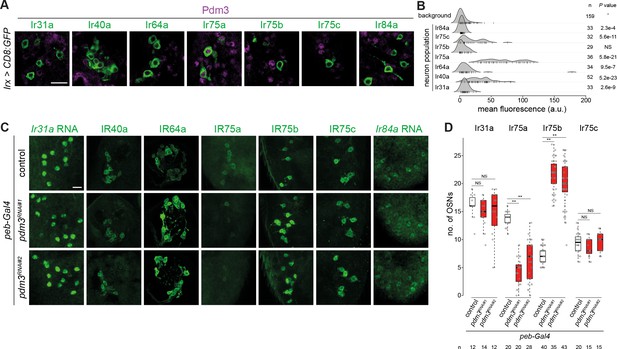
Heterogeneous expression and function of Pdm3 in Ir neurons.
(A) Immunofluorescence for Pdm3 and GFP on antennal sections of animals in which the indicated Ir neuron populations are labeled with a CD8:GFP reporter. Genotypes are of the form: Irxx-Gal4/+;UAS-mCD8:GFP/+ or, for Ir75a and Ir75c neurons, UAS-mCD8:GFP/+;Irxx-Gal4/+. Scale bar = 10 µm. (B) Density plots for Pdm3 immunofluorescence signals (arbitrary units, a.u.) quantified from Ir neuron nuclei (each represented by a vertical line) in the genotypes in (A) and from background signals pooled across genotypes (see Materials and methods). Ir75a neuron nuclei have the highest levels of immunofluorescence. n = sample size; p-value based on one-sided Wilcoxon tests, with Bonferroni correction for multiple comparisons. (C) Immunofluorescence or RNA FISH for the indicated IRs on whole-mount antennae (or antennal sections for IR40a and IR64a, due to poor antibody penetration of whole-mount tissue) of control animals (peb-Gal4,UAS-Dcr-2/+;+/CyO) or two independent pdm3 RNAi lines (peb-Gal4,UAS-Dcr-2/+;;UAS-pdm3JF02312/+ (RNAi#1) and peb-Gal4,UAS-Dcr-2/+;UAS-pdm3HMJ21205/+ (RNAi#2)). Scale bar = 10 µm. (D) Quantification of neuron number for the indicated Ir neuron populations for the genotypes shown in (C). In this and other panels, boxplots show the median, first and third quartile of the data, overlaid with individual data points. Comparisons to controls are shown for each neuron type (pairwise Wilcoxon rank-sum two-tailed test and p-values adjusted for multiple comparisons with the Bonferroni method, **p<0.001; NS p>0.05). Sample sizes are shown below the plot. Ir40a and Ir64a OSN population sizes could not be quantified because the sections visualized necessarily contain a variable number of neurons; similarly, we could not confidently count Ir84a neuron number because of the weak signal. Nevertheless, when visualized blindly, the control and RNAi genotypes were not distinguishable for any of these neuron populations (assessing the phenotype in antennae of at least 10 animals from two independent genetic crosses).
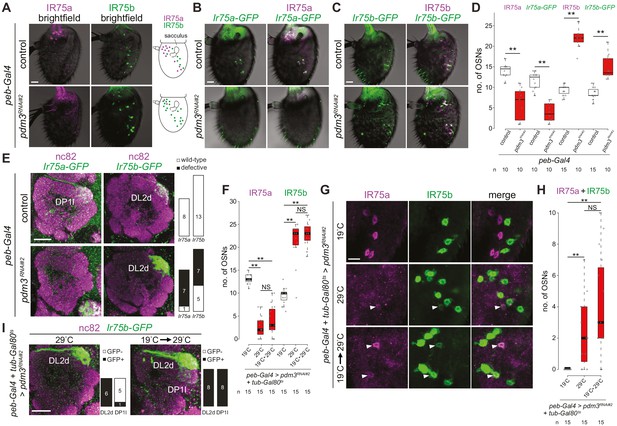
Pdm3 is required to distinguish Ir75a and Ir75b neuron fate.
(A) Immunofluorescence for IR75a and IR75b on whole-mount antennae of control (peb-Gal4,UAS-Dcr-2/+) and pdm3RNAi#2 (peb-Gal4,UAS-Dcr-2/+;UAS-pdm3HMJ21205/+) animals. Scale bar = 10 µm. The schematics on the right summarize the distribution of labeled neurons. (B) Immunofluorescence for GFP and IR75a on whole-mount antennae of control (peb-Gal4,UAS-Dcr-2/+;Ir75a-GFP/+) and pdm3RNAi#2 (peb-Gal4,UAS-Dcr-2/+;Ir75a-GFP/UAS-pdm3HMJ21205) animals. Scale bar = 10 µm. (C) Immunofluorescence for GFP and IR75b on whole-mount antennae of control (peb-Gal4,UAS-Dcr-2/+;Ir75b-GFP/+) and pdm3RNAi#2 (peb-Gal4,UAS-Dcr-2/+;Ir75b-GFP/UAS-pdm3HMJ21205) animals. Scale bar = 10 µm. (D) Quantification of the number of neurons that express Ir75a-GFP or Ir75b-GFP in the genotypes shown in (B–C). Comparisons to the controls are shown (pairwise Wilcoxon rank-sum two-tailed test and p-values adjusted for multiple comparisons with the Bonferroni method, **p<0.001). The increase in number of Ir75b-GFP labeled neurons in pdm3RNAi is lower than the increase in IR75b-expressing neurons, potentially because the transgenic reporter is not fully faithful in this genetic background. (E) Immunofluorescence for nc82 and GFP on whole-mount antennal lobes of control and pdm3RNAi#2 animals. Genotypes are as in (B–C). Scale bar = 20 µm. Quantification of phenotypes are shown on the right. (F) Quantification of the number of Ir75a and Ir75b neurons in animals (peb-Gal4,UAS-Dcr-2/+;pdm3RNAi#2/+;UAS-Gal80ts/+) in which peb-Gal4-driven pdm3RNAi is continuously suppressed (19°C, the permissive temperature for the Gal4 inhibitor Gal80ts), continuously allowed (29°C, the restrictive temperature for Gal80ts) or induced only in adults (19°C → 29°C temperature shift after eclosion). Comparisons between conditions are shown for each neuron type (pairwise Wilcoxon rank-sum two-tailed test with Bonferroni correction for multiple comparisons, **p<0.001, NS p>0.05). (G) Immunofluorescence for IR75a and IR75b on whole-mount antennae of the genotypes shown in (F). Single optical sections are shown, to reveal the weak co-expression of IR75a and IR75b in a subset of cells (arrowheads). Scale bar = 10 µm. (H) Quantification of the number of neurons that co-express IR75a and IR75b. Comparisons to the controls are shown (pairwise Wilcoxon rank-sum two-tailed test with Bonferroni correction for multiple comparisons, **p<0.001, NS p>0.05). (I) Immunofluorescence for nc82 and GFP on whole-mount antennal lobes of pdm3RNAi#2 animals (peb-Gal4,UAS-Dcr-2/+;UAS-pdm3HMJ21205/Ir75b-GFP;UAS-Gal80ts/+) in which RNAi is allowed throughout development (29°C) or limited only to adults (19°C → 29°C temperature shift after eclosion). Quantification of glomerular labeling pattern by the Ir75b-GFP reporter is shown on the right of each image.
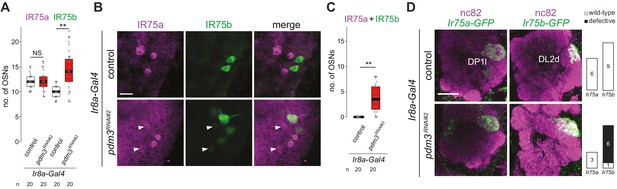
Phenotypic analysis of Ir75a and Ir75b neurons with late developmental induction of pdm3 RNAi.
(A) Quantification of the number of Ir75a and Ir75b neurons in control (Ir8a-Gal4/+) and pdm3RNAi#2 (Ir8a-Gal4/UAS-pdm3HMJ21205) animals. Comparisons to the control are shown for each neuron type (pairwise Wilcoxon rank-sum two-tailed test, **p<0.001, NS p>0.05). Although the number of IR75a-expressing cells was unchanged compared to controls, receptor protein levels were lower in several cells (see (B)). (B) Immunofluorescence for IR75a and IR75b on whole-mount antennae of control and pdm3RNAi#2 animals (genotypes as in (A)). Single optical sections are shown, to reveal the weak co-expression of IR75a and IR75b in a subset of cells (arrowheads). Scale bar = 10 µm. (C) Quantification of the number of neurons that co-express IR75a and IR75b. Comparison to the control is shown (pairwise Wilcoxon rank-sum two-tailed test). (D) Immunofluorescence for nc82 and GFP on whole-mount antennal lobes of pdm3RNAi#2 animals. Genotypes: Ir75a-GFP control (Ir75a-GFP/+;Ir8a-Gal4/+), Ir75a-GFP pdm3RNAi#2 (Ir75a-GFP/UAS-pdm3HMJ21205;Ir8a-Gal4/+), Ir75b-GFP control (Ir75b-GFP/+;Ir8a-Gal4/+), Ir75b-GFP pdm3RNAi#2 (Ir75b-GFP/UAS-pdm3HMJ21205;Ir8a-Gal4/+). Scale bar = 20 µm. Quantification of projection pattern phenotypes are shown on the right. In contrast to observations with adult-only pdm3 RNAi driven by peb-Gal4 (Figure 6I), we did not detect labeling of DP1l in these experiments. This difference is likely to reflect the weaker RNAi induced by Ir8a-Gal4 (and/or the absence of UAS-Dcr-2 in these genotypes), as is apparent from the quantification of Ir75a and Ir75b neuron numbers (compare panel (A) with Figure 6F).
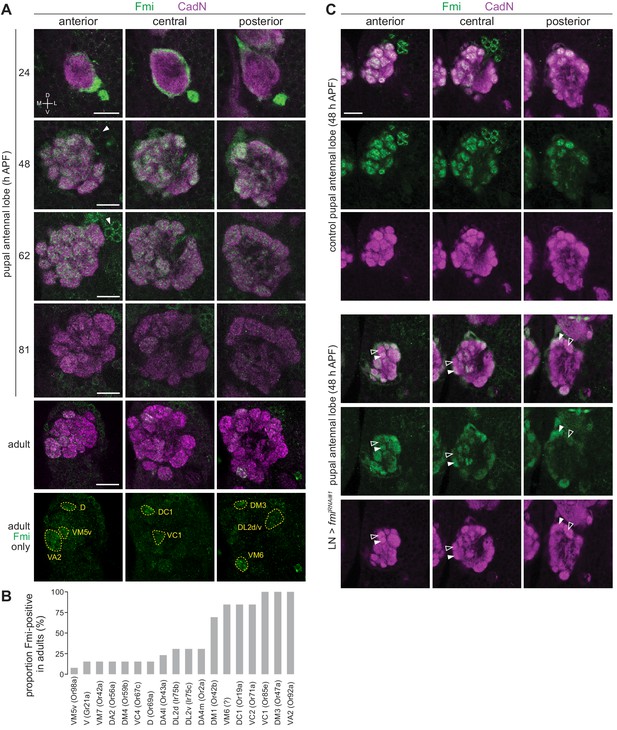
Expression analysis of Fmi in OSNs.
(A) Immunofluorescence for Fmi and the ubiquitously-expressed neuronal cadherin (CadN) on whole-mount antennal lobes of wild-type (w1118) animals of the indicated age. Three optical sections are shown to reveal the morphology of most glomeruli. Bottom row: Fmi-positive glomeruli were identified in adult antennal lobes based upon their stereotyped position and morphology. Dorsal-ventral (D-V) and medial-lateral (M-L) axes are indicated. The arrowheads point to the soma of Fmi-expressing LNs that are detectable from 48 hr APF. Scale bar = 20 μm. (B) Histogram of frequency of detectable Fmi immunoreactivity in individual glomeruli of the adult antennal lobe (n = 14 antennal lobes). (C) Immunofluorescence for Fmi and CadN on whole-mount antennal lobes of control (1081-Gal4/+;449-Gal4/UAS-Dcr-2) and LN>fmiRNAi#1 (1081-Gal4/UAS-fmiKK100512;449-Gal4/UAS-Dcr-2) 48 hr APF animals. Open and filled arrowheads highlight adjacent glomeruli with low and high levels, respectively, of Fmi immunoreactivity. Scale bar = 20 μm.
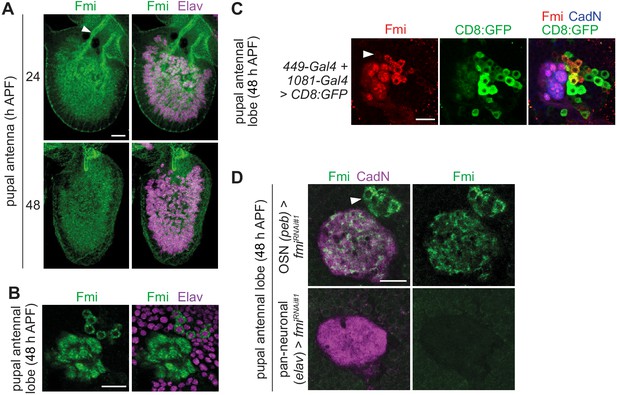
Characterization of Fmi expression in antennae and local interneurons.
(A) Immunofluorescence for Fmi and Elav on whole-mount antennae of control animals (w;Ir8a-Gal4,UAS-RedStinger/CyO; note the RedStinger reporter is not shown in these images) at 24 and 48 hr APF. Partial optical stacks are shown. The arrowhead points to the antennal nerve. Scale bar = 20 μm. (B) Immunofluorescence for Fmi and Elav on whole-mount antennal lobes of 48 hr APF control (w1118) animals. A partial optical stack is shown, revealing expression of Elav in the Fmi-positive soma adjacent to the antennal lobe. Scale bar = 20 μm. (C) Immunofluorescence for Fmi, GFP, and CadN on a whole-mount antennal lobe of a 48 hr APF animal in which Fmi-expressing LNs are labeled with the combined 449-Gal4 and 1081-Gal4 drivers (1081-Gal4/+;449-Gal4/UAS-mCD8:GFP), which were identified by screening a panel of LN drivers (Liou et al., 2018). A partial optical stack is shown. We distinguish two types of Fmi-positive neurons: 100% of strongly expressing neurons (n = 194, seven brains) are GFP-positive; 75% of weakly expressing neurons (e.g. the neuron indicated by the arrowhead) are GFP-positive (n = 213, seven brains). Note the drivers are expressed in several additional Fmi-negative cells. Scale bar = 20 μm. (D) Immunofluorescence for Fmi and CadN on whole-mount antennal lobes of 48 hr APF animals with OSN>fmiRNAi#1 (peb-Gal4,UAS-Dcr-2/+;UAS-fmiKK100512/+) or pan-neuronal>fmiRNAi#1 (elav-Gal4,UAS-Dcr-2/+;UAS-fmiKK100512/+;). In OSN>fmiRNAi#1 animals the remaining, relatively homogeneous glomerular signal is derived from its expression in LNs (soma labeled with an arrowhead); we cannot rule out that the homogeneity is a secondary consequence of the glomerular segregation defects. Pan-neuronal>fmiRNAi#1 is semi-lethal, but we recovered a few adult escapers in which all Fmi immunoreactivity in the antennal lobe is lost. Scale bar = 20 μm.
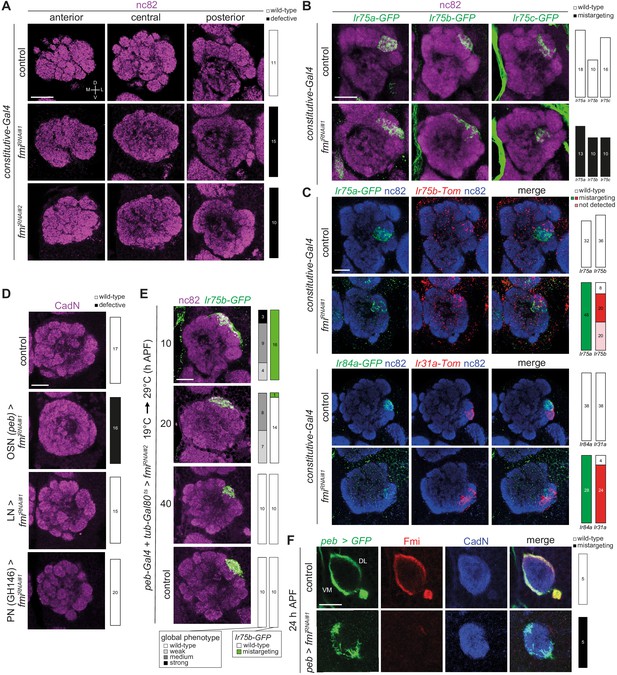
Fmi is required in OSNs for glomerular segregation.
(A) Immunofluorescence for nc82 on whole-mount antennal lobes of control (w;ey-Flp/+;act>stop>Gal4/+), fmiRNAi#1 (w;ey-Flp/UAS-fmiKK100512;act>stop>Gal4/+) and fmiRNAi#2 (w;ey-Flp/+;act>stop>Gal4/UAS-fmiJF02047) animals. Three optical sections are shown. Scale bar = 20 μm. Quantification of wild-type or defective antennal lobe glomerular architectures are shown on the right; the n for each phenotypic category are indicated on the plots. (B) Immunofluorescence for nc82 and GFP on whole-mount antennal lobes of control and fmiRNAi#1 animals. Genotypes: Ir75a-GFP (w;ey-Flp,Ir75a-GFP/[+ or UAS-fmiKK100512];act>stop>Gal4/+), Ir75b-GFP (w;ey-Flp,Ir75b-GFP/[+ or UAS-fmiKK100512];act>stop>Gal4/+), Ir75c-GFP (w;ey-Flp,Ir75c-GFP/[+ or UAS-fmiKK100512];act>stop>Gal4/+). Scale bar = 20 μm. (C) Immunofluorescence for nc82, GFP, RFP (detects Tomato (Tom)) on whole-mount antennal lobes of control and fmiRNAi#1 animals. Genotypes: w;Ir75a-GFP,Ir75b-Tom/[+ or UAS-fmiKK100512];ey-FLP,act>stop>Gal4/Ir75b-RFP (top panels) and eyFlp,act>stop>Gal4/[+ or UAS-fmiKK100512];Ir31a-Tom,Ir84a-GFP/+ (bottom panels). Scale bar = 20 μm. Quantification of wild-type or defective targeting phenotypes are shown on the right, as indicated in the key at the top-right. (D) Immunofluorescence for CadN on whole-mount antennal lobes of control (peb-Gal4), OSN>fmiRNAi#1 (peb-Gal4/+;UAS-fmiKK100512/+;UAS-Dcr-2/+), LN>fmiRNAi#1 (1081-Gal4/UAS-fmiKK100512;449-Gal4/UAS-Dcr-2), PN>fmiRNAi#1 (GH146-Gal4/+;UAS-fmiKK100512/+) animals. Scale bar = 20 μm. (E) Immunofluorescence for nc82 and GFP on whole-mount antennal lobes of peb-Gal4,UAS-Dcr-2/+;Ir75b-GFP/tub-Gal80ts;UAS-fmiJF02047/+ animals in which Gal4-driven RNAi was suppressed until shifting from the permissive to restrictive temperature (19°C → 29°C) for Gal80ts at the indicated time points. Scale bar = 20 μm. (F) Immunofluorescence for GFP, Fmi and CadN on whole-mount pupal antennal lobes (24 hr APF) of control (peb-Gal4/+;;UAS-mCD8::GFP/+) and OSN>fmiRNAi#1 (peb-Gal4/+;UAS-fmiKK100512/+;UAS-mCD8::GFP/+) animals. The merged channels are shown on the right. The dorsolateral (DL) and ventromedial (VM) bundles of OSN axons are indicated. Scale bar = 20 μm.
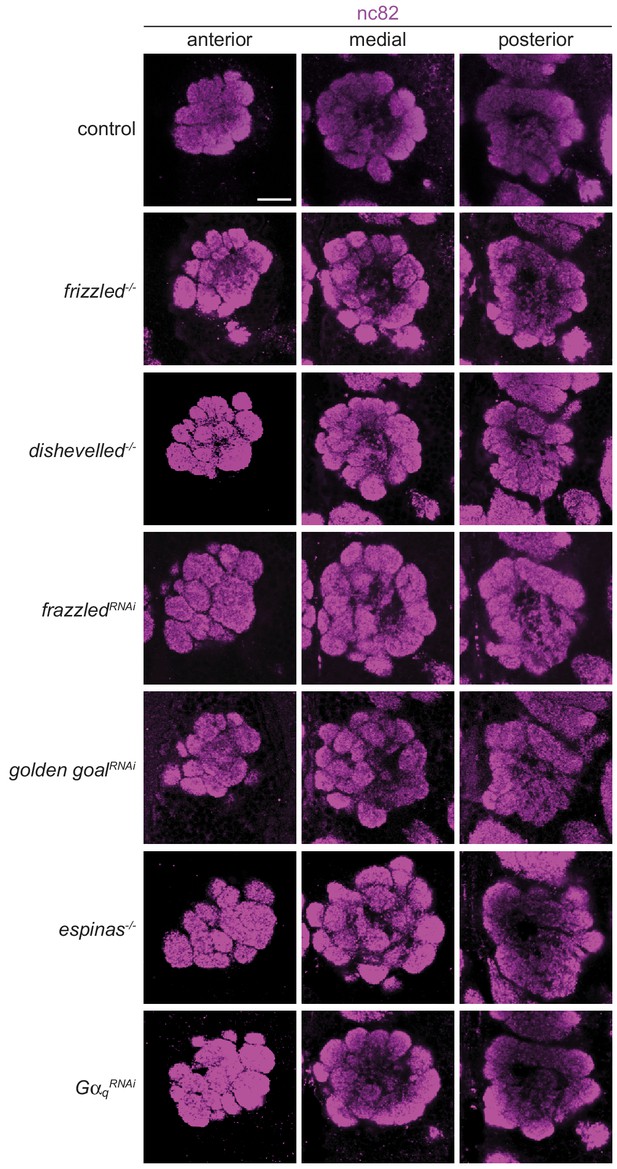
Fmi does not act in OSNs with known genetic and physical interaction partners.
Representative images of nc82 immunofluorescence on whole-mount antennal lobes (three optical sections) of animals of the following genotypes (top-to-bottom; other loss-of-function alleles or RNAi lines tested, but not shown, for certain genes are also listed): control (peb-Gal4,UAS-Dcr-2/+), frizzled-/- (fzP21/fzKD4a; also tested fzP21/fzH51), dishevelled-/- (dsh1), frazzledRNAi (peb-Gal4,UAS-Dcr-2;Ir75b-GFP/+;fraJF01457/+; also tested fraHMS01147 and fraJF01231), golden goalRNAi (peb-Gal4,UAS-Dcr-2;Ir75b-GFP/gogoGD3616; also tested gogoHMC05937), espinas-/- (esnKO6), GαqRNAi (peb-Gal4,UAS-Dcr-2/+;GαqdsRNA.UAS.1f1/+; also tested GαqGL01048 and GαqJF01209). The Ir75b-GFP reporter present in some of the genotypes is not shown in these images, but was indistinguishable from controls. Scale bar = 20 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | w1118 | Bloomington Drosophila Stock Center | RRID:BDSC_3605 | |
| Genetic reagent (D. melanogaster) | Ir8a-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_41731 | |
| Genetic reagent (D. melanogaster) | Ir31a-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_41726 | |
| Genetic reagent (D. melanogaster) | Ir40a-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_41727 | |
| Genetic reagent (D. melanogaster) | Ir64a-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_41732 | |
| Genetic reagent (D. melanogaster) | Ir75a-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_41748 | |
| Genetic reagent (D. melanogaster) | Ir75b-Gal4 | Prieto-Godino et al., 2017 | ||
| Genetic reagent (D. melanogaster) | Ir75c-Gal4 | Prieto-Godino et al., 2017 | ||
| Genetic reagent (D. melanogaster) | Ir84a-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_41734 | |
| Genetic reagent (D. melanogaster) | peb-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_80570 | |
| Genetic reagent (D. melanogaster) | ey-Flp (II) | Bloomington Drosophila Stock Center | RRID:BDSC_5576 | |
| Genetic reagent (D. melanogaster) | ey-Flp (III) | Bloomington Drosophila Stock Center | RRID:BDSC_5577 | |
| Genetic reagent (D. melanogaster) | act>stop>Gal4 (II) | Bloomington Drosophila Stock Center | RRID:BDSC_3953 | |
| Genetic reagent (D. melanogaster) | act>stop>Gal4 (III) | Bloomington Drosophila Stock Center | RRID:BDSC_4780 | |
| Genetic reagent (D. melanogaster) | GH146-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_30026 | |
| Genetic reagent (D. melanogaster) | 449-Gal4 | Liou et al., 2018 | ||
| Genetic reagent (D. melanogaster) | 1081-Gal4 | Liou et al., 2018 | ||
| Genetic reagent (D. melanogaster) | elav-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_458 | |
| Genetic reagent (D. melanogaster) | tub-Gal80ts | Bloomington Drosophila Stock Center | RRID:BDSC_7018 | |
| Genetic reagent (D. melanogaster) | UAS-Dcr-2 (X) | Bloomington Drosophila Stock Center | RRID:BDSC_24648 | |
| Genetic reagent (D. melanogaster) | UAS-Dcr-2 (II) | Bloomington Drosophila Stock Center | RRID:BDSC_24650 | |
| Genetic reagent (D. melanogaster) | UAS-Dcr-2 (III) | Bloomington Drosophila Stock Center | RRID:BDSC_24651 | |
| Genetic reagent (D. melanogaster) | UAS-LT3-Dam (III) | Southall et al., 2013 | ||
| Genetic reagent (D. melanogaster) | UAS-LT3-Dam:RpII15 (III) | Southall et al., 2013 | ||
| Genetic reagent (D. melanogaster) | UAS-mCD8:GFP | Bloomington Drosophila Stock Center | RRID:BDSC_5130 | |
| Genetic reagent (D. melanogaster) | UAS-RedStinger | Bloomington Drosophila Stock Center | RRID:BDSC_8546 | |
| Genetic reagent (D. melanogaster) | Ir75b-CD4:tdGFP | Prieto-Godino et al., 2017 | ||
| Genetic reagent (D. melanogaster) | UAS-fmiKK100512 (RNAi #1) | Vienna Drosophila Resource Center | VDRC v107993 | |
| Genetic reagent (D. melanogaster) | UAS-fmiJF02047 (RNAi #2) | Bloomington Drosophila Stock Center | RRID:BDSC_26022 | |
| Genetic reagent (D. melanogaster) | UAS-fraHMS01147 (RNAi) | Bloomington Drosophila Stock Center | RRID:BDSC_40826 | |
| Genetic reagent (D. melanogaster) | UAS-fraJF01231 (RNAi) | Bloomington Drosophila Stock Center | RRID:BDSC_31469 | |
| Genetic reagent (D. melanogaster) | UAS-fraJF01457 (RNAi) | Bloomington Drosophila Stock Center | RRID:BDSC_31664 | |
| Genetic reagent (D. melanogaster) | UAS-GaqdsRNA.UAS.1f1 (RNAi) | Bloomington Drosophila Stock Center | RRID:BDSC_30735 | |
| Genetic reagent (D. melanogaster) | UAS-GaqJF01209 (RNAi) | Bloomington Drosophila Stock Center | RRID:BDSC_31268 | |
| Genetic reagent (D. melanogaster) | UAS-GaqGL01048 (RNAi) | Bloomington Drosophila Stock Center | RRID:BDSC_36820 | |
| Genetic reagent (D. melanogaster) | UAS-gogoGD3616 (RNAi) | Vienna Drosophila Resource Center | VDRC v43928 | |
| Genetic reagent (D. melanogaster) | UAS-gogoHMC05937 (RNAi) | Bloomington Drosophila Stock Center | RRID:BDSC_65193 | |
| Genetic reagent (D. melanogaster) | UAS-pdm3JF02312 (RNAi #1) | Bloomington Drosophila Stock Center | RRID:BDSC_26749 | |
| Genetic reagent (D. melanogaster) | UAS-pdm3HMJ21205 (RNAi #2) | Bloomington Drosophila Stock Center | RRID:BDSC_53887 | |
| Genetic reagent (D. melanogaster) | dsh1 | Krasnow et al., 1995 | ||
| Genetic reagent (D. melanogaster) | esnKO6 | Matsubara et al., 2011 | ||
| Genetic reagent (D. melanogaster) | fzH51 | Jones et al., 1996 | ||
| Genetic reagent (D. melanogaster) | fzKD4a | Adler et al., 1990 | ||
| Genetic reagent (D. melanogaster) | fzP21 | Jones et al., 1996 | ||
| Genetic reagent (D. melanogaster) | Ir31a-CD4:tdTom | This work | ||
| Genetic reagent (D. melanogaster) | Ir75a-CD4:tdGFP | This work | ||
| Genetic reagent (D. melanogaster) | Ir75b-CD4:tdTom | This work | ||
| Genetic reagent (D. melanogaster) | Ir75c-CD4:tdGFP | This work | ||
| Genetic reagent (D. melanogaster) | Ir84a-CD4:tdGFP | This work | ||
| Antibody | Anti-En (mouse monoclonal) | DSHB 4D9 | RRID:AB_528224 | (1:100) |
| Antibody | Anti-Pdm3 (guinea pig polyclonal) | Chen et al., 2012 | RRID:AB_2567243 | (1:200) |
| Antibody | Anti-Fmi (mouse monoclonal) | DSHB #74 | RRID:AB_2619583 | (1:20) |
| Antibody | Anti-IR40a (guinea pig polyclonal) | Silbering et al., 2011 | (1:200) | |
| Antibody | Anti-IR64a (rabbit polyclonal) | Ai et al., 2010 | RRID:AB_2566854 | (1:1000) |
| Antibody | Anti-IR75a (rabbit polyclonal) | Prieto-Godino et al., 2017 | RRID:AB_2631091 | (1:200) |
| Antibody | Anti-IR75b (guinea pig polyclonal) | Prieto-Godino et al., 2017 | RRID:AB_2631093 | (1:500) |
| Antibody | Anti-IR75c (rabbit polyclonal) | Prieto-Godino et al., 2017 | RRID:AB_2631094 | (1:200) |
| Antibody | Anti-Bruchpilot (mouse monoclonal) | DSHB nc82 | RRID:AB_2314866 | (1:10) |
| Antibody | Anti-Cadherin-N (rat monoclonal) | DSHB Ex#8–2 | RRID:AB_528121 | (1:25) |
| Antibody | Anti-Elav (rat monoclonal) | DSHB 7E8A10 | RRID:AB_528218 | (1:10) |
| Antibody | Anti-GFP (chicken polyclonal) | Abcam 13970 | (1:2000) | |
| Antibody | Anti-GFP (mouse monoclonal) | Invitrogen A11120 | (1:1000) | |
| Antibody | Anti-RFP (rabbit polyclonal) | Abcam 62341 | (1:1000) | |
| Antibody | Alexa488 (goat anti-guinea pig) | Invitrogen A11073 | (1:500) | |
| Antibody | Alexa488 (goat anti-chicken) | Abcam 150169 | (1:1000) | |
| Antibody | Cy3 (goat anti-rabbit) | Milan Analytica AG 111-165-144 | (1:1000) | |
| Antibody | Cy3 (goat anti-mouse) | Milan Analytica AG 115-165-166 | (1:1000) | |
| Antibody | Cy5 (donkey anti-rat) | Jackson ImmunoResearch 712-175-153 | (1:250) | |
| Antibody | Anti-DIG-POD | Roche Diagnostics 11 207 733 910 | (1:300) | |
| Software algorithm | Fiji | Fiji | RRID:SCR_002285 |
Additional files
-
Supplementary file 1
Full set of genes with significant OSs in Ir neuron populations.
OSs correspond to those summarized in Figure 2A.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp1-v2.txt
-
Supplementary file 2
List of gene IDs only from Supplementary file 1.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp2-v2.txt
-
Supplementary file 3
Genes with significant OSs unique to each Ir neuron population.
IDs correspond to the counts displayed in Figure 2A.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp3-v2.txt
-
Supplementary file 4
Set of chemosensory receptor and Obp genes.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp4-v2.txt
-
Supplementary file 5
Chemosensory receptor and Obp genes with significant OSs in Ir neuron populations.
Only significant OSs are listed. Values correspond to those plotted in Figure 1—figure supplements 1–4. NA = not applicable (i.e. where there is non-significant occupancy for a given gene in a particular neuron population).
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp5-v2.txt
-
Supplementary file 6
Set of neuropeptide genes.
Genes correspond to those in Figure 2C.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp6-v2.txt
-
Supplementary file 7
Neuropeptide genes with significant OSs in Ir neuron populations.
IDs correspond to the counts displayed in Figure 2C.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp7-v2.txt
-
Supplementary file 8
Neuropeptide genes with significant OSs unique to each Ir neuron population.
IDs correspond to the counts displayed in Figure 2C.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp8-v2.txt
-
Supplementary file 9
Set of G protein-coupled receptor genes.
Genes correspond to those in Figure 2D.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp9-v2.txt
-
Supplementary file 10
G protein-coupled receptor genes with significant OSs in Ir neuron populations.
IDs correspond to the counts displayed in Figure 2D.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp10-v2.txt
-
Supplementary file 11
G protein-coupled receptor genes with significant OSs unique to each Ir neuron population.
IDs correspond to the counts displayed in Figure 2D.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp11-v2.txt
-
Supplementary file 12
Set of ion channel genes.
Genes correspond to those in Figure 2E.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp12-v2.txt
-
Supplementary file 13
Ion channel genes with significant OSs in Ir neuron populations.
IDs correspond to the counts displayed in Figure 2E.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp13-v2.txt
-
Supplementary file 14
Ion channel genes with significant OSs unique to each Ir neuron population.
IDs correspond to the counts displayed in Figure 2E.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp14-v2.txt
-
Supplementary file 15
Set of neuron projection guidance molecule genes.
Genes correspond to those in Figure 2F. Note this list encompasses genes encoding molecules potentially directly or indirectly involved in neural guidance, and includes some neuronally-expressed transcription factors (such as pdm3, the unique marker of Ir75a neurons).
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp15-v2.txt
-
Supplementary file 16
Neuron projection guidance molecule genes with significant OSs in Ir neuron populations.
IDs correspond to the counts displayed in Figure 2F.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp16-v2.txt
-
Supplementary file 17
Neuron projection guidance molecule genes with significant OSs unique to each Ir neuron population.
IDs correspond to the counts displayed in Figure 2F.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp17-v2.txt
-
Supplementary file 18
Filtered set of transcription factor genes.
Genes correspond to those in Figure 2G.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp18-v2.txt
-
Supplementary file 19
Transcription factor genes with significant OSs in Ir neuron populations.
IDs correspond to the counts displayed in Figure 2G.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp19-v2.txt
-
Supplementary file 20
Transcription factor genes with significant OSs unique to each Ir neuron population.
IDs correspond to the counts displayed in Figure 2G.
- https://cdn.elifesciences.org/articles/63036/elife-63036-supp20-v2.txt
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63036/elife-63036-transrepform-v2.docx





