PP2A/B55α substrate recruitment as defined by the retinoblastoma-related protein p107
Figures
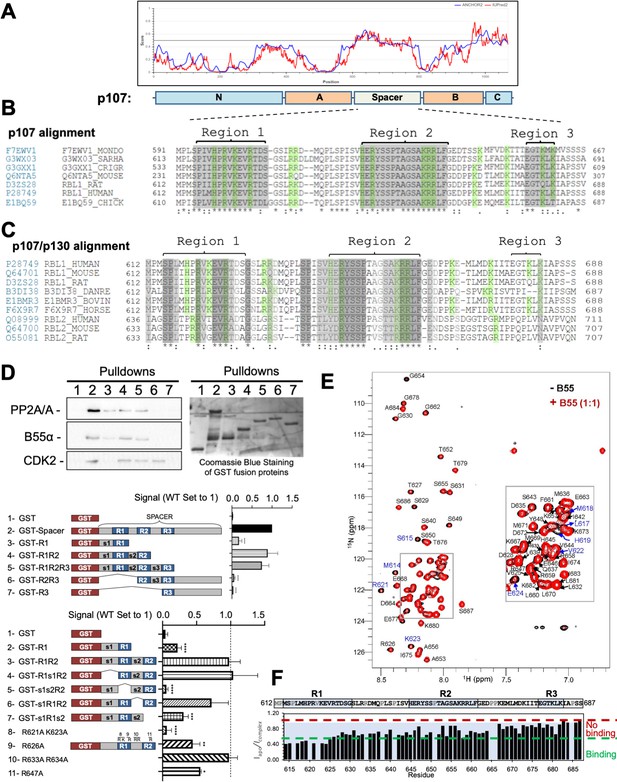
The intrinsically disordered spacer region of p107 contains three highly conserved regions (R1, R2, and R3), of which R1 is required for B55α binding and R2 enhances the binding interaction as determined via mutational analysis and NMR.
(A) The spacer and the C-terminus of p107 are intrinsically disordered (IUPred2a web interface, Sievers et al., 2011). (B) Clustal W alignment of conserved amino acid sequences of the p107 spacer from different species. Three highly conserved regions within the spacer were identified, which are highlighted in gray and named as region 1 (R1), region 2 (R2), and region 3 (R3). Positively charged residues are highlighted in green. (C) Clustal W alignment of conserved amino acid sequences of the p107 and p130 spacer from the indicated species. Conserved residues are highlighted in shades of gray. Positively charged residues are highlighted in green. (D) A GST-p107 spacer construct was used as a template to systematically delete indicated regions and mutate positively charged amino acids in R1 and R2. The p107 spacer spans amino acids 584–782 and the small spacers, s1 (584–615), s2 (631–644), and s3 (662–676), separate the beginning of the spacer from R1 and the three conserved regions R1, R2, and R3. Pull-down assays with the indicated fusion proteins from U2-OS lysates were performed, and binding of the indicated proteins was determined by western blot analysis. GST-p107 was determined by Coomassie Blue or Ponceau S staining. Experiments were performed in triplicate, and quantification values represent the mean B55α/p107 variant ratios ± standard deviation (SD). (E) Overlay of the 2D [1H, 15N] HSQC spectra of 15N-labeled p107 (M612-S687) in the presence (red) and absence (black) of purified monomeric full-length B55α (M1–N447) (see Materials and methods for B55α purification). (F) p107 (M612-S687) sequence is shown above the peak intensity ratios for data shown in (E). Prolines, residues not assigned (M612 and R634), and overlapping residue (V622) are labeled in grey. Residues corresponding to R1, R2, and R3 are highlighted in blue. Approximate binding cutoffs are marked by dashed lines.
-
Figure 1—source data 1
Uncropped replicates of western blot images and PVDF membranes stained with Coomassie Blue used to quantitate B55α binding to conserved regions in the p107 spacer (top bar graph).
B55α signal was normalized to the GST-fusion protein signal (selected bands are marked with dashed white boxes and corrected for background from a band-less identical area).
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig1-data1-v2.zip
-
Figure 1—source data 2
Uncropped replicates of western blot images and PVDF membranes stained with Coomassie Blue or Ponceau S used to quantitate binding to B55α of conserved regions in the p107 R1–R2 construct (top bar graph) and R/K point mutant variants of p107 R1–R2 (lower bar graph).
Only the R/K mutants labeled blue were included in the quantitation.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig1-data2-v2.zip
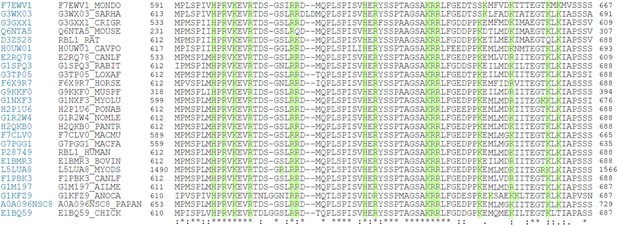
Clustal W alignment of conserved amino acid sequences of the p107 spacer from different species.
(A) Complete Clustal W alignment of conserved amino acid sequences of the p107 spacer from different species. Three highly conserved regions within the spacer were identified, which are highlighted in gray and named as region 1 (R1), region 2 (R2), and region 3 (R3). Positively charged residues are highlighted in green.
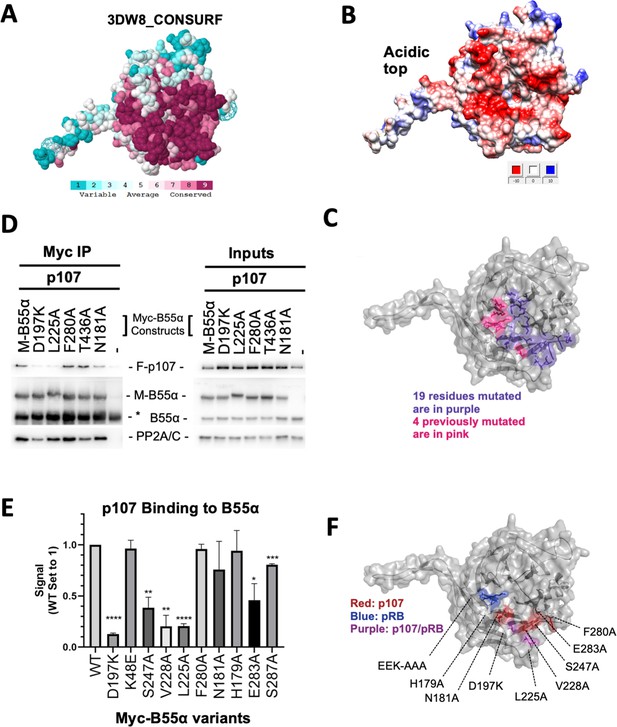
Mutation of highly conserved residues on the β-propeller top of B55α have substrate-specific effects on binding, supporting the notion that substrates contact different surfaces on B55α.
(A) ConSurf depiction of PP2A/B55α mapping amino acid conservation (where amino acids are color-coded by conservation). (B) Electrostatic predictions mapped to the surface of the PP2A/B55α structure indicate an acidic top (red, acidic; blue, basic). (C) Nineteen single-point mutations on the conserved top of the B55α β-propeller were generated. These are shown in purple. Four mutations generated and analyzed previously are shown in pink. (D) Representative immunoprecipitation experiment to test p107 binding requirements on Myc-B55α. Flag-p107 and wild-type and mutant Myc-B55α mutant constructs were co-transfected into 293T cells and used for IPs with anti-Myc agarose conjugate. These assays were resolved via SDS-PAGE, and proteins were detected using anti-Flag, anti-B55α, and anti-PP2A/C. (E) Mean values for cumulative immunoprecipitation assays for Flag-tagged p107 binding to Myc-B55α constructs are shown, with statistics indicated above. Experiments were performed in triplicate or duplicate, and quantification values represent the mean p107:Myc-B55α variant ratios ± standard error. (F) The surface structure of B55α is depicted, with residues that appear important for p107 and pRB binding color-coded (in red and blue, respectively). The two Myc-B55α mutants that affect binding of both p107 and pRB (D197K and L225A) are colored purple.
-
Figure 2—source data 1
Upper, middle, and lower western blot membranes for Figure 2D (replicate 1).
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig2-data1-v2.zip
-
Figure 2—source data 2
Western blot membranes for replicates 1–3.
All replicates were used for the quantitation shown in Figure 2E. The legends indicate the B55α variants used in this set of replicates. Relevant proteins and IgG (in the IP membranes) are indicated.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig2-data2-v2.zip
-
Figure 2—source data 3
Western blot membranes for replicates 1–2 used for the quantitation shown in Figure 2E.
The legend indicates the B55α variants used in this set of replicates. Relevant proteins and IgG (in the IP membranes) are indicated.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig2-data3-v2.zip
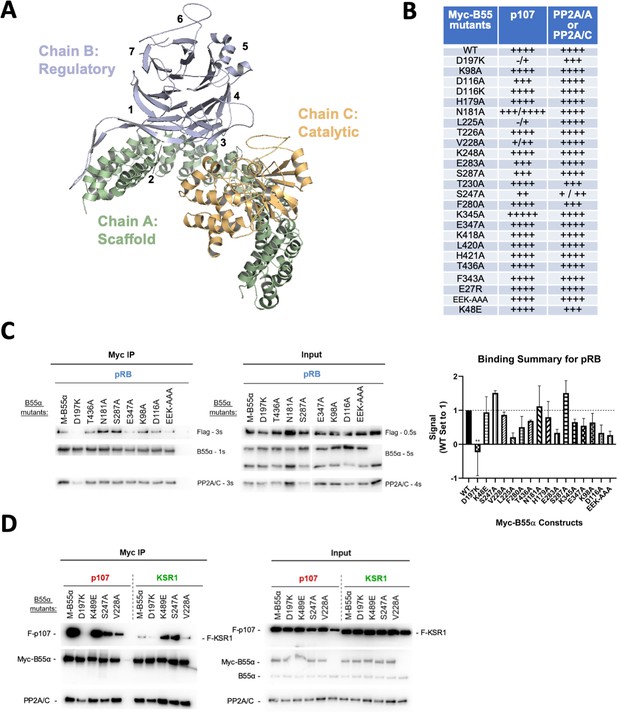
Effect of B55α mutations on p107, pRB and KSR1 binding.
(A) Structure of the PP2A/B55α holoenzyme (PDB:3DW8). (B) Table depicting complete list of Myc-tagged B55α mutants generated, as well as summarizing the effect on binding to p107 and PP2A/A or PP2A/C based on immunoprecipitation experiments. (C) Representative immunoprecipitation experiment to test pRB binding requirements on Myc-B55α. Flag-pRB and wild-type and mutant Myc-B55α mutant constructs were co-transfected into 293T cells and used for IPs with anti-Myc agarose conjugate. These assays were resolved via SDS-PAGE, and proteins were detected using anti-Flag, anti-B55α, and anti-PP2A/C. Mean values for cumulative immunoprecipitation assays for Flag-tagged pRB binding to Myc-B55α constructs are shown on right, with statistics indicated above. (D) Representative immunoprecipitation experiment to test KSR1 binding requirements on Myc-B55α. Experiments were performed as above. KSR1 signal-to-noise ratio was often low due to the apparent lower affinity of KSR1 for B55α. Thus, quantitation produced higher standard errors, and we only considered the data as qualitative.
-
Figure 2—figure supplement 1—source data 1
Western blot membranes for replicates used for the quantitation of Flag-pRB:Myc-B55α binding ratios shown in Figure 2—figure supplement 1 (middle).
The legends indicate the B55α variants used in this set of replicates. Relevant proteins and IgG (in the IP membranes) are indicated.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig2-figsupp1-data1-v2.zip
-
Figure 2—figure supplement 1—source data 2
Western blot membranes for replicates used for the quantitation of Flag-pRB:Myc-B55α binding ratios shown in Figure 2—figure supplement 1 (middle).
The legends indicate the B55α variants used in this set of replicates. Relevant proteins and IgG (in the IP membranes) are indicated.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig2-figsupp1-data2-v2.zip
-
Figure 2—figure supplement 1—source data 3
Western blot membranes for replicates used for the quantitation of Flag-pRB:Myc-B55α binding ratios shown in Figure 2—figure supplement 1 (middle).
The legends indicate the B55α variants used in this set of replicates. Relevant proteins and IgG (in the IP membranes) are indicated.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig2-figsupp1-data3-v2.zip
-
Figure 2—figure supplement 1—source data 4
Upper, middle, and lower western blot membranes for Figure 2—figure supplement 1, bottom.
Boxes indicate approximate area shown in the figure. Relevant proteins are indicated.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig2-figsupp1-data4-v2.zip
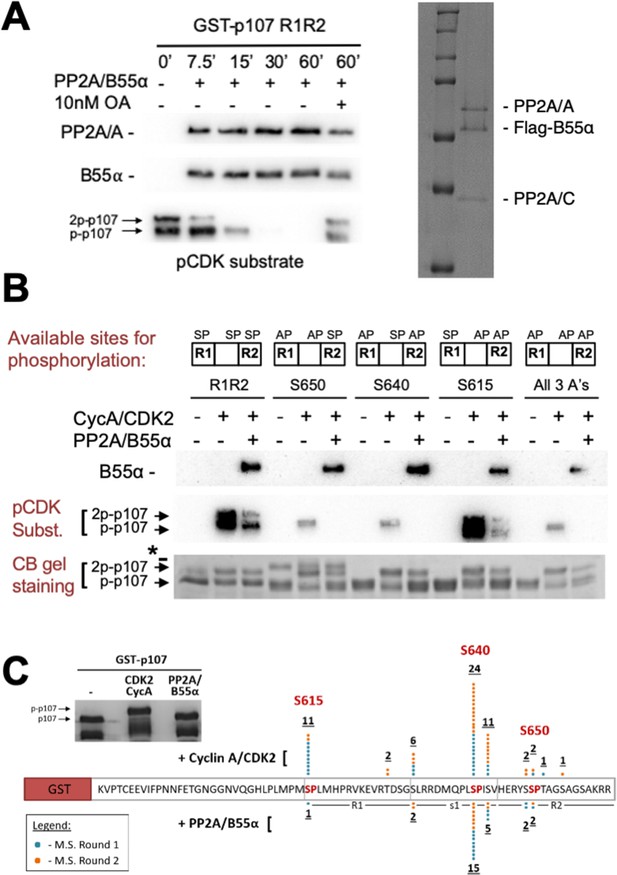
Combination of in vitro enzymatic assays and mass spectrometry identified S615 on R1 of p107 as the major site of PP2A/B55α-mediated dephosphorylation.
(A) Dephosphorylation of GST-p107 R1R2 was performed using approximately 10 ng purified PP2A/B55α. The indicated time points were collected and samples were resolved via SDS-PAGE. Proteins were detected with anti-PP2A/A, anti-B55α, and anti-pCDK substrate [(K/H)pSP]. Representative Coomassie Blue-stained gel depicting affinity-purified PP2A/B55α holoenzymes is shown on right. (B) In vitro phosphorylation and dephosphorylation of GST-p107 R1R2 with single SP sites available were performed using 0.25 μg recombinant cyclin A/CDK2 and approximately 10 ng PP2A/B55α (each for 1 hr, respectively). Proteins were resolved via SDS-PAGE and detected by Coomassie Blue gel staining and western blotting using anti-B55α and pCDK substrate antibodies. Note basal levels of pCDK substrate signal in all ‘+ CycA/CDK2’ lanes, which indicates phosphorylation on CDK2 itself. Relevant proteins and p107 phosphorylated species are indicated (a bacterial contaminant in the S650 MT is indicated with an asterisk). (C) Representative Coomassie Blue-stained PAGE used for mass spectrometry, with schematic summarizing the findings from two independent rounds of mass spectrometry analyses, is shown.
-
Figure 3—source data 1
Uncropped blots and Coomassie-stained gels for Figure 3A-C.
Images in Figure 3A were generated from the boxed regions in each PVDF membrane. Comparable experiments are shown in Figure 4. Images in Figure 3B were generated from the boxed regions in replicate 1. Replicates 1–3 show comparable dephosphorylation of WT and MT p107 R1R2 by Coomassie Blue staining. The gel image in Figure 3C was generated form representative replicate 1, and the bands cut out for mass spectrometry are boxed.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig3-data1-v2.zip

Experiments performed to determine optimal GST-p107 R1R2 phosphorylation parameters using purified cyclin A/CDK2.
(A) Phosphorylation of purified GST-p107 R1R2 was performed using 0.25 μg recombinant cyclin A/CDK2 and 5 μCi γ–32P. The indicated time points were collected and samples were resolved via SDS-PAGE. Proteins were detected via Coomassie gel staining and 15 s exposure to a Phosphorimager screen, respectively. (B) Phosphorylation of purified GST-p107 R1R2 was performed using 0.25 μg recombinant cyclin A/CDK2 and 100 μM ATP. The indicated time points were collected and samples were resolved via SDS-PAGE. Proteins were detected by anti-cyclin A and anti-pCDK substrate [(K/H)pSP], as well as Coomassie gel staining.
-
Figure 3—figure supplement 1—source data 1
Uncropped Coomassie-stained gel, Phosphorimager exposure and western blots for Figure 3—figure supplement 1.
Dashed boxes correspond to the areas shown in Figure 3—figure supplement 1 (top). The 7.5’ sample was loaded after the 15” sample in error. The lane was omitted in the figure for clarity and the omitted lane marked with an asterisk.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig3-figsupp1-data1-v2.zip
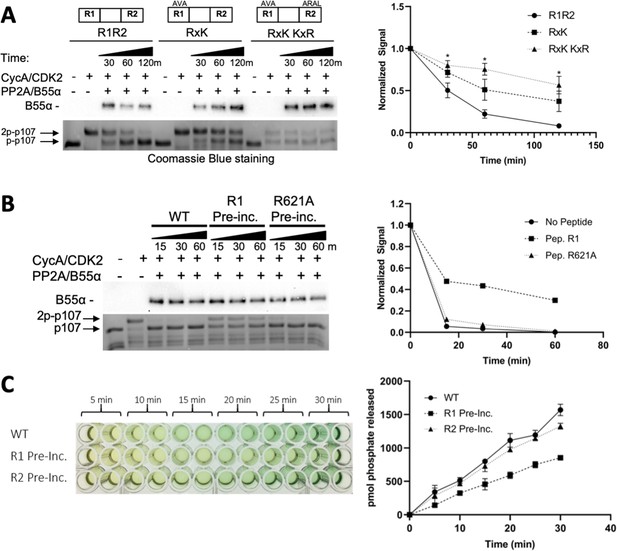
Residues critical for B55α/PP2A binding to p107 are critical for p107 spacer dephosphorylation.
(A) Approximately 10 ng of purified PP2A/B55α was used to dephosphorylate wildtype GST-p107 R1R2 or mutant constructs (GST-p107 R621A K623A and GST-p107 RxK K657A R659A) in a time-course experiment. Proteins were resolved via SDS-PAGE and detected by Coomassie Blue staining or western blotting using anti-B55α antibodies. Quantifications of the ‘phospho’-p107 band for each construct tested were performed using ImageJ and plotted as a function of time, with statistics shown above. (B) Representative assay in which purified PP2A/B55α was preincubated with either wildtype R1 peptide or R621A mutant R1 peptide and then used in time-course dephosphorylation assays using GST-p107 R1R2 (native enzyme was used as a positive control for dephosphorylation). Proteins were resolved via SDS-PAGE and detected by Coomassie Blue. Quantification is shown on right. (C) Purified PP2A/B55α was preincubated with either wildtype R1 or R2 peptide and then used in Malachite Green Phosphatase Assay with a p107-derived phosphopeptide (in which S615 is the available phosphosite). The indicated time points were collected and absorbance was read at 600 nm by microplate reader (quantification is shown on right).
-
Figure 4—source data 1
Uncropped Coomassie-stained gels and blots for Figure 4A and B.
Images in Figure 4A were generated from the boxed regions in replicate 1 PVDF membrane and the Coomassie Blue-stained gel. Quantifications shown in Figure 4A of the ‘phospho’-p107 band were obtained from Coomassie Blue-stained gel replicates 1–3. Images in Figure 4B were generated from the boxed regions in replicate 1. A comparable experiment using an R1 peptide (R1-627TDS-AAA) variant that binds B55α (Figure 5B) showed delayed phosphorylation as R1, while R1-R621A and R2, which do not bind B55α (Figure 5A and C), did not inhibit dephosphorylation.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig4-data1-v2.zip
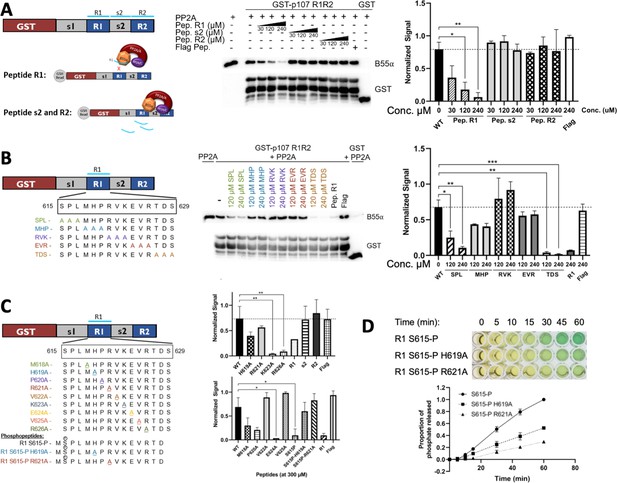
Identification of critical residues within the central 9-mer stretch of the ‘R1’ region of p107 for binding to B55α/PP2A (SPxxHxRVxxV).
(A) PP2A/B55α purified from 293T-Flag-B55α cells was preincubated with synthetic p107 peptides and then used in pull-down assays with GST-p107 R1R2 constructs. Proteins were resolved via SDS-PAGE and detected via western blotting using anti-B55α and GST antibodies. An independent replicate experiment was performed using 293T cell lysates as source of PP2A/B55α with comparable results. Quantification of B55α pulled down relative to the wildtype pulldown was performed for both replicates using ImageJ, with statistics shown above. (B, C) Pulldowns were performed using purified PP2A/B55α and GST-p107 R1R2 as above with preincubations using mutant p107-derived synthetic peptides (both scanning triple-mutants and point mutants) as well as wildtype phosphopeptides. Proteins were resolved via SDS-PAGE and detected via western blotting using anti-B55α and GST antibodies. Quantifications were performed as above. (D) Purified PP2A/B55α was used in time-course Malachite Green Phosphatase Assay using wildtype or mutant p107-derived phosphopeptides as substrates of dephosphorylation. The indicated time points were collected and absorbance was read at 600 nm by microplate reader (quantification of duplicate assays is shown below).
-
Figure 5—source data 1
Uncropped upper membrane (anti-B55α) and lower membrane (anti-GST) western blots for Figure 5A and B.
Images in Figure 4A and B were generated from the boxed regions in replicate 1 PVDF membranes. B55α band intensities were normalized to the corresponding full-length GST-R1R2 band intensities.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig5-data1-v2.zip
-
Figure 5—source data 2
Uncropped upper membrane (anti-B55α) and lower membrane (anti-GST) western blots for Figure 5B, C (three replicates) were used to quantitate peptide competition of B55α binding to GST-R1R2.
B55α band intensities were normalized to the corresponding full-length GST-R1R2 band intensities.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig5-data2-v2.zip
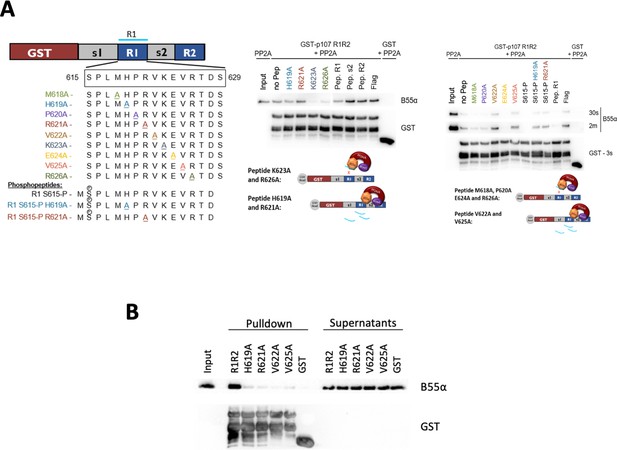
Identifying esential rediues in the R1 region of the p107 spacer that affect binding to B55α, using peptide competition and GST puldown assays.
(A) Representative pulldowns were performed using purified PP2A/B55α (PP2A) and GST-p107 R1R2 with preincubations using mutant p107-derived synthetic peptides, phosphopeptides, and control peptides, as indicated. Proteins were resolved via SDS-PAGE and detected via western blotting using anti-B55α and GST antibodies. (B) Pull-down assay using GST-R1R2 mutants substituting critical short linear motif (SLiM) residues for Ala. Input shown is ~3% of total volume of PP2A/B55α eluate used in pulldowns.
-
Figure 5—figure supplement 1—source data 1
Uncropped western blot images for panel B.
All the replicates for panel A are shown in Figure 5—source data 2.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig5-figsupp1-data1-v2.zip
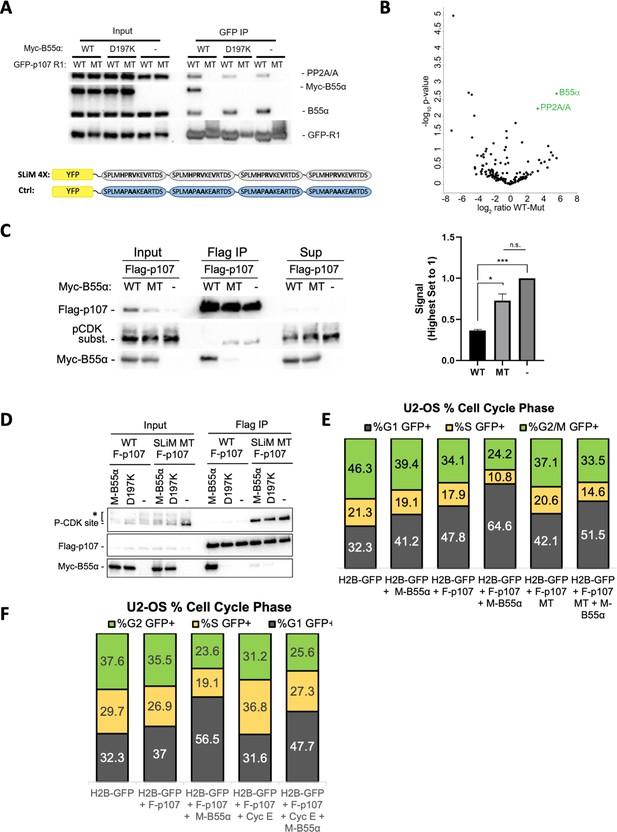
p107 R1 interaction with the B55α holoenzyme in cells depends on sites required for binding and dephosphorylation in vitro.
(A) GFP-p107 R1 wildtype and mutant constructs were co-transfected with Myc-B55α wildtype and B55α-D197K mutant constructs into 293T cells and used for IPs with anti-GFP agarose conjugate. Input lysates and IPs were resolved via SDS-PAGE, and proteins were detected using anti-PP2A/A, anti-B55α, and anti-GFP. Schematic of WT and MT GFP-p107 R1 constructs is shown below. (B) Volcano plot depicts the differences of protein abundances in YFP-SLIM4X and YFP-Ctrl pulldowns (Figure 6—source data 1). (C) Flag-p107 was co-transfected with Myc-B55α wildtype and B55α-D197K mutant constructs into 293T cells and used for IPs with anti-Flag agarose conjugate. IPs and input/supernatant lysates were resolved via SDS-PAGE, and proteins were detected using anti-Flag and anti-pCDK substrate antibodies. (Right) Quantification of pCDK substrate signal (relative to Flag) is shown, with statistics shown above. (D) (Left) WT and short linear motif (SLiM) MT Flag-p107 were co-transfected with Myc-B55α wildtype and B55α-D197K mutant constructs into 293T cells and analyzed as in (C). (E) U-2 OS cells were co-transfected with the indicated plasmids, and the percent of cells in G1, S, and G2/M were determined by measuring DNA content via FACS/PI analyses of GFP-positive cells. (F) U-2 OS cells were co-transfected with the indicated plasmids and analyzed as in (D, right).
-
Figure 6—source data 1
Uncropped upper membranes for Figure 6A and replicate experiments of endogenous B55α interaction with wildtype, but not mutant, GFP-R1-SLiM.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig6-data1-v2.zip
-
Figure 6—source data 2
Mass spectrometry dataset used to generate the volcano plot shown in Figure 6.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig6-data2-v2.xlsx
-
Figure 6—source data 3
Uncropped upper and lower membranes for Figure 6C and D (representative replicate 1 was selected for the C and D panels).
In (C), B55α-mediated dephosphorylation was quantitated using the pCDK substrate vs. Flag-p107 signal using the corresponding bands in the dashed boxes.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig6-data3-v2.zip

U-2 OS cells were co-transfected with the indicated plasmids, lysates were immunoprecipitated with anti-Flag agarose conjugate, and the indicated co-immunoprecipitated proteins were detected by western blot analyses.
(Right) Quantification of Flag-p107/HA-E2F4 rations is shown.
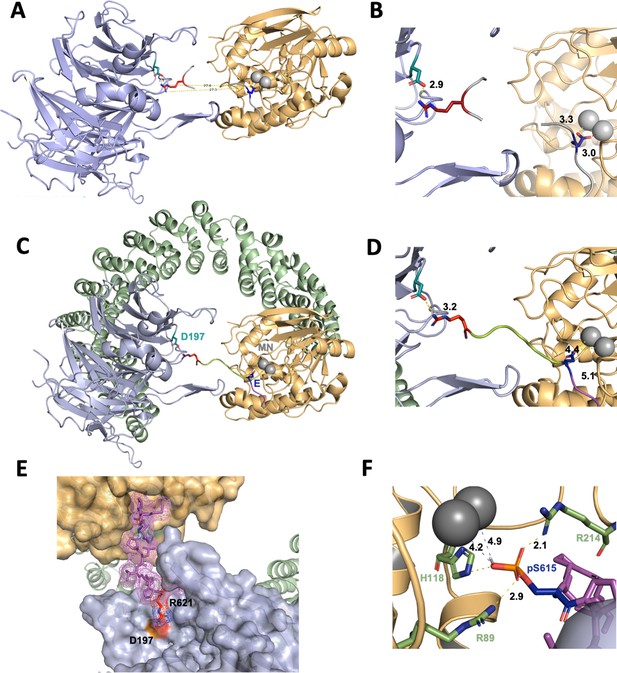
A computational model of the p107 phosphopeptide (613–622) binding B55α and the active site of PP2A/C is consistent with p107 spacer contacts to B55α as determined by NMR spectroscopy.
(A) The ‘two-fragment’ model depicting B55α and PP2A/C with two modeled peptide fragments. Distances between the OE1/OE2 atoms of Glu and the NH1/NH2 atoms of Arg are shown in center. (B) A closer view of ‘two-fragment’ model highlighting the distances between Arg and D197 of B55α and between the Glu residue and the Mn2+ ions within PP2A/C active site. (C, D) Peptide model depicting the best alignments with the two reference fragments and the best distances to the Mn2+ ions and D197. A closer view highlighting the distances between the NH2 of Arg residue and D197 of B55α, and the Glu residue with the Mn2+ ions, is shown in (C). (E) Close-up of model in (C, D) showing the peptide side chains and contacts to B55α surface. (F) Close-up of pSer-621 (substituted for the Glu residue) and residues critical for phosphate coordination (R630, H559, R655). Distances between residues where H-bonding is predicted to occur are shown.
-
Figure 7—source data 1
PyMOL session source data for Figure 7A.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-data1-v2.zip
-
Figure 7—source data 2
PyMOL session source data for Figure 7B.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-data2-v2.zip
-
Figure 7—source data 3
PyMOL session source data for Figure 7C and D.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-data3-v2.zip
-
Figure 7—source data 4
PyMOL session source data for Figure 7E.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-data4-v2.zip
-
Figure 7—source data 5
PyMOL session source data for Figure 7F.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-data5-v2.zip

Step details to generate a computational model of the p107 phosphopeptide (613-622) binding B55α and the active site of PP2A/C.
(A) Density plot of distances between OE1/2 of Glu and NH1/2 of Arg in 520 peptide structures with the consensus sequence EPXXXPR. The red dot is the distance between OE1/2 of Glu and NH1/2 of Arg from two reference fragments. (B) Pair fit OE1/2 of Glu residues and NH1/2 of Arg residues of 217 peptide structures to OE1/2 of Glu in MPMEPL fragment and NH1/2 of Arg residue in HPRV fragment. Asp197 is colored cyan and shown in sticks. Arg in HPRV fragment is colored magenta and shown in sticks. Glu in MPMEPL fragment is colored blue and shown in sticks. Peptides with EPXXXPR consensus sequence are shown in ribbon, with their Glu residues colored blue and Arg residues colored magenta. (C) Density plot of sum of minimum distances (OE1/2 of Glu residues and NH1/2 Arg residues) between each peptide and two reference fragments. The red points are the minimum and maximum values (1.123294 and 8.840146, respectively). (D) Superposing human PPP catalytic subunit structures to PP2A enzyme and the chain of 5HPE, OE1/OE2 atoms of Glu residue are in same positions as oxygen atoms from phosphate groups. (E, right) The top 20 peptide structures were aligned to the reference fragments by pair fitting OE1/2 of Glu residues and NH1/2 of Arg residues in PyMOL. (Left) The p107 peptide substrate was built in PyMOL by mutating residues XXX to LMH, and adding MPM residues before the EP and V residue after PR.
-
Figure 7—figure supplement 1—source data 1
Distances between OE1/2 and NH1/2 for 520 peptide structures with the consensus sequence EPXXXPR retrieved from PDB.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-figsupp1-data1-v2.txt
-
Figure 7—figure supplement 1—source data 2
R script file to generate density plots for Figure 7—figure supplement 1A and B.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-figsupp1-data2-v2.txt
-
Figure 7—figure supplement 1—source data 3
PyMOL session source data for Figure 7—figure supplement 1B.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-figsupp1-data3-v2.zip
-
Figure 7—figure supplement 1—source data 4
Distances of OE1/2 and NH1/2 between each peptide and two reference fragments for Figure 7—figure supplement 1C.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-figsupp1-data4-v2.zip
-
Figure 7—figure supplement 1—source data 5
PyMOL session source data for Figure 7—figure supplement 1D.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-figsupp1-data5-v2.zip
-
Figure 7—figure supplement 1—source data 6
PyMOL session source data for Figure 7—figure supplement 1E.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-figsupp1-data6-v2.zip
-
Figure 7—figure supplement 1—source code 1
Model code.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-figsupp1-code1-v2.zip

PP2A/A scaffold flexibility upon binding to the catalytic subunit and B subunits of the four distinct holoenzymes.
(A) Comparison of the flexibility of the PP2A/A scaffold upon binding to the catalytic subunit and B subunits of the four distinct holoenzymes. B55/B is colored red, B56/B′ is colored yellow, PR70/B′′ is colored lime, and STRN3/B′′′ is colored blue. (B) Comparison of the flexibility of the PP2A/A scaffold upon binding to TIPRL (colored teal) and small-T antigen (colored orange). B55/B (red) is shown for reference. (C, D) Average distance over 25 residues of scaffold protein in PDB:3DW8, which are in contact with the PP2A enzyme (PDB:3DW8 chain A as reference). (C) PP2A/A structures with no regulatory subunits (PP2A/A noreg). 6EF4A corresponds to PP2A/A scaffold without catalytic subunit. (D) Scaffold structures with B56 and microcystin (PP2A-B56_micro).
-
Figure 7—figure supplement 2—source data 1
PyMOL session source data for Figure 7—figure supplement 2A.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-figsupp2-data1-v2.txt
-
Figure 7—figure supplement 2—source data 2
PyMOL session source data for Figure 7—figure supplement 2B.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-figsupp2-data2-v2.txt
-
Figure 7—figure supplement 2—source data 3
Scaffold variation source data without regulatory subunit for Figure 7—figure supplement 2C.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-figsupp2-data3-v2.zip
-
Figure 7—figure supplement 2—source data 4
Scaffold variation source data with B56 subunits for Figure 7—figure supplement 2D.
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig7-figsupp2-data4-v2.zip
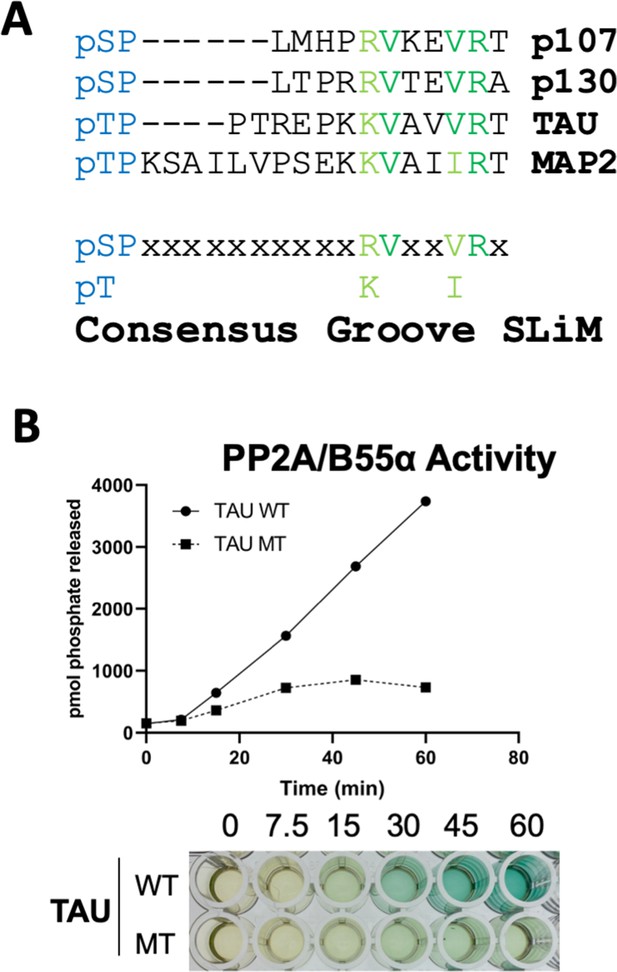
A derived p107 -pSPxxHxRVxxV- short linear motif (SLiM) is conserved in other substrates and functional validated in TAU.
(A) Schematic of our proposed hypothetical consensus groove SLiM, p[ST]-P-x(4,10)-[RK]-V-x-x-[VI]-R, for TAU, MAP2, and the conserved p107 family member, p130, each of which contain residues that align with our defined p107 SLiM. (B) Time-course Malachite Green Phosphatase Assay using a wildtype phospho-TAU peptide encompassing the putative conserved SLiM and a variant peptide with the conserved residues mutated to Ala. (Above) Quantification of phosphatase assay is shown below.

Degenerate short linear motif (SLiM) search in the proteome vs. datasets enriched for B55 interactor and potential substrates.
The bar graph represents the percent of proteins with a degenerate version of the SLiM, [ST]-P-x(4,10)-[RK]-[VIL]-x(2)-[VILM] in the proteome and in a dataset of B55α interactors (Hertz et al., 2016) and potential in vitro B55α mitotic substrates (Kruse et al., 2020). This enrichment indicates that many substrates and B55 interactors potentially use this SLiM-based mechanism. Proteins identified in these two datasets are listed in Figure 8—figure supplement 1.
-
Figure 8—figure supplement 1—source data 1
Table of proteins containing the [ST]-P-x(4,10)-[RK]-[VIL]-x(2)-[VILM] sequence in a dataset of B55α interactors (Hertz et al., 2016) and potential in vitro B55α mitotic substrates (Kruse et al., 2020).
- https://cdn.elifesciences.org/articles/63181/elife-63181-fig8-figsupp1-data1-v2.xlsx
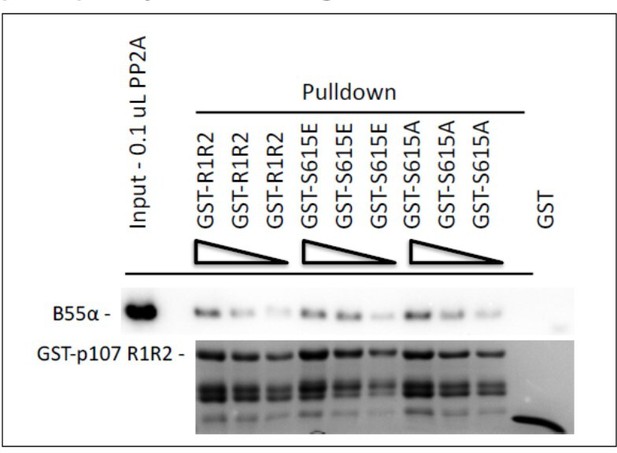
Pull down assays with the indicated fusion proteins from HEK-293 lysates were performed and binding of B55a was determined by western blot analysis.
Two replicates of the experiment showed comparable results.
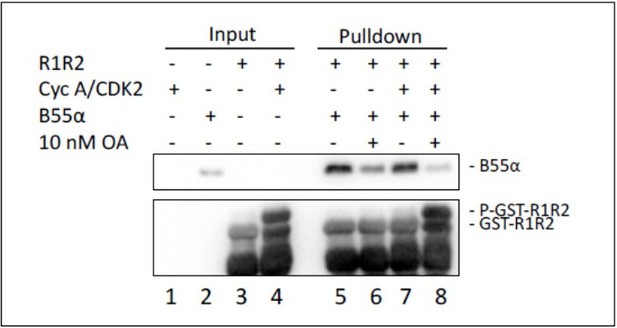
GST-R1R2 was phosphorylated with Cyclin A/CDK2 where indicated as described in the Manuscript.
B55a binding to GST-R1R2 was determined in the presence or absence of OA to prevent dephosphorylation. Proteins were resolved via SDS-PAGE and detected by western blotting using anti-B55α and GST antibodies. Two replicates of the experiment showed comparable results.
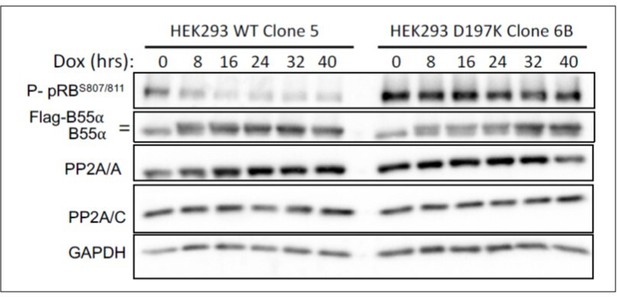
HEK-293 cells with doxicycline (Dox) inducible WT B55a and B55aD197K stable transgenes where treated with Dox for the indicated times and expression of indicated proteins and phospho pRB were determined by western blot analysis.
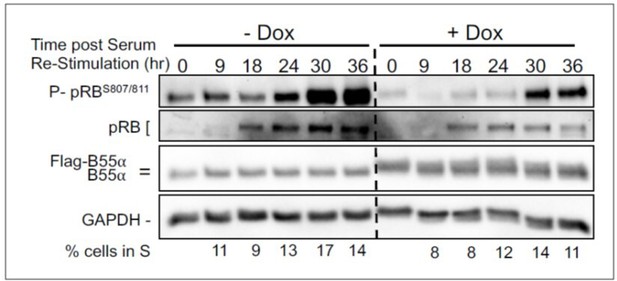
BJ-TERT fibroblasts with doxicycline (Dox) inducible WT B55a and B55aD197K stable transgenes where grown to confluency treated with Dox for for 48 h, serum starved for 24 hours and then restimulated with Medium conaning 10% FBS for the indicated times.
Expression of indicated proteins and phospho pRB was determined by western blot analysis.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | CDK2(rabbit polyclonal) | Santa Cruz | sc-163 | WB(1:1000) |
| Antibody | Cyclin A(mouse monoclonal) | Santa Cruz | sc-271682 | WB(1:1000) |
| Antibody | Flag(mouse monoclonal) | Sigma | A8592-.2MG | WB(1:2500, 1:10,000) |
| Antibody | Flag(mouse monoclonal) | GenScript | A00187 | IP (1 µg/mL) |
| Antibody | GFP(rabbit monoclonal) | CST | 2956S | WB(1:1000) |
| Antibody | GST(mouse monoclonal) | Santa Cruz | sc-138 | WB(1:1000) |
| Antibody | PP2A Aβ subunit(goat polyclonal) | Santa Cruz | sc-6113 | WB(1:2000) |
| Antibody | PP2A B subunit(100C1)(rabbit monoclonal) | CST | 2290S | WB(1:2000) |
| Antibody | PP2A C subunit (1D6) (mouse monoclonal) | Millipore | 05-421 | WB(1:5000) |
| Antibody | Phospho-CDK Substrate Motif(rabbit monoclonal) | CST | 9477S | WB(1:1000) |
| Antibody | ECL Rabbit IgG, HRP-linked whole Ab (from donkey) | GE Healthcare | NA934V | WB/secondary antibody(1:10,000) |
| Antibody | HA (12CA5)(mouse monoclonal) | Roche/Sigma | 11583816001 | WB(1:500) |
| Antibody | ECL Mouse IgG, HRP-linked whole Ab (from donkey) | GE Healthcare | NA931V | WB/secondary antibody(1:10,000) |
| Antibody | Mouse anti-goat IgG-HRP | Santa Cruz | sc-2354 | WB/secondary antibody(1:10,000) |
| Antibody | Monoclonal ANTI-FLAG M2 antibody produced in mouse, ANTI-FLAG M2 Affinity Agarose Gel | Sigma | A2220 | IP (10 μL) |
| Antibody | Anti-c-Myc Agarose Affinity Gel antibody produced in rabbit (polyclonal) | Sigma | A7470 | IP (10 μL) |
| Antibody | GFP-Trap agarose beads | Chromotek | gta-10 | IP (5 μL) |
| Peptide, recombinant protein | Synthetic peptides for competition assays | Biomatik | Custom | Sequence variant described in this paper |
| Peptide, recombinant protein | DYKDDDDK peptide | GenScript | RP10586 | |
| Peptide, recombinant protein | Recombinant cyclin A/CDK2 | Thermo Fisher | PV3267 | |
| Peptide, recombinant protein | Purified recombinant B55α/PP2A holoenzyme | Zhao et al., 2019 | Trimeric Flag-B55α/PP2A holoenzyme purified from 293T cells. | |
| Strain, strain background (Escherichia coli) | BL21-Gold (DE3) cells | Agilent | 230132 | To generate GST-Fusion proteins |
| Cell line (Homo sapiens) | 293T cells | ATCC | CRL-3216 | Transient transfections and source of cell lysates |
| Cell line (H. sapiens) | U-2 OS cells | ATCC | HTB-96 | Transient transfections |
| Cell line (H. sapiens) | Expi293F cells | Thermo Fisher | A14528 | Purification of recombinant B55α |
| Transfected construct (human) | Flag-B55α 293T cells | Zhao et al., 2019 | 293T cells stably transfected with pCPP-Flag-B55α and selected with puromycin | |
| Commercial assay or kit | (AminoLink Plus Immobilization Kit) | Thermo Fisher Scientific | 44894 | |
| Commercial assay or kit | Ser/Thr phosphatase assay kit | EMD Millipore | 17-127 | |
| Commercial assay or kit | QuikChange II | Agilent | 200521 | |
| Recombinant DNA reagent | pBOS GFP-H2B plasmid | Kanda et al., 1998 | ||
| Recombinant DNA reagent | pcDNA3.4-K-GFP-RP1B | This paper | His6-green fluorescent protein-tag, a TEV cleavage | |
| Recombinant DNA reagent | pTHMT | Peti and Page, Protein Expr. Purif. 51, 1–10 (2007) | N-terminal His6-Maltose Binding Protein (MBP)-tag, a TEV cleavage | |
| Recombinant DNA reagent | pCPP-Flag-B55α | Zhao et al., 2019 | ||
| Recombinant DNA reagent | pMSCV-puro-Myc-B55α | Jayadeva et al., 2010 | ||
| Recombinant DNA reagent | pMSCV-puro-Myc-B55α variants | This paper | Generated by site-directed mutagenesis (primer sequences provided in Appendix table) | |
| Recombinant DNA reagent | pCDNA5/FRT/TO-GFP-p107-R1 | This paper | Inserting wild-type or mutant p107-R1 gBlock Gene Fragments (IDT) into the pCDNA5/FRT/TO-GFP vector via the BamHI/Not sites | |
| Recombinant DNA reagent | pGEX-2T-p107 spacer | Jayadeva et al., 2010 | ||
| Recombinant DNA reagent | pGEX-2T-p107 spacer variants | This paper | Generated by site-directed mutagenesis (primer sequences provided in Appendix table) | |
| Recombinant DNA reagent | pCDNA5/FRT/TO-GFP-p107-R1 | This paper | Inserting a gblock in pCDNA5/FRT/TO-GFP containing 4 copies of R1 | |
| Recombinant DNA reagent | pCDNA5/FRT/TO-GFP-p107-R1-H/AxR/AV/AxxV/A | This paper | Inserting a gblock in pCDNA5/FRT/TO-GFP containing four mutant copies of R1. (gblock sequences provided in Appendix table) | |
| Recombinant DNA reagent | pSG5-puro-Flag-p107 | Kurimchak et al., 2013 | ||
| Recombinant DNA reagent | pCMV-Flag-p107 | Voorhoeve et al., 1999 | ||
| Recombinant DNA reagent | pCMV-HA-E2F4 | Ginsberg et al., 1994 | ||
| Recombinant DNA reagent | pRC-cyclin E | Addgene | #8963 | |
| Software, algorithm | ConSurf | ConSurfAshkenazy et al., 2010 | ||
| Software, algorithm | ImageJ software | |||
| Software, algorithm | FlowJo v10 software | BD Biosciences | v10.8 | |
| Software, algorithm | Clustal Omega | Sievers et al., 2011 |






