Control and regulation of acetate overflow in Escherichia coli
Figures
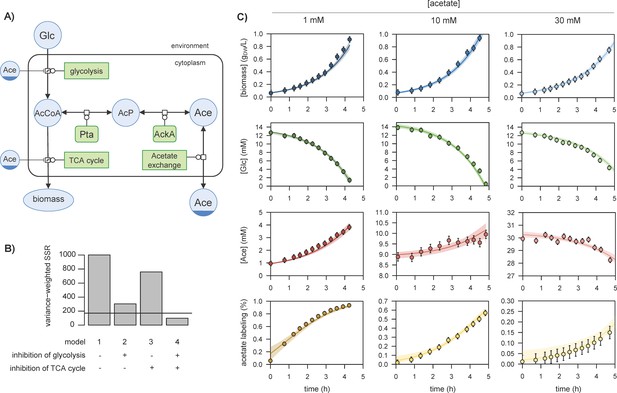
Representation of glucose and acetate metabolism in Escherichia coli (A), in Systems Biology Graphical Notation format (http://sbgn.org) (Le Novère et al., 2009).
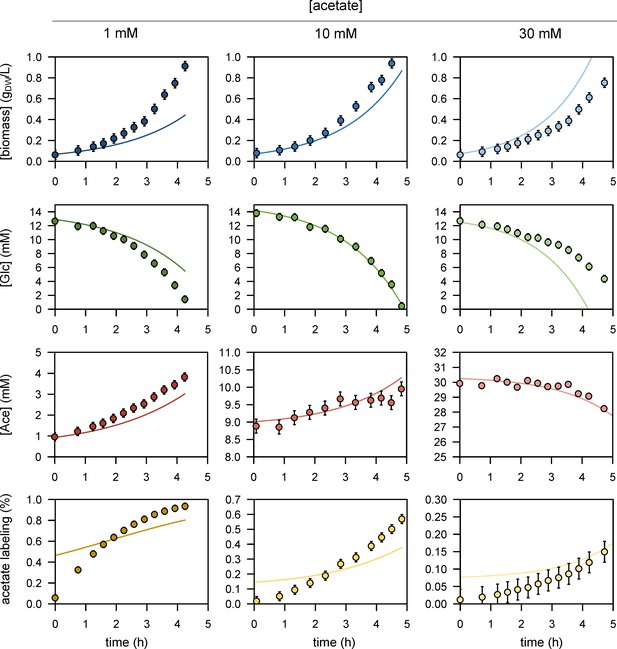
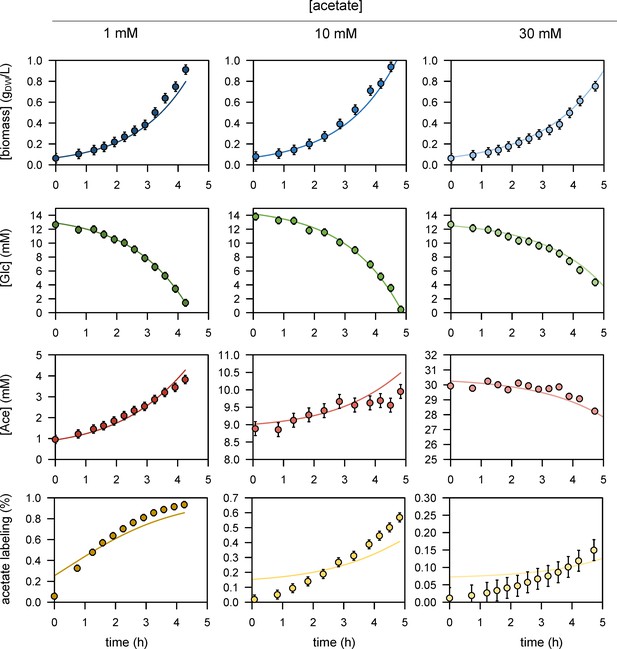
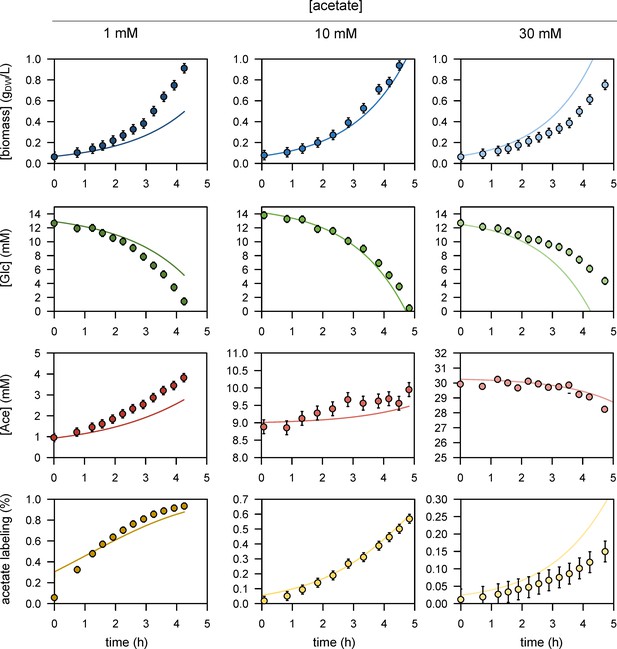
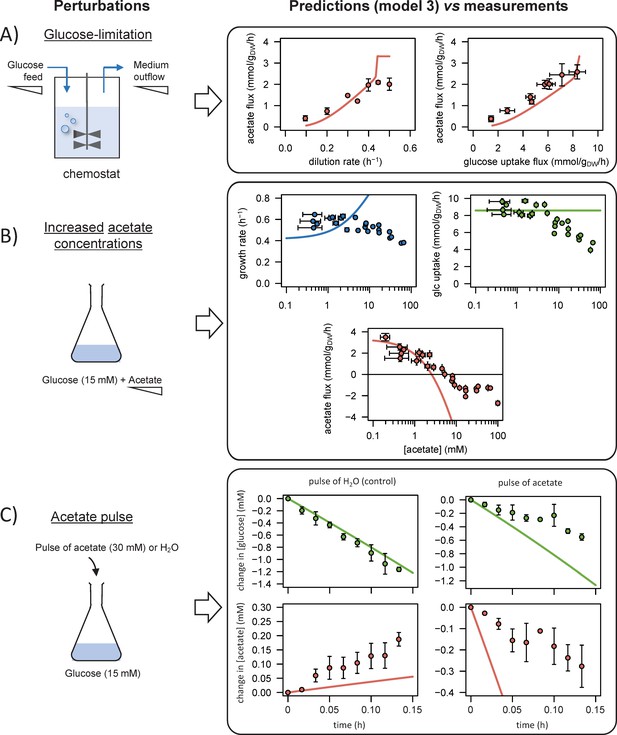
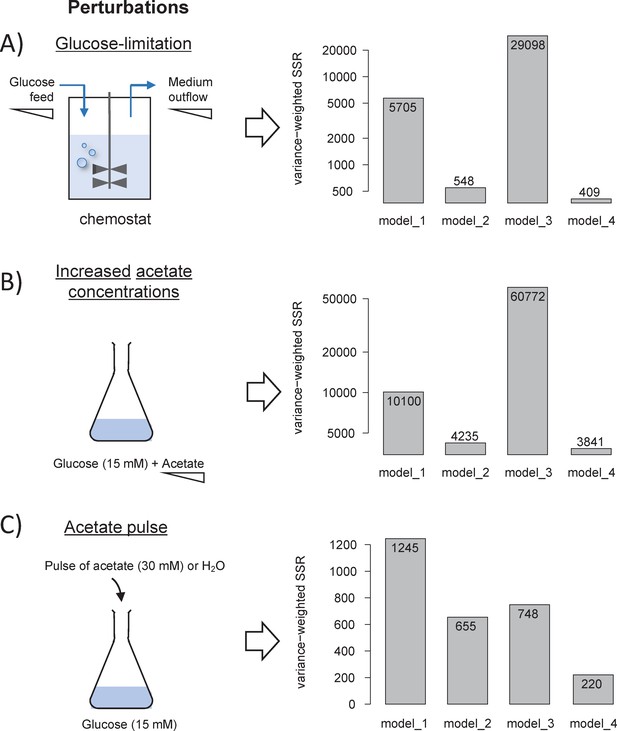
We performed 13C-labelling experiments to calibrate the model and evaluated the goodness of fit for different topologies (B). The initial model (model 1), which does not include inhibition of the glycolytic pathway and TCA cycle by acetate, did not fit the data satisfactorily. Adding inhibition by acetate of glycolysis (model 2) or of the TCA cycle (model 3) improved the fit, but both pathways had to be inhibited (model 4) for the goodness-of-fit criterion to be satisfied. In (B), the horizontal line represents the 95% confidence threshold for the variance-weighted sum of squared residuals (SSR). The best fits of the experimental data obtained with model 4 are shown in (C), where the shaded areas represent the 95% confidence interval on the fits. The best fits obtained with the alternative models (models 1–3) are shown in Figure 1—figure supplements 1–3.
-
Figure 1—source data 1
Experimental data used to calibrate the model.
- https://cdn.elifesciences.org/articles/63661/elife-63661-fig1-data1-v2.xlsx
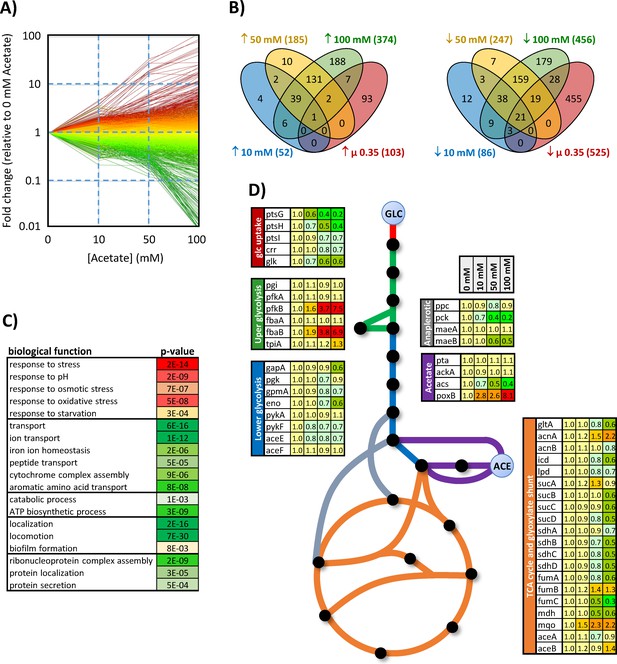
Response of the E. coli transcriptome to changes in acetate concentration (0, 10, 50, or 100 mM) during growth on glucose (15 mM).
The changes in gene expression are shown in (A). Each line represents the expression of a single gene relative to its expression level measured in the absence of acetate. Up- and downregulated genes are shown in red and green, respectively. The Venn diagrams (B) represent the total number of genes upregulated (left) and downregulated (right) by at least a factor of 2 under each condition and during growth on glucose in the absence of acetate but at the same growth rate as in the presence of 100 mM acetate (0.35 hr−1, extrapolated from the data from Esquerré et al., 2014). Biological functions modulated by the presence of acetate (based on Gene Ontology analysis) are shown in (C), with the corresponding p-values. The expression levels of central metabolic genes are shown in (D). The data were obtained from four independent biological replicates for each condition.
-
Figure 2—source data 1
Response of the E. coli transcriptome to changes in acetate concentration (0, 10, 50, or 100 mM) during growth on glucose (15 mM).
- https://cdn.elifesciences.org/articles/63661/elife-63661-fig2-data1-v2.xlsx
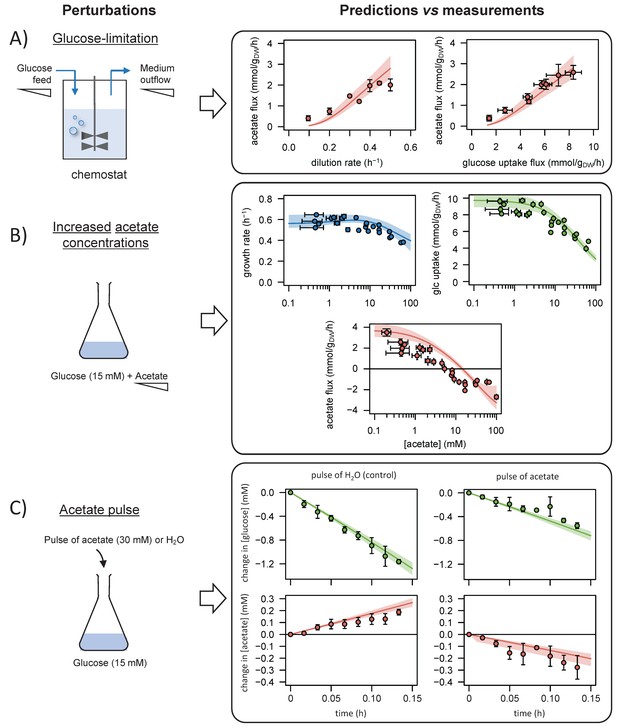
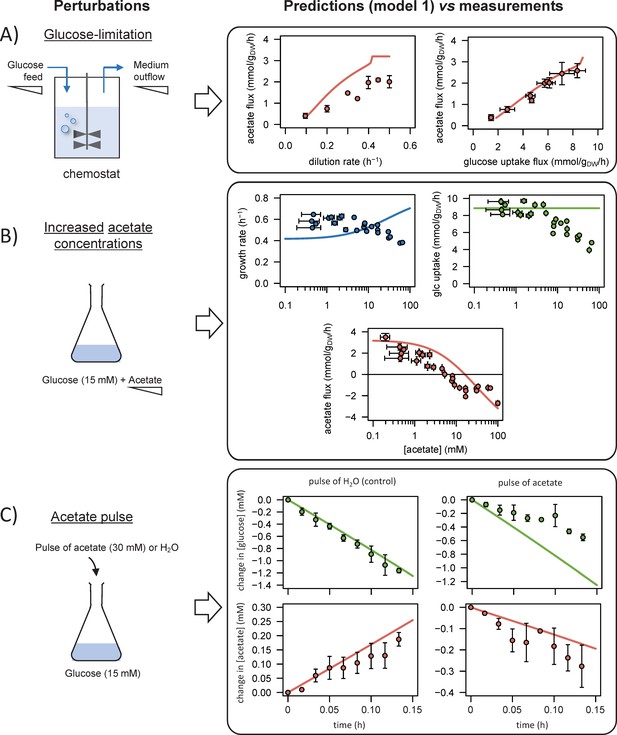
Comparison of model predictions with experimental data.
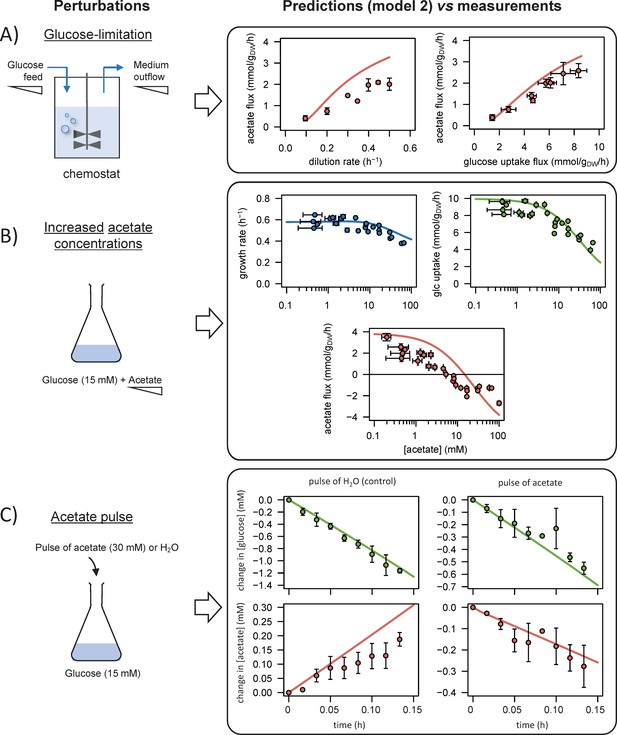
We used the model to simulate (i) steady-state glucose and acetate fluxes in glucose-limited chemostat cultures at dilution rates of 0.1–0.5 hr−1 (A), (ii) the growth rates and glucose and acetate fluxes during growth on glucose at various acetate concentrations (B), and (iii) the time courses of the changes in glucose and acetate concentrations during exponential growth on glucose after a pulse of either acetate or water (C). Model predictions are represented by lines and experimental data are shown as dots (the error bars represent one standard deviation), the shaded areas represent the 95% confidence intervals on the predictions. Predictions obtained with the alternative models (models 1–3) are shown in Figure 3—figure supplements 1–3. The predictive accuracy was compared between models based on the variance-weighted sum of squared residuals between simulated and experimental data (Figure 3—figure supplements 1–4).
-
Figure 3—source data 1
Experimental data used to validate the model.
- https://cdn.elifesciences.org/articles/63661/elife-63661-fig3-data1-v2.xlsx
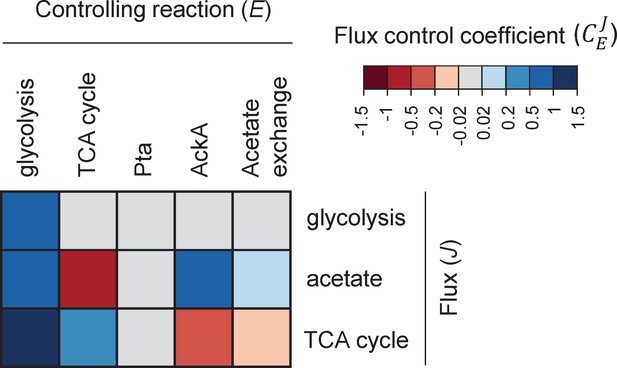
Heatmap of flux control coefficients during growth on glucose (15 mM) and acetate (0.1 mM).
Each column represents a controlling reaction (E) and each row, a flux (J). Red and blue cells represent negative and positive flux control coefficients (), respectively, with darker (lighter) tones indicating stronger (weaker) control.
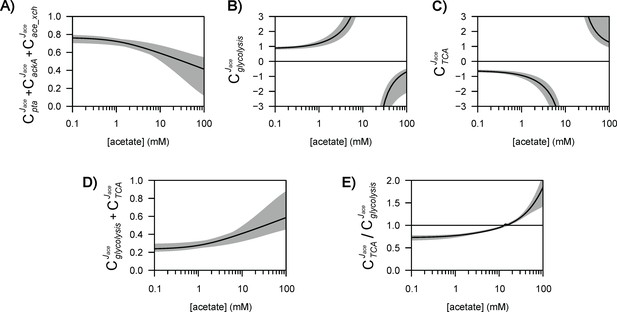
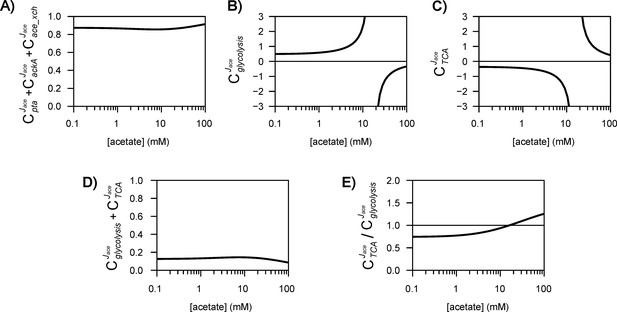
Control of acetate flux over a broad range of acetate concentrations.
The shaded areas represent the 95% confidence intervals. Flux control coefficients calculated with the alternative models (models 1–3) are shown in Figure 5—figure supplements 1–3.
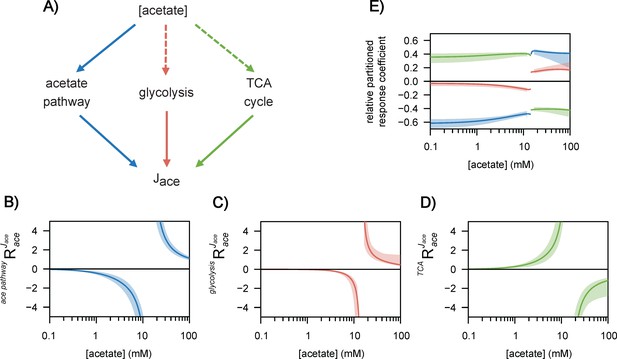
Regulation of acetate flux in Escherichia coli.
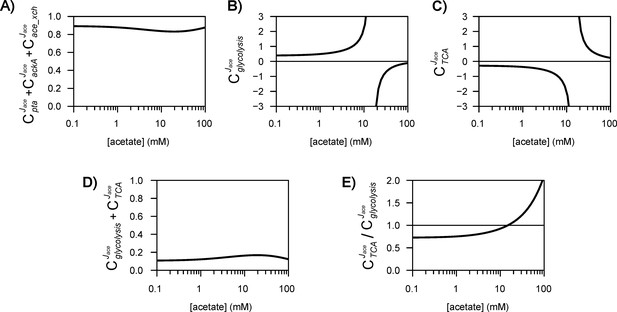
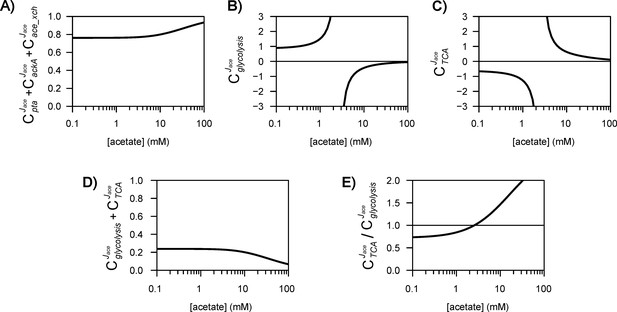
The different routes through which acetate flux can be regulated by the acetate concentration are shown in (A). Dotted lines represent indirect (hierarchical) regulation, and straight lines represent direct (metabolic) regulation. The strengths of the three regulatory routes are respectively shown in (B–D), and their relative contributions are shown in (E). The shaded areas represent the 95% confidence intervals.
Tables
Reactions included in the kinetic model of glucose and acetate metabolism of Escherichia coli.
| Name | Reaction | Rate law | Comment |
|---|---|---|---|
| Glucose_feed | ø → GLC | C | Glucose inflow and medium outflow to simulate chemostat experiments |
| Acetate_outflow | ACEenv → ø | MA | |
| Biomass_outflow | X → ø | MA | |
| Glucose_outflow | GLC → ø | MA | |
| Glycolysis | GLC → 1.4 × ACCOA | IMM | Stoichiometric coefficient taken from Millard et al., 2014 |
| TCA_cycle | ACCOA → ø | IMM | Utilisation of AcCoA by the TCA cycle |
| Pta | ACCOA ↔ ACP | RMM | Rate law from Enjalbert et al., 2017; Millard et al., 2017; Kadir et al., 2010 |
| AckA | ACP ↔ ACEcell | RMM | Rate law from Enjalbert et al., 2017; Millard et al., 2017; Kadir et al., 2010 |
| Acetate_exchange | ACEcell ↔ ACEenv | RMM | Rate law from Millard et al., 2017 |
| Growth | X → 2 × X | MA | Rate calculated from the TCA cycle flux, assuming a constant biomass yield (Enjalbert et al., 2017; Pinhal et al., 2019) |
-
Abbreviations: C: constant flux; MA: mass action; RMM: reversible Michaelis-Menten; IMM: irreversible Michaelis-Menten.
Values of kinetic parameters taken from the literature.
Values and 95% confidence intervals of the estimated parameters.
| Reaction | Parameter | Value | 95 % CI |
|---|---|---|---|
| AckA | Vmax | 3.4 × 105 | 2.8 × 105 – 5.5 × 105 |
| Pta | Vmax | 9.8 × 105 | 4.9 × 104 – 9.9 × 106 |
| Glycolysis | Vmax | 5.6 × 103 | 5.3 × 103 – 5.9 × 103 |
| Ki_ACE | 36.7 | 30.9 – 46.9 | |
| TCA cycle | Km_ACCOA | 24.8 | 8.4 – 615.4 |
| Vmax | 7.4 × 105 | 2.4 × 105 – 1.7 × 106 | |
| Ki_ACE | 2.3 | 1.8 – 3.4 | |
| Growth | Y | 1.0 × 10−4 | 9 × 10−5 – 1.1 × 10−4 |
| Acetate exchange | Vmax | 4.8 × 105 | 8 × 104 – 1.5 × 106 |
| Km_ACE | 33.2 | 1.5 – 99.8 |
Additional files
-
Supplementary file 1
R scripts used to construct the models, perform the simulations and generate the figures.
- https://cdn.elifesciences.org/articles/63661/elife-63661-supp1-v2.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63661/elife-63661-transrepform-v2.docx