A digital 3D reference atlas reveals cellular growth patterns shaping the Arabidopsis ovule
Figures
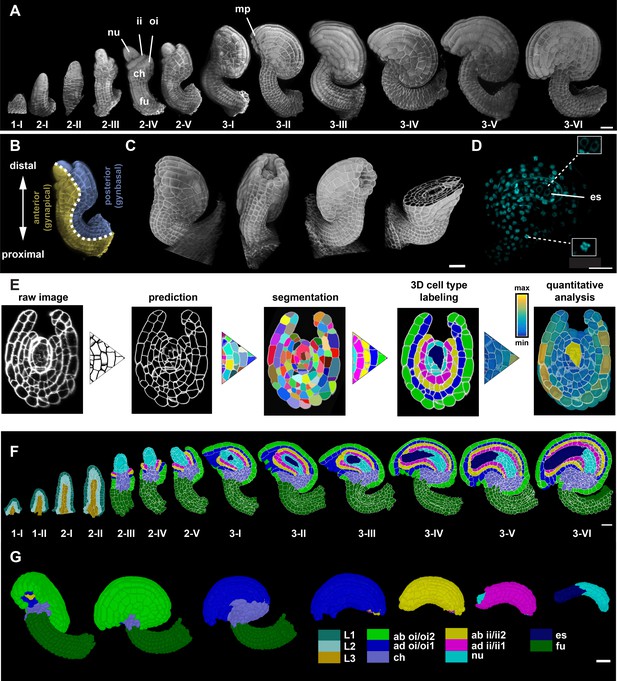
Stage-specific 3D digital ovules with cellular and tissue resolution.
(A) 3D rendering of confocal z-stacks of SR2200-stained cell walls of ovule depicting ovule development from initiation at stage 1-I to maturity at stage 3-VI. (B) The different polarities of the ovule: the proximal-distal axis and the anterior-posterior (gynapical-gynbasal) axis are indicated. (C) 3D rendering of confocal z-stacks with multi-view of an ovule depicting the quality of the raw microscopic image. (D) Mid-section clip plane from the TO-PRO-3 channel displaying a two-nuclear embryo sac and mitotic nuclei. (E) Pipeline generating 3D digital ovules: raw data, PlantSeg cell contour prediction, 3D GASP segmentation, cell type annotation and quantitative analysis. (F) Mid-sagittal section of ovules from stages 1-I to 3-VI showing the cell type organization in wild-type ovules. Stages 1-I to 2-II includes radial L1, L2, L3 labeling. From stage 2-III, individual cell type labels are assigned according to the specific tissue. (G) 3D view of a mature ovule with cell type labels. The inner tissues are extracted from the 3D ovule after removing the overlying tissues and visualized separately. Different colors represent different tissue type labels. ii1/ii2, oi1/oi2 designate the integument layers as described in Beeckman et al., 2000. Number of 3D digital ovules scored: 10 (stages 2-III, 2-IV, 2-V, 3-I, 3-II, 3-IV, 3-VI), 11 (3-III, 3-V), 13 (stage 2-II), 23 (stage 1-I), 49 (stage 2-I), 66 (stage 1-II). Abbreviations: ab, abaxial; ad, adaxial; ch, chalaza; es, embryo sac; fu, funiculus; ii, inner integument; mp, micropyle; nu, nucellus; oi, outer integument. Scale bars: 20 μm.
-
Figure 1—source data 1
Includes information of the available wild-type dataset of ovules at different developmental stages and their respective IDs.
- https://cdn.elifesciences.org/articles/63262/elife-63262-fig1-data1-v1.xlsx
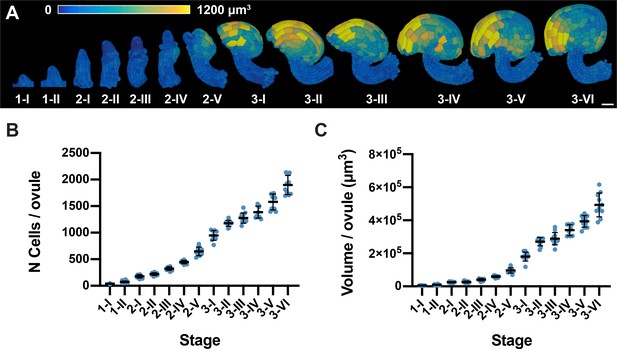
Ovule developmental stages and overall growth patterns.
(A) 3D cell mesh view of wild-type ovules at different stages displaying heatmaps of cell volume ranging from 0 to 1200 µm3. (B, C) Plots depicting the total number of cells and total volume of individual ovules from early to late stages of development, respectively. Number of 3D digital ovules scored: 10 (stages 2-III, 2-IV, 2-V, 3-I, 3-II, 3-IV, 3-VI), 11 (3-III, 3-V), 13 (stage 2-II), 23 (stage 1-I), 49 (stage 2-I), 66 (stage 1-II). Mean ± SD is shown. Scale bar: 20 μm.
-
Figure 2—source data 1
Includes the list of ovule IDs, stage, total number of cells and total volume of the available wild-type dataset.
- https://cdn.elifesciences.org/articles/63262/elife-63262-fig2-data1-v1.xlsx
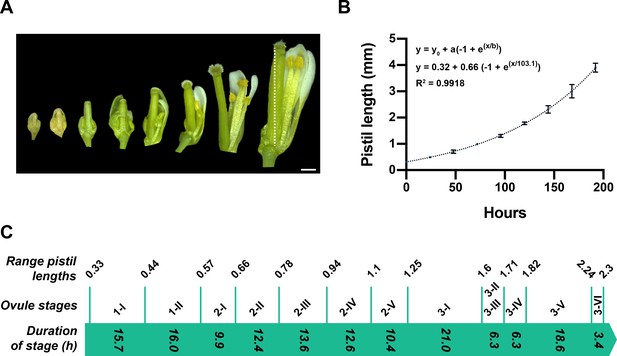
Temporal progression of ovule development.
(A) Micrograph depicting gynoecia (pistils) at different time points (day 0 to day 7). Number of pistils scored: 6. (B) Graph showing the increase in pistil length (mm) over time. Mean ± SD are represented as bars. The fitted curve is indicated by the dotted line. The model and best-fit parameters are indicated. (C) Correlation between pistil length and duration of ovule stages. The values at the top represent the range of pistil lengths spanning the given ovule stage indicated beneath. The respective time values were deduced from the fitted curve in (B). The values within the green arrow indicate the deduced durations of the respective ovule stages. Number of pistils scored: 64. Scale bar: 0.5 mm.
-
Figure 3—source data 1
Includes the information on pistil lengths measured for each stage and the calculated duration in hours of ovule stages.
- https://cdn.elifesciences.org/articles/63262/elife-63262-fig3-data1-v1.xlsx
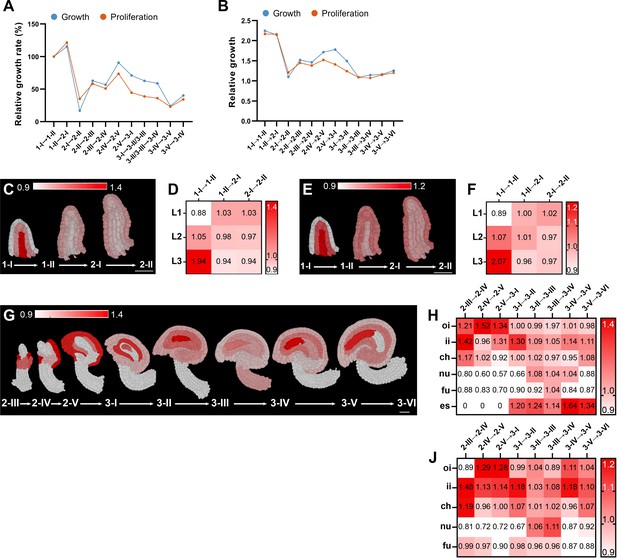
Growth dynamics of Arabidopsis ovule development.
(A, B) Plots indicate the global volume or cell number increases. (A) Plot shows growth rate normalized to the growth rate of the transition from stage 1-I to 1-II. (B) Plot depicts relative growth between two consecutive stages. (C–J) Optical mid-sections and heat maps depicting relative tissue growth across the different ovule stages. Stages are indicated. Heatmap values indicate ratios. (C, D, G, H) Tissue-specific growth rate. (E, F, I, J) Tissue-specific cell proliferation rate. (A,B) Number of 3D digital ovules scored: 10 (stages 2-III, 2-IV, 2-V, 3-I, 3-II, 3-IV, 3-VI), 11 (3-III, 3-V), 13 (stage 2-II), 23 (stage 1-I), 49 (stage 2-I), 66 (stage 1-II). (C,F) 10 (stages 2-III, 2-IV, 2-V, 3-I, 3-II, 3-IV, 3-VI), 11 (stages 2-I, 3-III, 3-V), 13 (stage 2-II), 14 (stage 1-I), 28 (stage 1-II). (G,J) Number of 3D digital ovules scored: 10 (stages 2-III, 2-IV, 2-V, 3-I, 3-II, 3-IV, 3-VI), 11 (3-III, 3-V). Abbreviations: ch, chalaza; es, embryo sac; fu, funiculus; ii, inner integument; nu, nucellus; oi, outer integument.
-
Figure 4—source data 1
Includes the relative growth and proliferation, the relative growth and proliferation rate and the tissues relative growth per each ovule developmental stage.
- https://cdn.elifesciences.org/articles/63262/elife-63262-fig4-data1-v1.xlsx
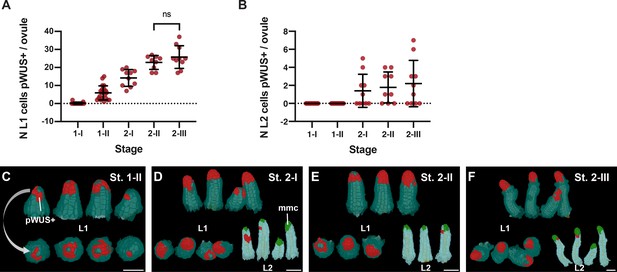
Expression pattern of the pWUS reporter.
(A) Plot showing the number of L1 cells per ovule expressing pWUS across stage 1-I to 2-III. (B) Plot showing the number of L2 cells per ovule expressing pWUS across stages 1-I to 2-III. (C–F) 3D cell meshes displaying L1 and L2 cells expressing pWUS in red from stage 1-II to stage 2-III. Data points indicate individual ovules. Number of 3D pWUS digital ovules scored: 17 (stage 1-I), 21 (stage 1-II), 10 (stage 2-I), 9 (stage 2-II), 10 (stage 2-III). Mean ± SD are represented as bars. Scale bars: 20 μm.
-
Figure 5—source data 1
Includes the list of ovule IDs, stage, total number of cells, number of cells per L1, L2, L3 layer, total number of WUS expressing cells and number of WUS expressing cells per L1, L2, L3 layer of the available pWUS dataset.
- https://cdn.elifesciences.org/articles/63262/elife-63262-fig5-data1-v1.xlsx
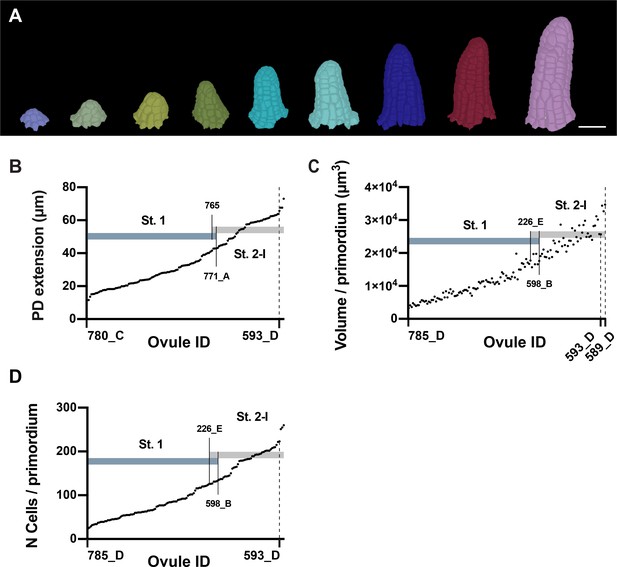
Ovule primordia grow in a continuous fashion.
(A) Developmental series of 3D cell meshes of ovule primordia. (B) Plot showing an ordered array of the PD extension of ovule primordia from early initiation to the end of stage 2-I. (C) Plot indicating the total volume of primordia ordered according to increasing volume. (D) Plot depicting the total number of cells in the ovules ordered according to the increasing number of cells. Number of 3D digital ovules scored: 23 (stage 1-I), 49 (stage 2-I), 66 (stage 1-II). Scale bar: 20 μm.
-
Figure 6—source data 1
Includes the list of ovule IDs, total number of cells, total volume and the PD extension of early stage wild-type ovule.
- https://cdn.elifesciences.org/articles/63262/elife-63262-fig6-data1-v1.xlsx
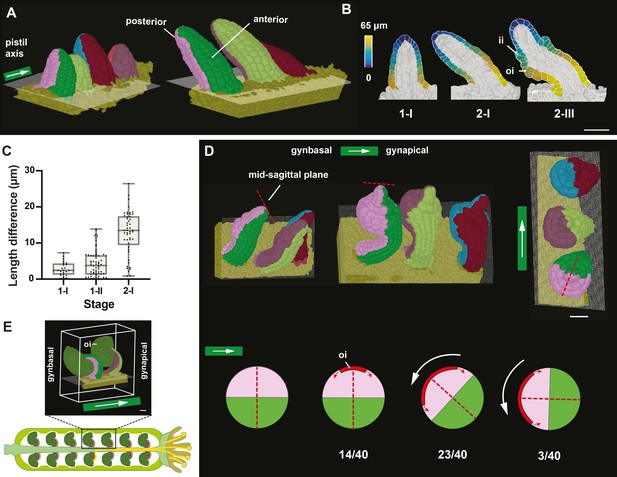
Ovule primordia slanting and early ovule polarity.
(A) 3D meshes with multiple ovules from the same carpel attached to the placenta showing unslanted ovules at stage 1-I and slanted ovules at stage 2-I. The 2D grid represents the surface of the placenta. Color labels depict the posterior and anterior cells, respectively. The pistil axis is indicated. (B) 2D section views of 3D cell meshes from early ovules. Stages are indicated. The heatmap on the surface cells of posterior and anterior halves depicts the quantified distance value between individual measured cells to the distal tip of primordia. (C) Plot depicting the extent of slanting, quantified by the difference in maximal length on the dorsal and ventral sides of ovule at stages 1-I, 1-II, and 2-I. Data points indicate individual ovules. Mean ± SD are represented as bars. (D) Top row: three 3D meshes with multiple ovules attached to the placenta and highlighting the reorientation of the anterior-posterior axis relative to the apical-basal axis of the gynoecium after outer integument initiation. The right-most mesh depicts a top view of the central mesh. The dotted red line indicates the mid-sagittal plane. The white arrows highlight the pistil axis. Bottom row: cartoon depicting transverse sections of stage 2 ovules and summarising the orientations of the anterior and posterior halves of the ovule during the turn along the PD funicular axis. The relative numbers of ovules per degree of turn are given. The dotted red line indicates the mid-sagittal plane. The horizontal white arrow marks the pistil axis. (E) 3D view of mature ovules inside the gynoecium with the micropyle facing the apex (stigma) of the gynoecium. The posterior and anterior sides of the ovules are oriented gynbasally and gynapically, respectively. The white arrow indicates the pistil axis. Number of 3D digital ovules scored for slanting: 23 (stage 1-I), 49 (stage 2-I), 66 (stage 1-II). Number of 3D digital ovules scored for degree of turn: 8 (stage 2-IV), 32 (stage 2-III). Scale bars: 20 μm.
-
Figure 7—source data 1
Includes the list of ovule IDs, the length along the anterior surface, length along the posterior surface and the length difference used for quantifying the slant of early stage wild-type ovule.
The additional excel file indicates the ovule IDs, stage and the orientation along the anterior-posterior axis of early stage wild-type ovule.
- https://cdn.elifesciences.org/articles/63262/elife-63262-fig7-data1-v1.xlsx
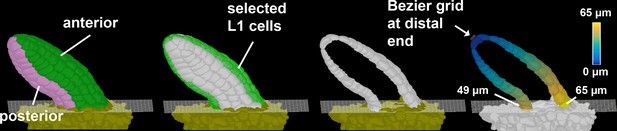
Primordium length and slant measurement.
3D surface view of primordia displaying the method for length measurement. The heatmap on the extracted cells depicts the quantified distance value between individual measured cells to the distal tip of primordia. Scale bars: 20 μm.

Morphologically discernible polarity within the funiculus.
Stage 3-IV wild-type ovule expressing the sieve element marker pPD1::GFP (blue signals) (Bauby et al., 2007). The anterior-posterior polarity in the funiculus is indicated. Note the posterior position of the pPD1::GFP signal. Number of 3D pPD1::GFP digital ovules scored: 8 (stage 3-IV). Scale bar: 20 μm.
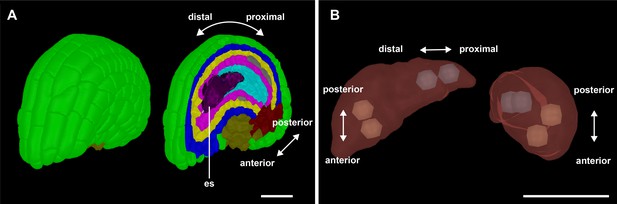
Morphologically discernible polarity in the four-nuclear embryo sac.
(A) Stage 3-IV 3D digital wild-type ovule. The different polarities are indicated. (B) Two views of the embryo sac depicted in (A). The central vacuole is omitted. The two distal nuclei are usually located on top of each other (7/10 embryo sacs scored) and at a right angle to the PD axis. A slight tilt is frequently observed (5/7). The two proximal nuclei always arrange along the PD axis (10/10). Number of 3D embryo sacs scored: 10 (stage 3-IV). Abbreviation: es, embryo sac. Scale bars: 20 μm.
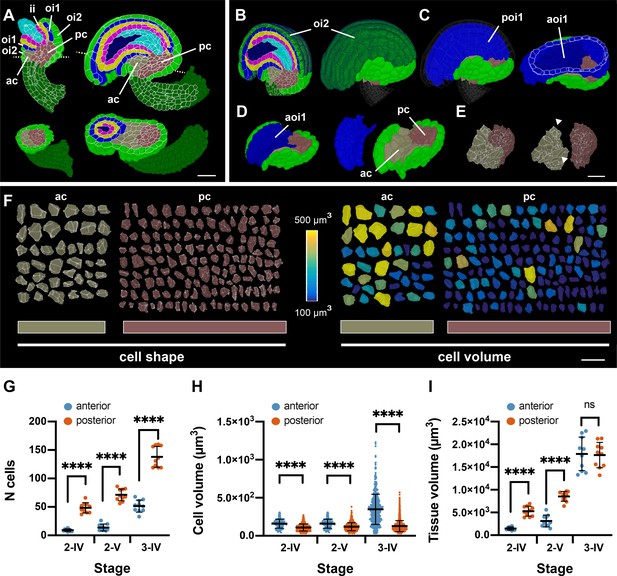
Organization of the proximal chalaza.
(A) Left: sagittal (top) and transverse (bottom) sections through a stage 3D digital wild-type ovule. Tissues are colored as in Figure 1. The anterior and posterior chalaza are highlighted. The dotted lines in the upper ovule indicate the plane of the transverse section shown in the bottom ovule. Right: sagittal (top) and oblique-transverse (bottom) sections through a stage 3-IV 3D digital ovule. The dotted lines in the upper ovule indicate the plane of the oblique-transverse section shown in the bottom ovule. Note how the inner integument and the nucellus are located on top of the posterior chalaza while the anterior chalaza forms a prominent bulge. (B–D) Sequential removal of various tissues of the stage 3-IV ovule shown in (A) to reveal the anterior and posterior chalaza. (B) The dark-green part of the outer integument marks the extent of the oi2 layer flanking the oi1 layer and the inner integument. (C) The highlighted (dark-green) part of the oi2 layer in (B) has been removed and the posterior side of the oi1 layer is visible in blue. In the right ovule, the nucellus and inner integument were removed. A transverse section through the slightly tilted ovule now allows inspection of the anterior oi1 layer (dark blue). (D) The posterior oi1 layer was removed. The remaining anterior oi1 layer covers the anterior chalaza. (E) The anterior oi1 layer and the remaining oi2 layer were detached. The arrowheads highlight the wing-like flaps present in the anterior chalaza. (F) Comparisons of cell shapes and cell volumes of individual cells of the anterior and posterior chalaza, respectively. Cells are from the stage 3-IV ovule depicted in (B). (G–I) Comparison of different parameters between the anterior and posterior chalaza of stage 2-IV, 2-V, and 3-IV ovules. Data points indicate individual ovules. Mean ± SD are represented as bars. Asterisks represent statistical significance (ns, not significant; ****, p<0.0001; unpaired two-tailed t-test). (G) Comparison of cell numbers. (H) Comparison of cell volumes. (I) Comparison of anterior chalaza and posterior chalaza tissue volumes. Number of 3D digital ovules scored for posterior and anterior chalaza analysis: 10 (stages 2-IV, 2-V, 3-IV). Abbreviations: ac, anterior chalaza; aoi1, anterior oi1 layer; ii, inner integument; oi1, adaxial (inner) layer of outer integument; oi2, abaxial (outer) layer of outer integument; pc, posterior chalaza; poi1, posterior oi1 layer. Scale bars: 20 μm.
-
Figure 8—source data 1
Includes the list of ovule IDs, stage, number of cells and tissue volume of posterior and anterior chalaza of wild-type ovule.
- https://cdn.elifesciences.org/articles/63262/elife-63262-fig8-data1-v1.xlsx
Anterior and posterior chalaza in a mature 3D ovule.
Video represents the 3D visualisation of the central region after subsequently removing outer tissues. Different colors represent 3D cells grouped according to respective tissue type labels. To the end, the 3D surfaces of anterior and posterior chalaza are extracted from the 3D ovule and visualised separately.
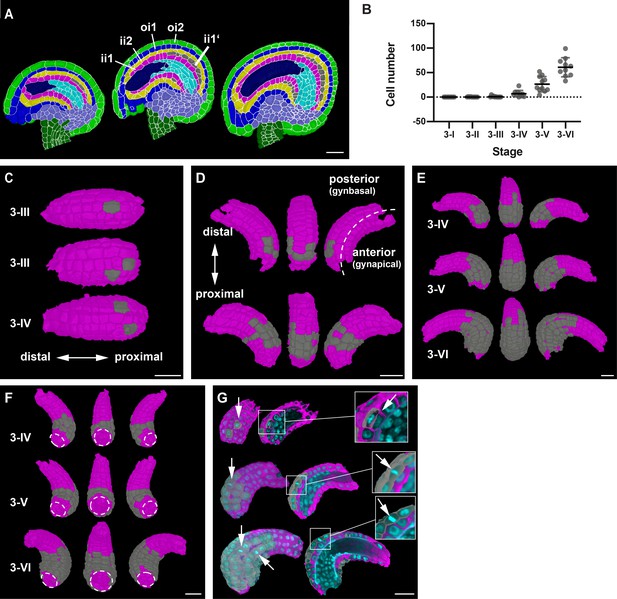
Formation of the parenchymatic inner integument layer (ii1').
(A) Mid-sagittal section of wild-type ovule at stages 3-IV, 3-V, and 3-VI, showing the initiation of a new cell layer (ii1’) in the adaxial inner integument (endothelium/ii1). (B) Plot depicting the number of cells of the developing ii1' layer. Data points indicate individual ovules. Mean ± SD are represented as bars. (C) 3D top surface view of the adaxial inner integument at stages 3-III and 3-IV showing the occurrence of the first few pioneer cells of the ii1' layer. (D) 3D side surface view of adaxial inner integument depicting the pattern of occurrence of ii1' cells. (E) 3D side surface view of adaxial inner integument at later stages of 3-IV, 3-V, and 3-VI where the emergent tissue layer is observed to be a patch of connected cells present only at the proximal region of the inner integument. (F) 3D bottom surface view of ii1' layer at different stages highlighting the formation of a ring-like structure of connected cells covering the proximal half of the inner integument. (G) Section view of the 3D cell meshes of adaxial inner integument with the overlaid nuclei z-stack displaying a periclinal division (top) and anticlinal divisions (center, bottom) in the ii1’ layer. Number of 3D digital ovules scored: 10 (stages 3-I, 3-II, 3-IV, 3-VI), 11 (stages 3-III, 3-V). Abbreviations: ii1, adaxial inner integument (endothelium); ii1’, parenchymatic inner integument layer; ii2, abaxial inner integument; oi1, adaxial outer integument; oi2, abaxial outer integument. Scale bars: 20 μm.
-
Figure 9—source data 1
Includes the list of ovule IDs, stage, number of cells and tissue volume of parenchymatic inner integument of wild-type ovule.
- https://cdn.elifesciences.org/articles/63262/elife-63262-fig9-data1-v1.xlsx
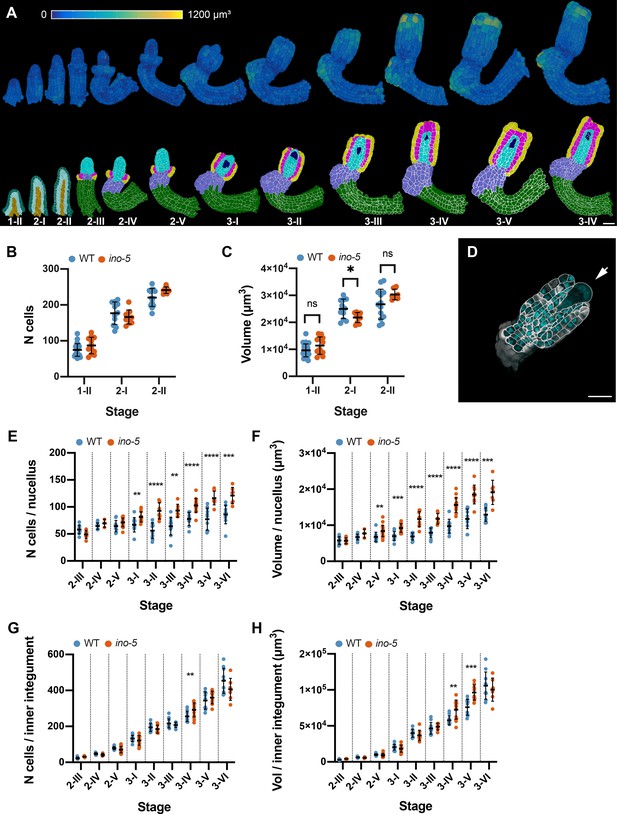
Quantitative analysis of the ino-5 digital ovule atlas.
(A) Surface view of 3D cell meshes showing heatmaps of cell volumes of ino-5 ovules from early to late stage of development. The bottom row shows 3D mid-sagittal section views depicting the cell type organization in ino-5 mutant ovules. (B) Plot showing the total number of cells in wild-type and ino-5 at early stages of ovule development. (C) Plot showing the total volume of wild-type and ino-5 ovules at early stages of ovule development. (D) Section view of the cell boundary z-stack (white) overlayed with the z-stack of stained nuclei showing a four-nuclear embryo sac in an ino-5 ovule. (E, F) Plots comparing total cell number and tissue volume of the nucellus between wild type and ino-5 at different stages. (G, H) Plots comparing total cell number and tissue volume of the inner integument between wild type and ino-5 at different stages. Data points indicate individual ovules. Mean ± SD are represented as bars. Asterisks represent statistical significance (ns, p≥0.5; *, p<0.05; **, p<0.01, ***, p<0.001; ****, p<0.0001; Student’s t-test). Number of 3D digital ino-5 ovules scored: 3 (stage 2-IV), 6 (stage 2-III), 7 (stages 2-II, 3-III, 3-VI), 9 (stages 3-II), 10 (stages 2-I, 3-V), 12 (stages 1-II), 14 (stage 3-I), 15 (stage 3-IV), 20 (stage 2-V). Scale bars: 20 μm.
-
Figure 10—source data 1
Includes the list of ovule IDs, stage, total number of cells and total volume of early stage ino-5 ovules.
The additional sheet includes the list of ovule IDs, stage, number of cells and tissue volume of nucellus and funiculus wild-type and ino-5 ovules.
- https://cdn.elifesciences.org/articles/63262/elife-63262-fig10-data1-v1.xlsx
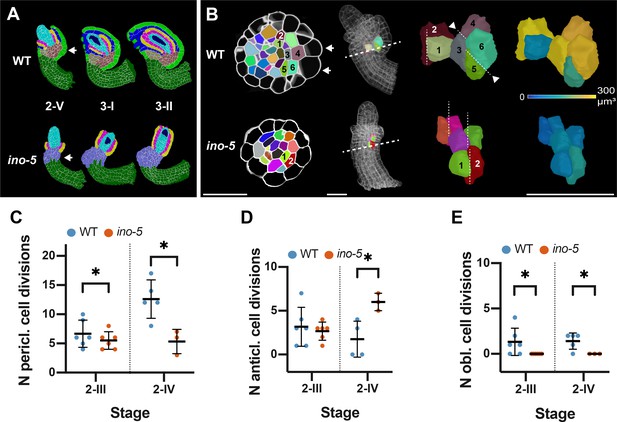
Growth patterns forming the subepidermal central region in wild-type and ino-5 ovules.
(A) Mid-sagittal section view of the cell-type-labeled 3D cell meshes of wild type and ino-5 showing the differences in tissue organization across stages 2-V to 3-II. The arrow indicates the posterior kink in wild type and its absence in ino-5. (B) Top row: Left side: transverse section view depicting the division patterns observed in the chalazal region in wild-type and ino-5 ovules. The two arrows indicate enlarged epidermal cells participating in outer integument formation. The dashed line in the ovule indicates the section plane shown on the left. Right side: Oblique 3D view of the cells numbered in the transverse section view. Dashed lines indicate the cell division plane. Arrowheads highlight oblique periclinal cell division planes. The heat map indicates the cell volumes of the pair of posterior oblique dividing cells resulting in asymmetric enlargement in wild-type and symmetric longitudinal anticlinal dividing cells in ino-5. (C) Plot comparing the number of periclinal divisions in wild-type and ino-5 ovules. (D) Plot comparing the number of longitudinal-anticlinal cell divisions in wild type and ino-5. (E) Plot comparing the number of oblique divisions in wild type and ino-5. Data points indicate individual cell division. Mean ± SD are represented as bars. Asterisks represent statistical significance (ns, p≥0.5; *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001; Student’s t-test). Number of 3D WT digital ovules scored: 6 (stage 2-III), 5 (stage 2-IV). Number of 3D ino-5 digital ovules scored: 3 (stage 2-IV), 6 (stage 2-III). Scale bars: 20 μm.
-
Figure 11—source data 1
Includes the list of ovule IDs, genotype, stage, number of different division planes scored and the volume of the pair of asymmetric enlarged oblique divided cells.
- https://cdn.elifesciences.org/articles/63262/elife-63262-fig11-data1-v1.xlsx
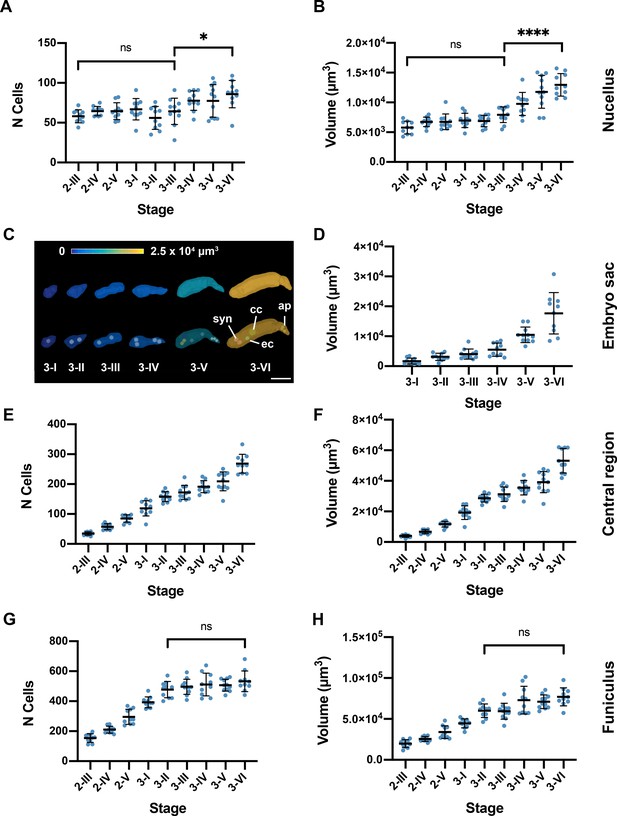
Tissue-specific quantitative analysis.
(A, B) Plots depicting the number of cells and volume at different developmental stages of the nucellus, respectively. (C) 3D mesh of the embryo sac, from stage 3-I to stage 3-IV, extracted from 3D ovule cell meshes. The volume is represented as a heat map. (D) Plot depicting the embryo sac volume from individual ovule datasets at different stages. (E, F) Plots showing the number of cells and volume of the central region, respectively. (G, H) Plots depicting the number of cells and volume of the funiculus, respectively. Data points indicate individual ovules. Mean ± SD are represented as bars. Asterisks represent statistical significance (ns, p≥0.5; *, p<0.05; ****, p<0.0001; Student’s t-test). Number of 3D digital ovules scored: 10 (stages 2-III, 2-IV, 2-V, 3-I, 3-II, 3-IV, 3-VI), 11 (stages 3-III, 3-V). Scale bar: 20 μm.
-
Appendix 1—figure 1—source data 1
Includes the list of ovule IDs, stage, number of cells and tissue volume of nucellus, embryo sac, chalaza and funiculus of wild-type ovule.
- https://cdn.elifesciences.org/articles/63262/elife-63262-app1-fig1-data1-v1.xlsx
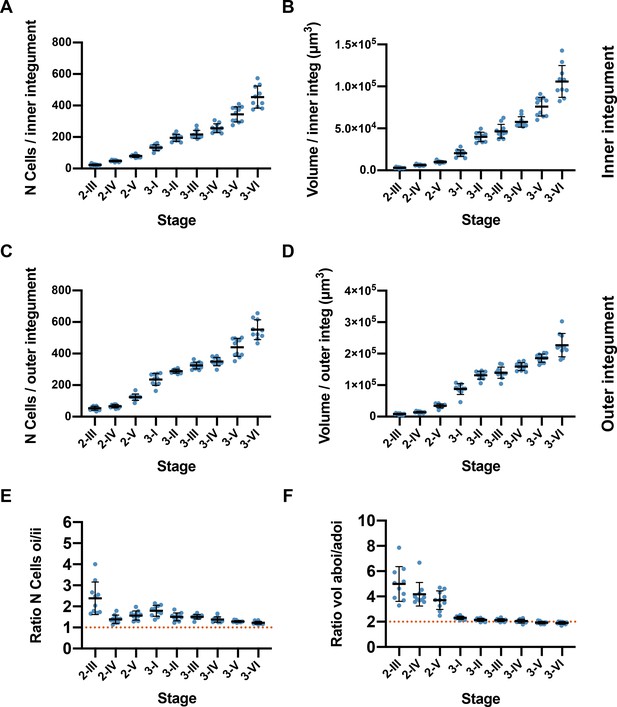
Quantitative analysis of cellular patterns in the integuments.
(A, B) Plots indicating the number of cells and volume of the inner integument from stage 2-III to stage 3-VI. (C, D) Plots indicating the number of cells and volume of the outer integument from stage 2-III to 3-VI stages. (E, F) Plot showing the ratio between the number of cells and tissue volume of the outer and inner integument. Data points indicate individual ovules. Number of 3D digital ovules scored: 10 (stages 2-III, 2-IV, 2-V, 3-I, 3-II, 3-IV, 3-VI), 11 (stages 3-III, 3-V). Mean ± SD are represented as bars.
-
Appendix 1—figure 2—source data 1
Includes the list of ovule IDs, stage, number of cells and tissue volume of outer and inner integument of wild-type ovule.
- https://cdn.elifesciences.org/articles/63262/elife-63262-app1-fig2-data1-v1.xlsx
Videos
Mature 3D digital ovule with cell and tissue resolution.
Video represents the summary of the workflow and multidimensional view of a 3D digital ovule of stage 3-IV generated from a z stack of cell wall images. Different colors represent 3D cells grouped according to respective tissue type labels. To the end, the 3D surfaces of inner tissues are extracted from the 3D ovule after removing the overlying tissues and visualized separately.
Tables
Cell numbers and total volumes of ovules at different stages.
| Stage* | N cells | Volume (x104 μm3) | N mitotic cells | % mitotic cells |
|---|---|---|---|---|
| 1-I | 39.6 ± 5.3 | 0.5 ± 0.09 | 1.0 ± 0.0 | 0.7 ± 1.2 |
| 1-II | 74.0 ± 17.1 | 1.0 ± 0.2 | 1.3 ± 0.5 | 0.7 ± 0.9 |
| 2-I | 176.9 ± 31.5 | 2.5 ± 0.4 | 3.1 ± 2.1 | 1.8 ± 1.2 |
| 2-II | 220.6 ± 24.9 | 2.7 ± 0.6 | 2.7 ± 1.6 | 1.1 ± 0.7 |
| 2-III | 324.1 ± 32.9 | 4.1 ± 0.7 | 3.6 ± 1.7 | 1.0 ± 0.7 |
| 2-IV | 447.1 ± 30.7 | 5.9 ± 0.6 | 4.1 ± 1.7 | 0.9 ± 0.4 |
| 2-V | 648.7 ± 81.5 | 9.7 ± 1.6 | 7.3 ± 3.0 | 1.1 ± 0.5 |
| 3-I | 948.1 ± 92.5 | 18.1 ± 2.7 | 6.4 ± 3.0 | 0.7 ± 0.3 |
| 3-II | 1178.0 ± 58.0 | 27.0 ± 2.5 | 10.4 ± 4.4 | 0.9 ± 0.4 |
| 3-III | 1276.0 ± 97.7 | 28.9 ± 3.8 | 10.7 ± 2.8 | 0.9 ± 0.2 |
| 3-IV | 1387 ± 111.9 | 34.0 ± 3.3 | 5.36 ± 1.8 | 0.4 ± 0.1 |
| 3-V | 1580.0 ± 150.7 | 39.4 ± 3.5 | 7.9 ± 5.3 | 0.5 ± 0.3 |
| 3-VI | 1897.0 ± 179.9 | 49.4 ± 7.2 | 11.1 ± 2.7 | 0.6 ± 0.2 |
-
* Number of 3D digital ovules scored: 10 (stages 2-II- 3-II, 3-IV, 3-VI), 11 (stages 2-I, 3-III, 3-V), 13 (stage 2-II), 14 (stage 1-I), 28 (stage 1-II).
Values represent mean ± SD.
Cell numbers and total volumes of the major ovule tissues.
| Tissue | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stage* | Nucellus | Central region | Inner integument | Outer integument | Funiculus | |||||
| N cells | Volume (x104 μm3) | N cells | Volume (x104 μm3) | N cells | Volume (x104 μm3) | N cells | Volume (x104 μm3) | N cells | Volume (x104 μm3) | |
| 2-III | 58.1 ± 8.1 | 0.6 ± 0.1 | 35.0 ± 5.7 | 0.4 ± 0.08 | 23.5 ± 5.5 | 0.30 ± 0.07 | 53.8 ± 12.1 | 0.8 ± 0.2 | 153.7 ± 28.5 | 2.0 ± 0.5 |
| 2-IV | 64.6 ± 5.9 | 0.7 ± 0.1 | 57.6 ± 9.7 | 0.7 ± 0.01 | 48.1 ± 5.9 | 0.62 ± 0.08 | 66.6 ± 10.6 | 1.4 ± 0.3 | 210.2 ± 23.6 | 2.5 ± 0.3 |
| 2-V | 64.5 ± 10.5 | 0.7 ± 0.1 | 85.0 ± 12.2 | 1.2 ± 0.2 | 79.3 ± 8.4 | 1.0 ± 0.1 | 124.0 ± 20.2 | 3.4 ± 0.6 | 295.9 ± 50.0 | 3.4 ± 0.8 |
| 3-I | 66.9 ± 13.4 | 0.7 ± 0.1 | 118.9 ± 25.4 | 1.9 ± 0.4 | 132.6 ± 18.2 | 2.04 ± 0.4 | 235.9 ± 37.6 | 8.8 ± 1.7 | 392.4 ± 36.4 | 4.5 ± 0.6 |
| 3-II | 56.0 ± 14.4 | 0.7 ± 0.1 | 158.1 ± 16.6 | 2.9 ± 0.3 | 194.7 ± 22.9 | 4.0 ± 0.6 | 289.1 ± 12.6 | 13.2 ± 1.3 | 477.1 ± 54.2 | 6.0 ± 0.8 |
| 3-III | 64.4 ± 16.6 | 0.8 ± 0.1 | 172.2 ± 23.4 | 3.1 ± 0.5 | 216.8 ± 25.4 | 4.7 ± 0.8 | 324.5 ± 21.4 | 13.9 ± 1.8 | 496.2 ± 50.6 | 6.0 ± 1.0 |
| 3-IV | 77.7 ± 12.2 | 1.0 ± 0.2 | 191.3 ± 20.1 | 3.6 ± 0.5 | 255.8 ± 28.4 | 5.8 ± 0.6 | 349.6 ± 26.6 | 15.9 ± 1.3 | 511.1 ± 75.8 | 7.3 ± 1.7 |
| 3-V | 77.4 ± 20.4 | 1.2 ± 0.3 | 209.2 ± 31.7 | 3.9 ± 0.7 | 343.5 ± 48.3 | 7.6 ± 1.1 | 439.8 ± 57.0 | 18.6 ± 1.3 | 506.8 ± 40.1 | 7.1 ± 0.8 |
| 3-VI | 85.9 ± 17.2 | 1.3 ± 0.2 | 268.5 ± 31.4 | 5.3 ± 0.8 | 453.9 ± 69.8 | 10.6 ± 1.9 | 551.6 ± 62.7 | 22.7 ± 3.7 | 533.0 ± 68.6 | 7.7 ± 1.1 |
-
*Number of 3D digital ovules scored: 10 (stages 2-II- 3-II, 3-IV, 3-VI), 11 (stages 3-III, 3-V).
Values represent mean ± SD.
Cellular parameters of the proximal chalaza.
| Tissue | ||||||
|---|---|---|---|---|---|---|
| Stage* | Anterior proximal chalaza | Posterior proximal chalaza | ||||
| N cells | Cell volume (μm3) | Tissue volume (x104 μm3) | N cells | Cell volume (μm3) | Tissue volume (x104 μm3) | |
| 2-III | - | - | - | 35 ± 5 | 113 ± 41.2 | 0.4 ± 0.08 |
| 2-IV | 9 ± 2 | 160 ± 59.6 | 0.14 ± 0.03 | 48 ± 9 | 109 ± 45.6 | 0.52 ± 0.1 |
| 2-V | 13 ± 6 | 226.4 ± 77.3 | 0.31 ± 0.1 | 71.2 ± 10 | 120 ± 52.7 | 0.85 ± 0.1 |
| 3-I | 32 ± 8 | 270.4 ± 121.3 | 0.87 ± 0.2 | 86 ± 20 | 120 ± 59.5 | 1 ± 0.2 |
| 3-II | 50 ± 8 | 298 ± 156 | 1.5 ± 0.2 | 108 ± 19 | 127 ± 70 | 1.3 ± 0.2 |
| 3-III | 51 ± 13 | 318.1 ± 192 | 1.6 ± 0.35 | 121 ± 32 | 124 ± 68 | 1.5 ± 0.3 |
| 3-IV | 52 ± 11 | 347.5 ± 198.2 | 1.8 ± 0.36 | 138 ± 18 | 128 ± 73 | 1.7 ± 0.2 |
| 3-V | 60 ± 14 | 336.8 ± 227.4 | 2 ± 0.4 | 148 ± 27 | 126 ± 70.5 | 1.8 ± 0.4 |
| 3-VI | 59 ± 6 | 428.7 ± 284.4 | 2.5 ± 0.4 | 209 ± 31 | 132 ± 90.7 | 2.7 ± 0.4 |
-
* Number of 3D digital ovules scored: 10 (stages 2-II- 3-II, 3-IV, 3-VI), 11 (stages 3-III, 3-V).
Values represent mean ± SD.
Cell numbers and total volumes of different tissues in ino-5.
| Tissue | ||||||||
|---|---|---|---|---|---|---|---|---|
| Stage* | Nucellus | Inner integument | Funiculus | Central region | ||||
| N cells | Volume (x104 μm3) | N cells | Volume (x104 μm3) | N cells | Volume (x104 μm3) | N cells | Volume (x104 μm3) | |
| 2-III | 49 ± 7.6 | 0.5 ± 0.08 | 31.3 ± 3.2 | 0.4 ± 0.03 | 189.0 ± 29 | 2.4 ± 0.4 | 35.8 ± 37.1 | 0.3 ± 0.3 |
| 2-IV | 69.7 ± 7.5 | 0.7 ± 0.1 | 41.6 ± 8.3 | 0.5 ± 0.1 | 202.7 ± 25 | 2.2 ± 0.4 | 68.3 ± 6.6 | 0.6 ± 0.2 |
| 2-V | 71.4 ± 8.1 | 0.8 ± 0.1 | 70.7 ± 17.7 | 0.9 ± 0.2 | 286.4 ± 35.2 | 3.4 ± 0.5 | 108.1 ± 19.3 | 1.2 ± 0.3 |
| 3-I | 81.4 ± 9.5 | 0.9 ± 0.1 | 122.0 ± 27.3 | 1.8 ± 0.5 | 379.0 ± 29.7 | 4.7 ± 0.5 | 134.0 ± 23.1 | 1.8 ± 0.4 |
| 3-II | 92.6 ± 15.0 | 1.1 ± 0.2 | 185.1 ± 21.9 | 3.9 ± 0.7 | 402.9 ± 32.6 | 5.1 ± 0.7 | 176.9 ± 24.6 | 2.6 ± 0.4 |
| 3-III | 93.0 ± 12.0 | 1.1 ± 0.1 | 208.4 ± 14.6 | 4.8 ± 0.5 | 451.6 ± 32.7 | 5.9 ± 0.7 | 202.9 ± 24.1 | 3.2 ± 0.5 |
| 3-IV | 102.7 ± 12.8 | 1.5 ± 0.2 | 296.9 ± 43.5 | 7.3 ± 1.3 | 481.3 ± 45.7 | 6.7 ± 0.5 | 215.4 ± 30.7 | 3.6 ± 0.7 |
| 3-V | 116.3 ± 12.5 | 1.8 ± 0.2 | 360.8 ± 35.2 | 9.6 ± 1.0 | 522.6 ± 47.4 | 7.2 ± 1.1 | 239.6 ± 31.2 | 4.2 ± 0.7 |
| 3-VI | 121.1 ± 15.2 | 1.9 ± 0.3 | 406.1 ± 61.3 | 10.0 ± 1.6 | 599.6 ± 40.6 | 8.8 ± 0.8 | 254.4 ± 48.5 | 4.5 ± 0.9 |
-
* Number of 3D digital ovules scored: 3 (stage 2-IV), 6 (stage 2-III), 7 (stages 3-III, 3-VI), 9 (stage 3-II), 10 (stage 3-V), 14 (stages 3-I, 3-IV), 20 (stage 2-V ). Values represent mean ± SD.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Arabidopsis thaliana) | ino-5 CRISPR mutant | This paper | In Col-0 background | |
| Genetic reagent (Arabidopsis thaliana) | pWUS::2xVenus:NLS | Zhao et al., 2017 | In Col-0 background | |
| Genetic reagent (Arabidopsis thaliana) | pPD1::GFP | Bauby et al., 2007 | In Col-0 background | |
| Recombinant DNA reagent | sgRNA | This paper | CRISPR/Cas9 system sgRNA | ACCATCTATTTGATCTGCCG |
| Chemical compound, drug | SR2200 | Renaissance Chemicals Musielak et al., 2015 | Cell wall stain | |
| Chemical compound, drug | TO-PRO-3 iodide | Thermo Fisher | T3605 | Nuclear stain |
| Chemical compound, drug | Vectashield | Vectashield Laboratories | VEC-H-1000 | Antifade agent |
| Software, algorithm | MorphographX | https://morphographx.org/ | Open source software for visualization and analysis of biological datasets | |
| Software, algorithm | PlantSeg | Wolny et al., 2020 | https://github.com/hci-unihd/plant-seg | Machine learning-based 3D image segmentation pipeline |
| Software, algorithm | GraphPad | https://www.graphpad.com/ | Statistical analysis software | |
| Software, algorithm | Adobe cc | https://www.adobe.com/ | Ai, Ps, Pr | Video and image rendering |
Additional files
-
Source code 1
Yaml file with PlantSeg parameters used in this study.
- https://cdn.elifesciences.org/articles/63262/elife-63262-code1-v1.zip
-
Supplementary file 1
Standardized cell type labels.
- https://cdn.elifesciences.org/articles/63262/elife-63262-supp1-v1.docx
-
Supplementary file 2
Pistil length of individual live pistils.
- https://cdn.elifesciences.org/articles/63262/elife-63262-supp2-v1.docx
-
Supplementary file 3
Correlation of pistil length with ovule stages.
- https://cdn.elifesciences.org/articles/63262/elife-63262-supp3-v1.docx
-
Supplementary file 4
Cell numbers and total volume of ino-5.
- https://cdn.elifesciences.org/articles/63262/elife-63262-supp4-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63262/elife-63262-transrepform-v1.docx
-
Appendix 1—figure 1—source data 1
Includes the list of ovule IDs, stage, number of cells and tissue volume of nucellus, embryo sac, chalaza and funiculus of wild-type ovule.
- https://cdn.elifesciences.org/articles/63262/elife-63262-app1-fig1-data1-v1.xlsx
-
Appendix 1—figure 2—source data 1
Includes the list of ovule IDs, stage, number of cells and tissue volume of outer and inner integument of wild-type ovule.
- https://cdn.elifesciences.org/articles/63262/elife-63262-app1-fig2-data1-v1.xlsx





