Torsin ATPases influence chromatin interaction of the Torsin regulator LAP1
Figures
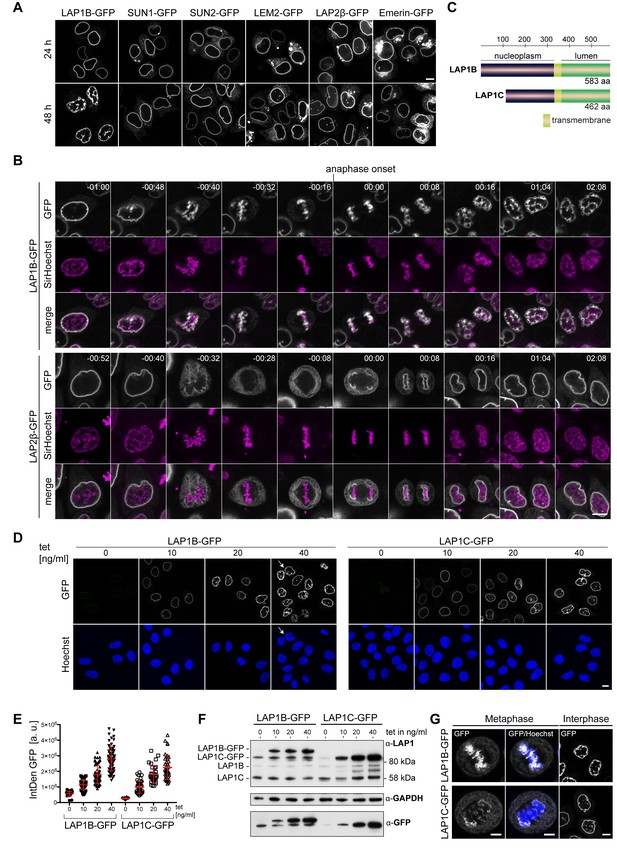
Overexpression of LAP1B and LAP1C causes post-mitotic nuclear envelope (NE) aberrations.
(A) Vectors encoding the indicated inner nuclear membrane (INM) proteins were transfected into HeLa cells. Cells were fixed after 24 or 48 hr and analyzed by confocal microscopy. Scale bar, 10 μm. (B) Time-lapse images of LAP1B-GFP or LAP2β-GFP expressing HeLa cells progressing through mitosis. Expression of the constructs was induced 24 hr prior to imaging. DNA was visualized by SirHoechst and used to define anaphase onset (t = 00:00 hr:min). Scale bar, 5 μm. (C) Scheme depicting the two human LAP1 isoforms, LAP1B and LAP1C. (D) Representative images of stable HeLa cells lines expressing LAP1B-GFP and LAP1C-GFP after induction with different tetracycline (tet) concentrations for 48 hr. Please note that we observed some ‘twin’ nuclei upon overexpression of LAP1B (white arrow), which can be a sign of binucleation, as further analyzed in Figure 5. Scale bar, 10 μm. (E) LAP1 levels at the NE of cells from the experiment shown in panel D were analyzed by quantification of the integrated GFP density (IntDen GFP) per nucleus. (F) LAP1 levels in cell lysates derived from the experiment shown in panel D were analyzed by immunoblotting. Note that the LAP1B-GFP cell line expressed LAP1C-GFP independent of tetracycline addition, due to an alternative transcriptional start site used for the production of the shorter LAP1 isoform (Santos et al., 2014). (G) Left: Maximum intensity z-projections (5 × 0.63 μm) of confocal images from fixed metaphase HeLa cells expressing LAP1B-GFP or LAP1C-GFP after 48 hr of induction. Scale bar, 5 μm. Right: Confocal images of fixed HeLa cells in interphase. Scale bar, 10 μm.
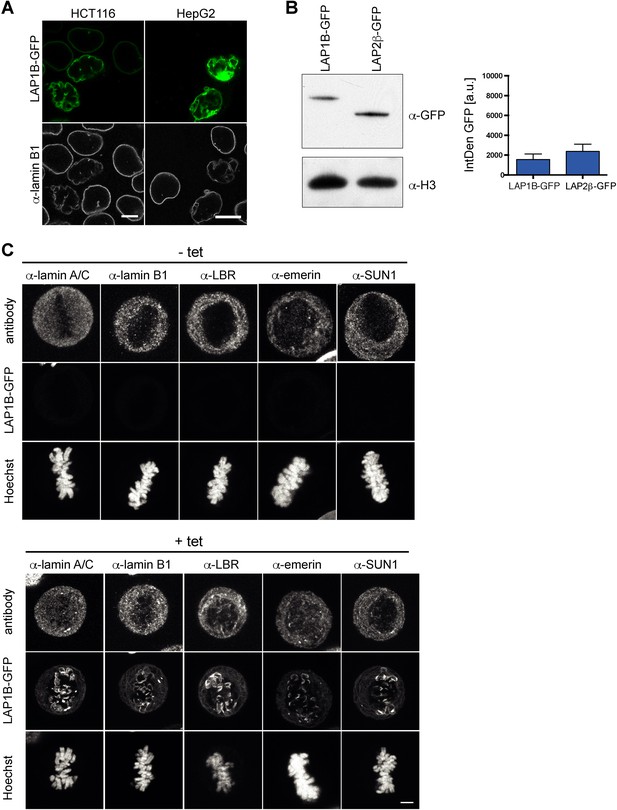
Interphase and mitotic localization of LAP1B-GFP and endogenous nuclear envelope (NE) proteins.
(A) Vector encoding LAP1B-GFP was transiently transfected into HCT116 or HepG2 cells. Cells were fixed after 48 hr and analyzed by confocal microscopy. Scale bars, 10 μm. (B) Western blot analysis (left) and integrated GFP fluorescence intensities (right) in LAP1B-GFP and LAP2β-GFP-expressing cells of the experiment depicted in Figure 1B. (C) Comparison of the metaphase localization of nuclear lamins and select inner nuclear membrane (INM) proteins in an inducible LAP1B-GFP cell line without and with tetracycline (tet) induction. Maximum intensity projections (5 × 0.63 μm). Scale bar, 5 μm.
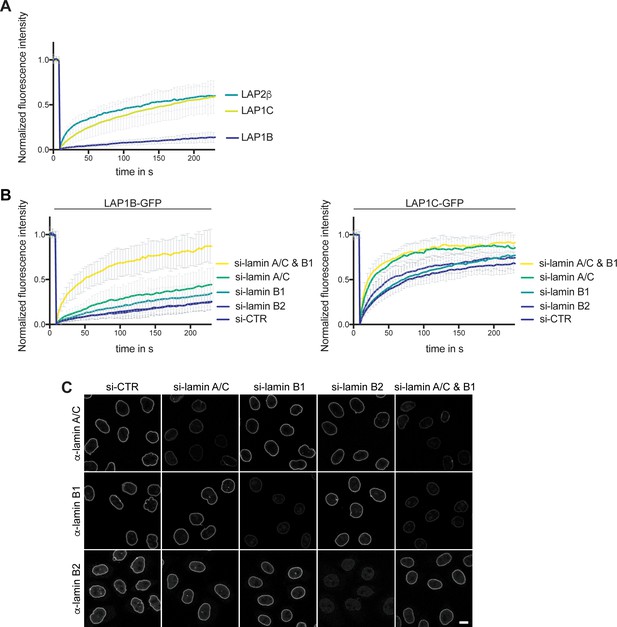
Lamin A/C and lamin B1 contribute to the retention of LAP1B at the nuclear envelope (NE).
(A) FRAP analysis of LAP1B-GFP, LAP1C-GFP and LAP2β-GFP at the NE (N = 5, n > 18, mean +/- SD). (B) Diffusional mobility of LAP1B-GFP (N = 5, n > 26, mean +/- SD) and LAP1C-GFP (N = 4, n > 21, mean +/- SD) after RNAi-mediated depletion of lamins analyzed by FRAP (N = 5, n > 21, mean +/- SD). (C) Depletion of lamins by RNAi was efficient, as shown by immunofluorescence analysis of lamin A/C, lamin B1 and lamin B2. Scale bar, 10 μm.
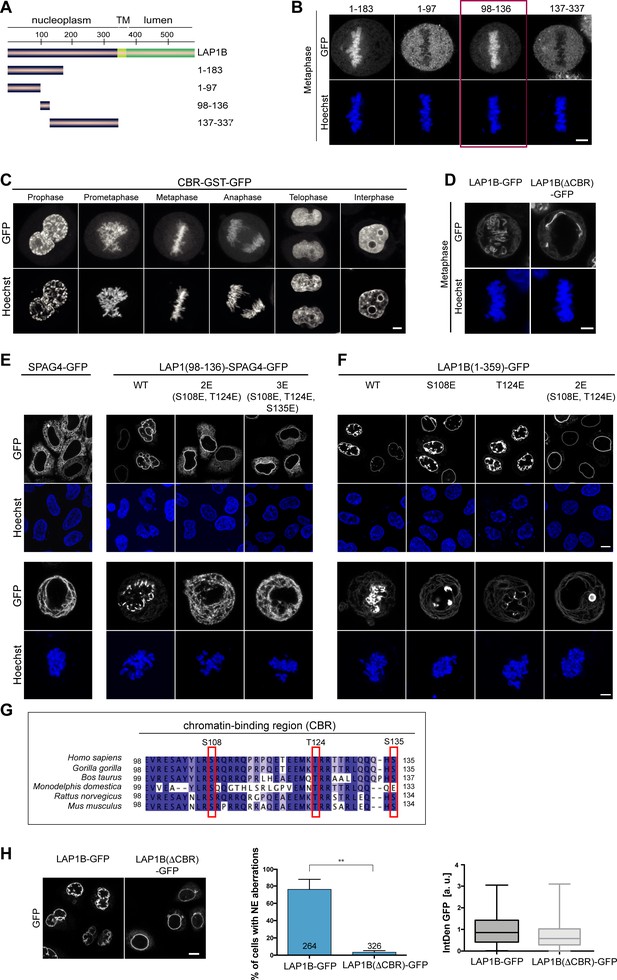
The nucleoplasmic domain of LAP1 contains a central chromatin-binding region (CBR) that can confer chromatin association during mitosis.
(A) Scheme depicting the generated fragments of the nucleoplasmic domain of LAP1B. (B) Metaphase localization of the depicted LAP1 fragments, transiently expressed in synchronized HeLa cells. Maximum intensity z-projections (3 × 1 μm). Scale bar, 5 μm. (C) Localization of LAP1(CBR)-GST-GFP during mitosis in HeLa cells. Scale bar, 5 μm. (D) LAP1B-GFP and LAP1B(ΔCBR)-GFP localization during metaphase. Scale bar, 5 μm. (E) Localization of wild-type SPAG4-GFP and (F) LAP1B(98-136)-SPAG4-GFP derivatives in interphase and prometaphase cells (arrested with nocodazole for 3 hr) 48 hr after transfection. Scale bars, 10 μm (upper panels) and 5 μm (lower panels). (G) Alignment of sequences of the CBR of LAP1B from the indicated mammalian species. The three mutated residues are boxed in red. (H) Left: Interphase HeLa cells expressing LAP1B-GFP or LAP1B(ΔCBR)-GFP for 48 hr. Scale bar, 10 μm. Middle and right: Percentage of nuclei with nuclear envelope (NE) aberrations, quantified as in Figure 3, in cells expressing LAP1B-GFP or LAP1B(ΔCBR)-GFP at similar levels, based on the integrated GFP density (IntDen GFP) (N = 3, n > 264, mean +/- SEM).
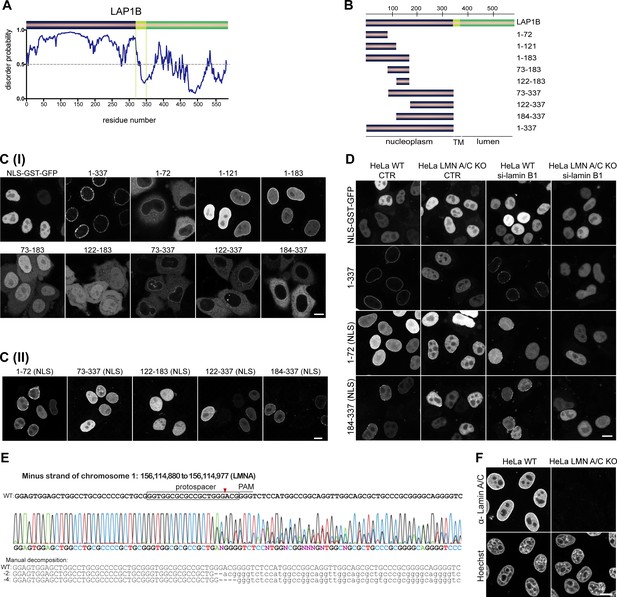
Delineation of the lamina-binding regions in LAP1B.
(A) Scheme illustrating the disorder probability of LAP1. (B) LAP1B fragments used to delineate lamina-binding regions in the nucleoplasmic domain of LAP1B. (C) Localization of the indicated LAP1B derivatives fused to GST-GFP (I) or NLS-GST-GFP (II) in interphase HeLa cells. An NLS-GST-GFP fusion served as control. Note that enrichment of protein fragments at the nuclear rim reflects lamina association, as confirmed in panel D. Scale bars, 10 μm. (D) Localization of LAP1 fragments fused to GST-GFP-NLS in either wild-type or LMN A/C KO HeLa cells, with or without downregulation of lamin B1. Note that nuclear envelope (NE) localization of LAP1(1–337) is strongly affected in LMN A/C KO cells, whereas the N-terminal fragment LAP1(1–72) is more strongly impaired in NE localization upon downregulation of lamin B1. Scale bar, 10 μm. (E) Characterization of LMN A/C knockout HeLa cells generated by CRISPR/Cas9 by DNA sequencing. (F) Analysis of LMN A/C knockout HeLa cells by immunofluorescence against lamin A/C. Scale bar, 10 μm.
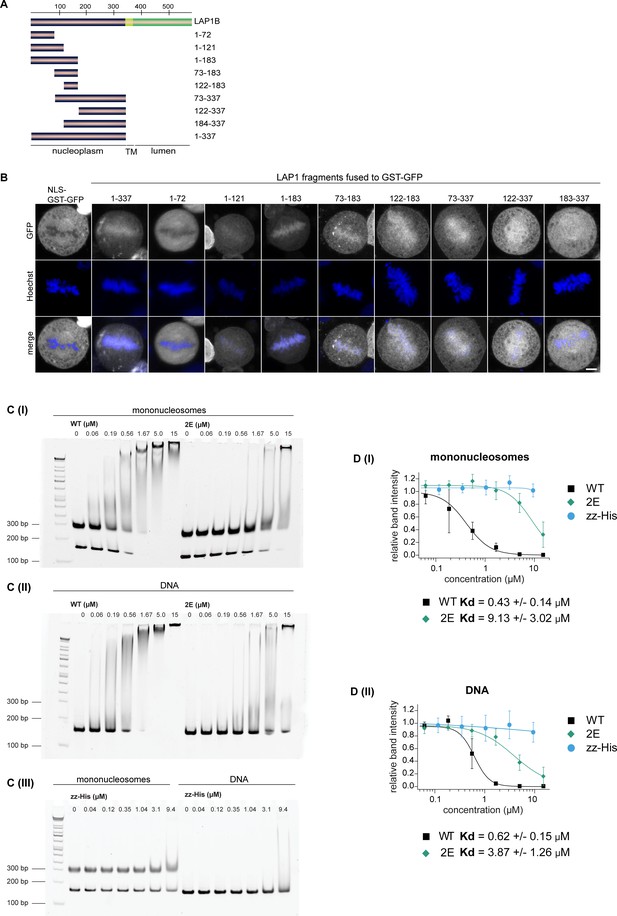
Delineation of the chromatin-binding region (CBR) of LAP1B.
(A) LAP1B fragments used to map the minimal CBR in the nucleoplasmic domain of LAP1. (B) Localization of the indicated GST-GFP fusion proteins in synchronized HeLa cells during metaphase. DNA was stained with Hoechst. Maximum intensity projections (5 × 0.63 μm). Scale bar, 5 μm. (C) Electrophoretic mobility shift assays used for quantitative analysis of DNA and nucleosome binding of the isolated recombinant CBR of LAP1B. Increasing concentrations of zz-LAP1(98–136), zz-LAP1(98–136; S108E, T124E (‘2E’)) (I and II) or zz-His (III) were incubated with either ‘601’ DNA or reconstituted mononucleosomes and subjected to native gel electrophoresis. DNA was stained with GelRed. (D) Quantification of experiments in C based on integration of gel-band intensities from three independent experiments (N = 3), (I) for binding to mononucleosomes, (II) to DNA. Normalized band intensities (mean +/- SD) were plotted over the protein concentration and Kd values calculated using the Hill equation.
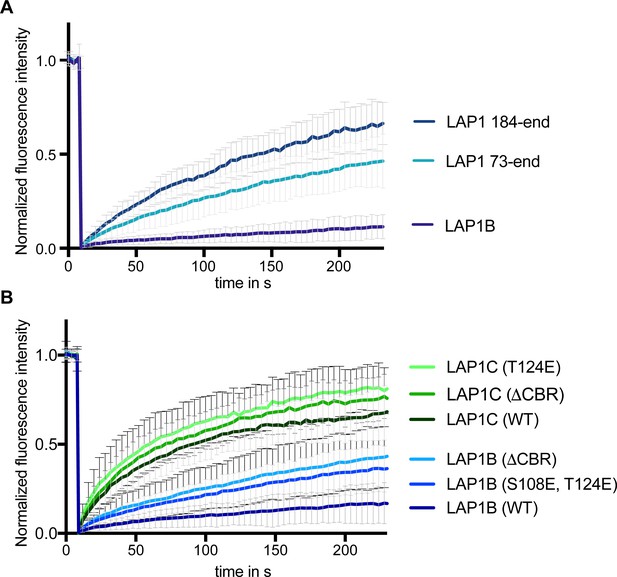
The chromatin-binding region (CBR) of LAP1 contributes to its retention at the inner nuclear membrane.
(A) FRAP analysis of LAP1B-GFP wild-type in comparison to truncation constructs lacking the N-terminal lamin-binding domain (LAP1(73-end)) or an extended part including the CBR (LAP1(184-end)); (N = 3, n > 20, mean +/- SD). Scale bar, 5 µm. (B) FRAP analysis of LAP1B and LAP1C-GFP and the indicated mutants of the CBR (N = 3, n > 14, mean +/- SD).
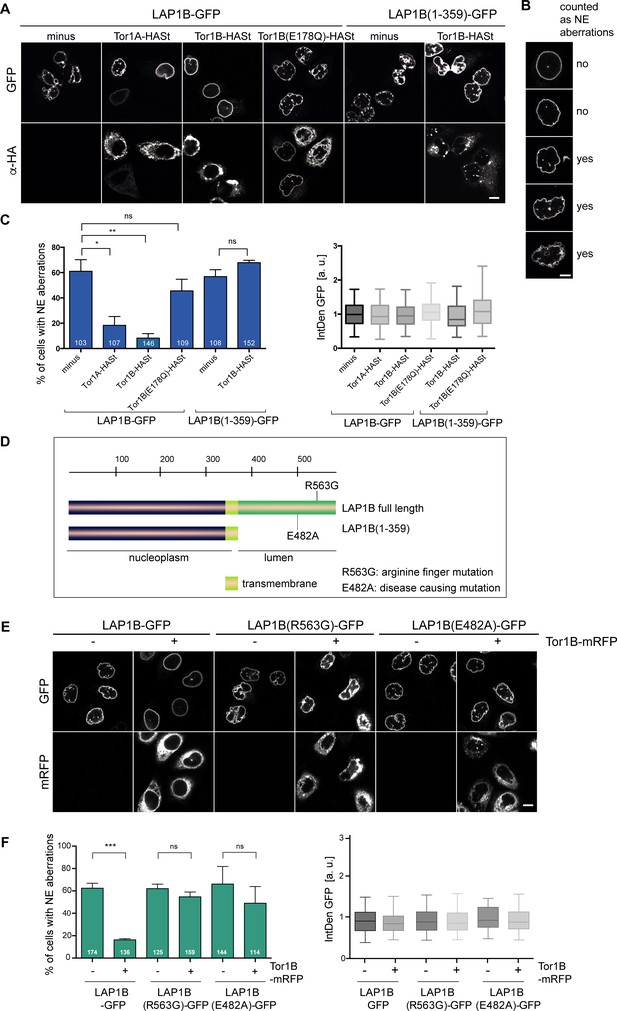
LAP1B-induced nuclear envelope (NE) aberrations can be rescued by co-expression of Torsins.
(A) HeLa cells were transiently transfected with constructs encoding for LAP1B-GFP or LAP1B(1-359), either alone or together with the depicted Torsin constructs. After 48 hr, cells were fixed, subjected to immunostaining of Torsins using an anti-HA antibody and analyzed by confocal microscopy. Scale bar, 10 μm. (B) Representative images of pre-defined classes of cell nuclei used for the classification of NE aberration phenotypes. Based on nuclear shape, cells were assigned into one of the five shown categories for the quantification in C. The two upper classes were regarded as normal (no; no NE aberration), and the three lower classes as aberrant (yes). Scale bar, 5 μm. (C) The percentage of cells with NE aberrations was quantified by assigning cells into one of five predefined phenotypic classes shown in B. (left; N = 3, n > 103, mean +/- SEM). Only cells with similar LAP1B-GFP expression levels, based on quantification of the integrated GFP density per cell (IntDen GFP, normalized to the LAP1B minus Torsin control; right) were considered. (D) Depiction of the mutant LAP1B constructs; R563G: Torsin activation deficient-mutant; E482A, disease-causing mutant. (E) HeLa cells were transiently transfected with constructs encoding for the indicated LAP1B-GFP derivatives, either alone or together with Tor1B-mRFP. After 48 hr, cells were fixed and analyzed by confocal microscopy. Scale bar, 10 μm. (F) The fraction of cells with NE aberrations was quantified as in C (N = 3, n > 114, mean +/- SEM).
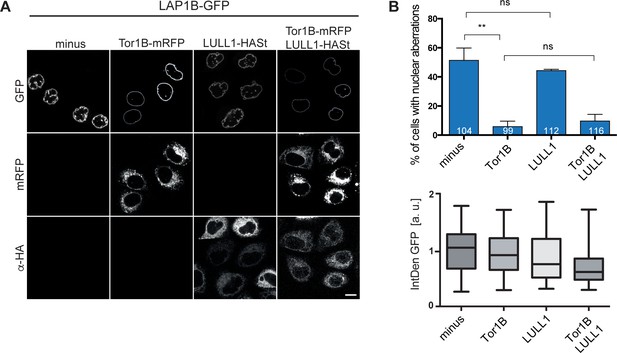
Nuclear envelope (NE) aberrations induced by LAP1B cannot be rescued by LULL1 co-expression.
(A) HeLa cells were transfected with constructs encoding for LAP1B-GFP, either alone or together with LULL1-HASt and Tor1B-mRFP expression vectors. Cell were fixed 48 hr after transfection and nuclear morphology analyzed by confocal microscopy. Scale bar, 10 μm. (B) Quantification of NE aberrations in cells with similar LAP1B-GFP expression levels as in Figure 3 (N = 3, n > 99, mean +/- SEM).
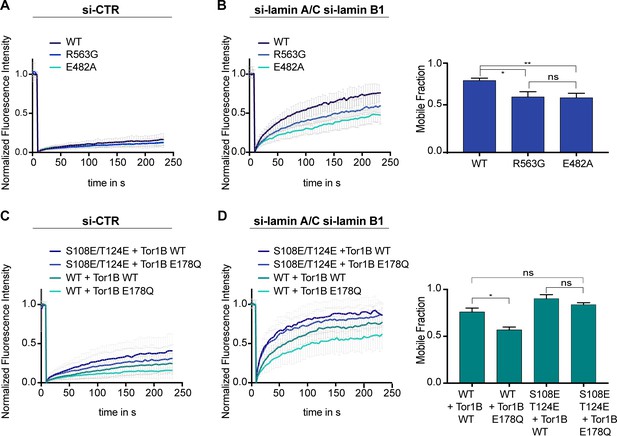
Mobility of LAP1B at the nuclear envelope (NE) of interphase cells is influenced by Torsins.
(A) FRAP analysis of HeLa cells expressing either wildtype LAP1B-GFP or the indicated mutant variants and treated with a control siRNA (N = 5, n > 18, mean +/- SD). (B) FRAP analysis as in B after RNAi-mediated co-depletion lamin A/C and lamin B1 (N = 5, n > 21, mean +/- SD). Corresponding mobile fractions (mean +/- SEM, *p<0.05). (C) FRAP analysis of LAP1B-GFP and the chromatin-binding deficient variants in cells expressing either Tor1B wild-type or the dominant-negative Tor1B(E178Q) mutant both tagged with mRFP, and treated with a control siRNA (N = 4, n > 18, mean +/- SD). (D) FRAP analysis in lamin-depleted cells as in C (N = 4, n > 25, mean +/- SD). Corresponding mobile fractions (mean +/-, *p<0.05).
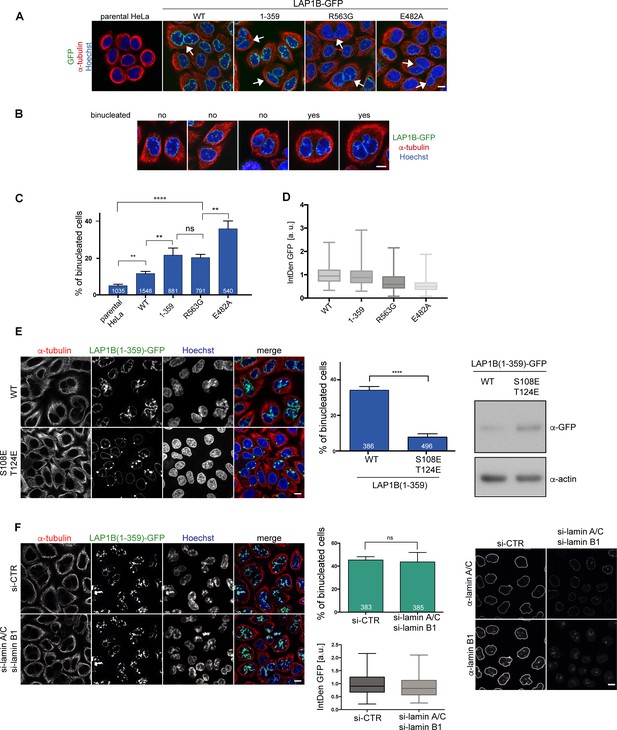
LAP1B mutants deficient in Torsin activation lead to increased binucleation dependent on chromatin interaction of LAP1.
(A) Expression of LAP1B(1-359)-GFP was induced with 0.01 μg/ml tetracycline and expression of LAP1B-GFP wild-type, LAP1(R563G)-GFP and LAP1(E482A)-GFP with 1 μg/ml tetracycline for 48 hr. The parental HeLa cell line was used as a negative control. Then, cells were fixed, immunostained for tubulin and analyzed by confocal microscopy. Scale bar, 10 μm. White arrows denote binucleated cells. (B) Images representing the categories used for classification of binucleation. (C) Quantification of the fraction of binucleated cells (N = 3, n > 540, mean +/- SEM). (D) The integrated density (IntDen) of the GFP signal was measured and normalized to LAP1B-GFP. (E) Binucleation caused by LAP1B is prevented by point mutations that abolish chromatin-binding. Expression of LAP1B(1-359)-GFP or LAP1B(1-359-2E)-GFP was induced with tetracycline (0.01 μg/ml and 1 μg/ml, respectively) in stable HeLa cell lines for 48 hr. Cells were immunostained for tubulin and counterstained with Hoechst. Binucleated cells were quantified (N = 3, n > 300, mean +/- SEM; ****p<0.0001). Expression levels of LAP1B(1-359)-GFP and LAP1B(1-359-2E)-GFP were compared by immunoblotting. (F) LAP1B(1-359)-GFP expression was induced for 48 hr as in E in either mock-treated or in lamin A/C and lamin B1-co-depleted cells (RNAi for 72 hr, induction of LAP1B constructs for the last 48 hr). Cells fixed and analyzed as in E. The number of binucleated cells was quantified (N = 3, n > 383, mean +/- SEM). LAP1B(1-359)-GFP expression levels were determined based on the integrated GFP intensity (IntDen). Lamin RNAi was controlled by immunofluorescence. Scale bars, 10 μm.
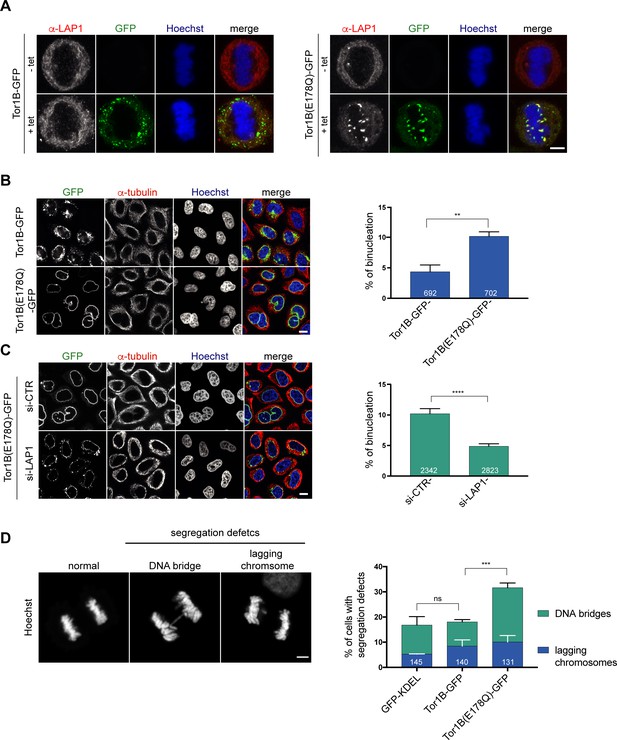
A dominant-negative, ATPase-deficient Torsin1B mutant increases binucleation in a LAP1-dependent manner.
(A) Endogenous LAP1 remains associated with chromatin in metaphase upon overexpression of Tor1B(E178Q)-GFP. Representative maximum intensity projections (5 × 0.63 μm) of confocal images of metaphase HeLa cells upon induction of either Tor1B-GFP and Tor1B(E178Q)-GFP expression with 1 μg/ μl tetracycline for 48 hr. Localization of endogenous LAP1 was analyzed by immunofluorescence staining. Scale bar, 5 μm. (B) Confocal images of Tor1B-GFP and Tor1B(E178Q)-GFP-expressing cells stained with Hoechst and immunostained for tubulin. Scale bar, 10 μm. Quantification of binucleated cells (N = 3, n > 835, mean +/- SD). (C) Expression of Tor1B(E178Q)-GFP was induced for 48 hr in LAP1-depleted or mock-treated cells. Binucleation was analyzed by confocal microscopy and quantified (N = 3, n > 1915, mean+/-SEM). Scale bar, 10 μm. (D) Quantification of chromosome segregation defects (DNA bridges and lagging chromosomes) in fixed GFP-KDEL, Tor1B-GFP, and Tor1B(E178Q)-GFP expressing anaphase cells after 24 hr of induction with tetracycline. Segregation errors were manually quantified (N = 3, n > 130, mean+/-SEM).
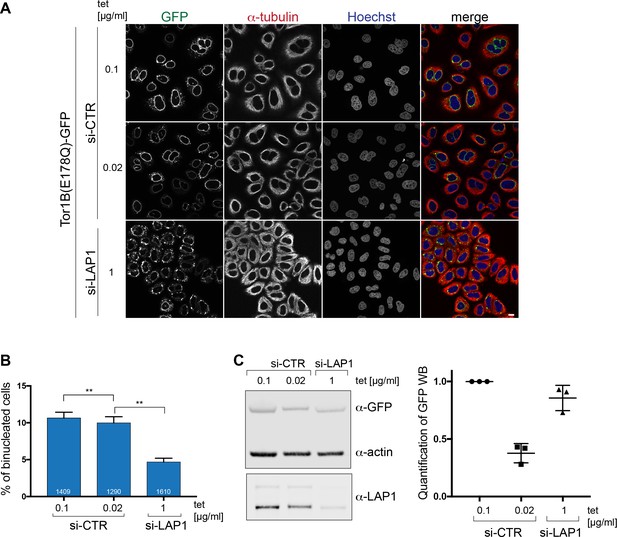
Expression of a dominant-negative Torsin1B variant leads to an increase in binucleated cells in a LAP1-dependent manner.
(A) Expression of Tor1B(E178Q)-GFP was induced for 48 hr in LAP1-depleted cells (1 μg/ml tet) and control siRNA-treated cells (0.1 μg/ml and 0.02 μg/ml tet). Scale bar, 10 μm. (B) Quantification of binucleated cells (N = 3, n > 631, mean+/-SEM). (C) Quantitative western blot analysis of Tor1B(E178Q)-GFP levels relative to actin for depicted conditions.
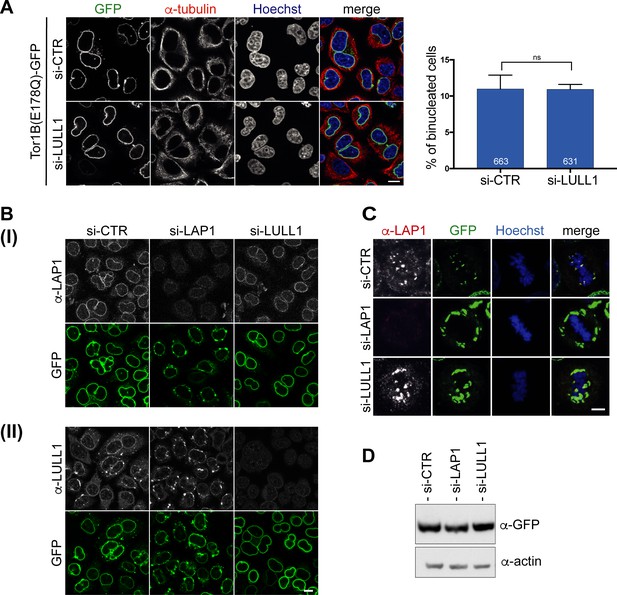
Downregulation of LULL1 does not prevent Tor1B(E178Q)-induced binucleation.
(A) Expression of Tor1B(E178Q)-GFP was induced for 48 hr in control or LULL1-depleted cells. Binucleation was quantified as in Figures 5–6 (N = 3, n > 631, mean+/-SEM). Scale bar, 10 μm. (B) Depletion of LAP1 (I) and LULL1 (II) was controlled by immunofluorescence. Tor1B(E178Q)-GFP was expressed at comparable levels as shown by immunoblotting in panel D. (C) Localization of endogenous LAP1 at metaphase chromatin upon overexpression of Tor1B(E178Q)-GFP disappears upon LAP1 depletion, but is not influenced by LULL1 depletion. Representative confocal images (maximal z-projections: 5 × 0.63 μm) of metaphase HeLa cells with TorsinB-E178Q-GFP expression. Localization of LAP1 was analyzed by immunofluorescence staining. Scale bar, 5 μm. (D) Western blot analysis of Tor1B(E178Q)-GFP levels after LAP1 or LULL1 depletion.
Videos
Time-lapse imaging of a LAP1B-GFP expressing cell progressing through mitosis.
Expression of LAP1B-GFP was induced 24 hr prior to imaging. DNA was visualized by SirHoechst. The movie starts 68 min before anaphase onset. Note that nuclear envelope (NE) aberrations manifest upon NE reformation after mitosis and persist.
Time-lapse imaging of a LAP2β-GFP expressing cell progressing through mitosis.
Expression of LAP2β-GFP was induced 24 hr prior to imaging. DNA was visualized by SirHoechst. The movie starts 68 min before anaphase onset. Note that the nuclear envelope (NE) rapidly adopts a smooth topology soon after NE reformation.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HeLa FlpIn T-REx LAP1B-EGFP | This paper | See Materials and methods, Cell lines, antibodies, and reagents | |
| Cell line (Homo sapiens) | HeLa FlpIn T-REx LAP2β-EGFP | This paper | See Materials and methods, Cell lines, antibodies, and reagents | |
| Cell line (Homo sapiens) | HeLa FlpIn T-REx LAP1C-EGFP | This paper | See Materials and methods, Cell lines, antibodies, and reagents | |
| Cell line (Homo sapiens) | HeLa FlpIn T-REx LAP1B(1-359)-EGFP | This paper | See Materials and methods, Cell lines, antibodies, and reagents | |
| Cell line (Homo sapiens) | HeLa FlpIn T-REx LAP1B(E482A)-EGFP | This paper | See Materials and methods, Cell lines, antibodies, and reagents | |
| Cell line (Homo sapiens) | HeLa FlpIn T-REx LAP1B(R563G)-EGFP | This paper | See Materials and methods, Cell lines, antibodies, and reagents | |
| Cell line (Homo sapiens) | HeLa FlpIn T-REx LAP1B(1-359, S108E, T124E)-EGFP | This paper | See Materials and methods, Cell lines, antibodies, and reagents | |
| Cell line (Homo sapiens) | HeLa FlpIn T-REx Tor1B-EGFP | This paper | See Materials and methods, Cell lines, antibodies, and reagents | |
| Cell line (Homo sapiens) | HeLa FlpIn T-REx Tor1B(E178Q)-EGFP | This paper | See Materials and methods, Cell lines, antibodies, and reagents | |
| Cell line (Homo sapiens) | HeLa FlpIn T-REx | (Hafner et al., 2014) DOI:10.1038/ncomms5397 | Obtained from T. Maier (University Konstanz). | |
| Cell line (Homo sapiens) | HeLa Kyoto | other | RRID:CVCL_1922 | Obtained from D. Gerlich (IMBA, Vienna). |
| Cell line (Homo sapiens) | HeLa Gromeier LMN A/C KO | This paper | See Materials and methods, Cell lines, antibodies, and reagents antibodies, and reagents | |
| Cell line (Homo sapiens) | HCT116 | other | RRID:CVCL_0291 | Obtained from B. Vogelstein (Johns Hopkins, Baltimore, USA) |
| Cell line (Homo sapiens) | HepG2 | other | RRID:CVCL_0027 | Obtained from S. Werner (Institute of Molecular Health Science, Zurich) |
| Transfected construct (human) | si-Control | Qiagen | Cat# 1027281 | Allstars siRNA |
| Transfected construct (human) | si-lamin A/C | Microsynth (Hasan et al., 2006) DOI: 10.1016/j.febslet.2006.01.039 | 5’- CUGGACUUCCAGAAGAACA -3’ | |
| Transfected construct (human) | si-lamin B1 | Sigma (Hasan et al., 2006) DOI: 10.1016/j.febslet.2006.01.039 | 5’- UUCCGCCUCAGCCACUGGAAAU -3’ | |
| Transfected construct (human) | si-lamin B2 | Microsynth | 5’- ACAACUCGGACAAGGAUC -3’ | |
| Transfected construct (human) | si-LAP1 | Qiagen/Microsynth | 5’- CUCACUAAGUUUCCUGAGUUA- 3’ | |
| Transfected construct (human) | si-LULL1 | Qiagen/Microsynth | 5’- CTGGTCCTGACTGTTCTGCTA -3’ | |
| Transfected construct (human) | si-Tor1A | Qiagen/Microsynth | 5’- CACCAAGTTAGATTATTACTA-3’ | |
| Transfected construct (human) | si-Tor1B | Qiagen/Microsynth | 5’- CTGTCGGAGTCTTCAATAATA-3’ | |
| Antibody | anti-β-actin (mouse monoclonal) | Sigma | Cat# A1978 RRID:AB_476692 | WB(1:40’000) |
| Antibody | anti-emerin (rabbit polyclonal) | Abcam | Cat# ab40688 RRID:AB_2100059 | WB(1:1000) |
| Antibody | anti-HA (mouse monoclonal) | Covance | Cat# MMS-101P RRID:AB_2314672 | IF(1:3000) WB(1:3000) |
| Antibody | anti-lamin A/C (mouse monoclonal) | ImmuQuest | Cat# IQ332 RRID:AB_10660272 | IF(1:200) WB (1:200) |
| Antibody | anti-lamin A/C (rabbit polyclonal) | Proteintech | Cat# 10298-1-AP RRID:AB_2296961 | IF(1:500) |
| Antibody | anti-lamin B1 (rabbit polyclonal) | Abcam | Cat# ab16048 RRID:AB_443298 | IF(1:2000) WB(1:1000) |
| Antibody | anti-lamin B2 (rabbit monoclonal) | Abcam | Cat# ab151735 RRID:AB_2827514 | IF (1:1000) WB(1:1000) |
| Antibody | anti-LAP1 (rabbit polyclonal) | Abcam | Cat# ab86307 RRID:AB_2206124 | IF(1:500) WB(1:300) |
| Antibody | anti-LBR (rabbit polyclonal) | Abnova | Cat# PAB15583 RRID:AB_10696691 | IF(1:1000) |
| Antibody | anti-mAB414 (mouse monoclonal) | Abcam | Cat# ab 24609 RRID:AB_448181 | IF(1:20000) |
| Antibody | anti-H3 (rabbit polyclonal) | Abcam | Cat# ab1791 RRID:AB_302613 | WB(1:5000) |
| Antibody | anti-Tor1A (rabbit polyclonal) | Abexxa | Cat# abx001683 | WB(1:500) |
| Antibody | anti-Tor1B (rabbit polyclonal) | antibodies-online | Cat# ABIN1860834 | WB(1:500) |
| Antibody | anti-α-tubulin (mouse monoclonal) | Sigma | Cat# T5168 RRID:AB_477579 | WB(1:20000) IF(1:20000) |
| Antibody | anti-GAPDH (mouse monoclonal) | Abcam | Cat# ab8245 RRID:AB_2107448 | WB(1:10000) |
| Antibody | anti-LULL1 (rabbit polyclonal) | This paper | IF (1:500) See Materials and methods, Cell lines, antibodies, and reagents | |
| Antibody | anti-SUN1 (rabbit polyclonal) | (Sosa et al., 2012) DOI: 10.1083/jcb.200904048 | IF(1:1000) | |
| Antibody | anti-SUN2 (rabbit polyclonal) | (Turgay et al., 2010) DOI: 10.1261/rna.2325911 | IF(1:2000) | |
| Antibody | anti-GFP (rabbit polyclonal) | (Turgay et al., 2010) DOI: 10.1261/rna.2325911 | IF(1:1000) WB(1:1000) | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/LAP1B-EGFP | This paper | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/LAP1C-EGFP | This paper | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1B(1-72)-GST-EGFP | This paper | LAP1: aa 1-72 See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1B(1-183)-GST-EGFP | This paper | LAP1: aa 1-183 See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1B(98-136)-GST-EGFP | This paper | LAP1: aa 98-136 See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1B(184-337)-GST-EGFP | This paper | LAP1: aa 184-337 See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ LAP1B(Δ98-136)-EGFP | This paper | LAP1: aa Δ98-136 See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ LAP1B(1-359)-2E-EGFP | This paper | LAP1: aa 1-359, S108E, T124E See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ LAP1B(1-359)-EGFP | This paper | LAP1: aa 1-359, See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ Tor1A-cTAP | This paper | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ Tor1A-cTAP | This paper | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ Tor1B-cTAP | This paper | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ Tor1B(E178Q)-cTAP | This paper | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ LAP1B(R563G)-EGFP | This paper | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/LAP1B-E482A-EGFP | This paper | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ Tor1B-mRFP | This paper | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ Tor1B(E178Q)-mRFP | This paper | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ Tor1B-EGFP | This paper | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ Tor1B(E178Q)-EGFP | This paper | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ LAP1B(S108E,T124E)-EGFP | This paper | S108E, T124E See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ LAP1B(S108E, T124E, R563G)-EGFP | This paper | S108E, T124E, R563G See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7-GST-GFP | (Ungricht et al., 2015a) DOI: 10.1007/978-1-4939-3530-7_28 | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 NLS-GST-EGFP | (Erkmann et al., 2005) DOI: 10.1091/mbc.e04-11-1023 | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1(1-121)-GST-EGFP | This paper | LAP1: aa 1-121, See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1B(73-183)-GST-EGFP | This paper | LAP1: aa 73-183, See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1(122-183)-GST-EGFP-NLS | This paper | LAP1: aa 122-183, See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1B(73-337)-GST-EGFP | This paper | LAP1: aa 73-337, See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1(122-337)-GST-EGFP | This paper | LAP1: aa 122-337, See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1B(1-337)-GST-EGFP | This paper | LAP1: aa 1-337, See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1(1-72) GST-EGFP-NLS | This paper | LAP1: aa 1-72, See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1(122-183)-GST-EGFP-NLS | This paper | LAP1: aa 122-183, See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1(73-337) GST-EGFP-NLS | This paper | LAP1: aa 73-337, See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1(184-337) GST-EGFP-NLS | This paper | LAP1: aa 184-337, See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pEGFPN3 LAP1B -EGFP | This paper | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pEGFPN3 EMD-EGFP | This paper | See Materials and methods, Molecular cloning | |
| Recombinant DNA reagent | pEGFPN3 LAP2β-EGFP | (Ungricht et al., 2015a) DOI: 10.1007/978-1-4939-3530-7_28 | See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pEGFPN3 LEM2-EGFP | This paper | See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pEGFPN3 SUN1-EGFP | (Turgay et al., 2010) DOI: 10.1261/rna.2325911 | See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pEGFPN3 SUN2-EGFP | (Turgay et al., 2010) DOI: 10.1261/rna.2325911 | See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pEGFPN3-SPAG4-GFP | (Turgay et al., 2010) DOI: 10.1261/rna.2325911 | See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | LAP1(98-136)-SPAG4-EGFP | This paper | See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | LAP1(98-136_2E)-SPAG4-EGFP | This paper | See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | LAP1(98-136_3E)-SPAG4-EGFP | This paper | See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | LULL1-HASt | This paper | See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pCMV/ER/myc EGFP-KDEL | (Ungricht et al., 2015a) DOI: 10.1007/978-1-4939-3530-7_28 | See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pEGFPN3 LAP1B(72-end)-EGFP | This paper | LAP1: aa 72-583, See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pEGFPN3 LAP1B(184-end)-EGFP | This paper | LAP1: aa 184-583, See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pEGFPN3 LAP1C(Δ122-136) EGFP | This paper | LAP1: aa Δ122-136, See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ LAP1C(T124E)-EGFP | This paper | See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pcDNA5/FRT/TO/ LAP1B(184-end R563G)-EGFP | This paper | LAP1: aa 184-583, See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pEGFPN3 LAP1(R563G)-EGFP | This paper | See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1(1-97)-GST-GFP | This paper | LAP1: aa 1-97, See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pK7 LAP1(137-337)-GST-GFP | This paper | LAP1: aa 137-337, See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pQE60zz-His6 | (Sosa et al., 2012) DOI: 10.1016/j.cell.2012.03.046 | See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pQE60zz-LAP1B(98-136)-His6 | This paper | LAP1: aa 98-136, See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pQE60zz-His6 zz-LAP1B(98-136)-2E His6 | This paper | LAP1: aa 98-136, S108E, T124E See Materials and Methods, Molecular cloning | |
| Recombinant DNA reagent | pC2P | (Welte et al., 2019) DOI: 10.1101/gad.328492.119 | ||
| Recombinant DNA reagent | pC2P-gLMNA/C | This paper | Protospacer: 5’- CACCGGGTGGCGCGCCGCTGGGACG -3’ See Materials and Methods, Generation of LMN A/C knockout cells | |
| Chemical compound, drug | SIR-Hoechst | Spirochrome | Cat# SC007 | |
| Chemical compound, drug | Hoechst | Invitrogen | Cat# 63493 | |
| Chemical compound, drug | DTT | Applichem | Cat# A1101 | |
| Chemical compound, drug | nocodazole | Sigma-Aldrich | Cat# M1404 | |
| Chemical compound, drug | thymidine | Sigma-Aldrich | Cat #T1895 | |
| Chemical compound, drug | tetracycline | Invitrogen | Cat# 550205 |





