Association and dissociation between the mitochondrial Far complex and Atg32 regulate mitophagy
Figures
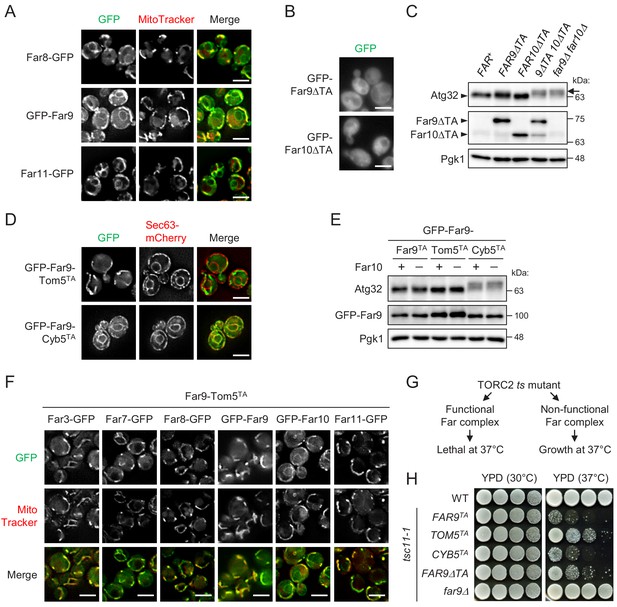
Mitochondria-localized, but not ER-localized, Far complex is required for Atg32 dephosphorylation.
(A, B, D, and F) Cells expressing the indicated GFP-fused Far proteins were cultured in YPD medium until the early log growth phase and analyzed by fluorescence microscopy. Sec63-mCherry and MitoTracker Red CMXRos were used to visualize the ER and mitochondria, respectively. Representative images of at least 100 cells are shown. Scale bar, 4 µm. (C and E) The indicated cells were cultured in YPL medium until the mid-log growth phase. Atg32 status was analyzed by western blot (WB) using an anti-Atg32 antibody. Far9ΔTA and Far10ΔTA were detected using the anti-HA antibody. GFP-Far9 derivatives were detected with an anti-Far9 antibody. Pgk1 was detected as a loading control (throughout this study). For Atg32 detection, arrowhead and arrow indicate the dephosphorylated and phosphorylated Atg32, respectively. (G) Functional and non-functional Far complexes cause opposite growth phenotypes in the TORC2 ts mutant background. (H) Wild-type cells, tsc11-1 cells expressing GFP-Far9 derivatives, and tsc11-1 far9Δ cells were cultured in YPD medium until the early log growth phase. Serial dilutions of each culture were spotted on YPD agar plates and cultured at 30°C for 24 hr or 37°C for 48 hr (three independent replicates). WB experiments were independently replicated three (E) or four times (C).
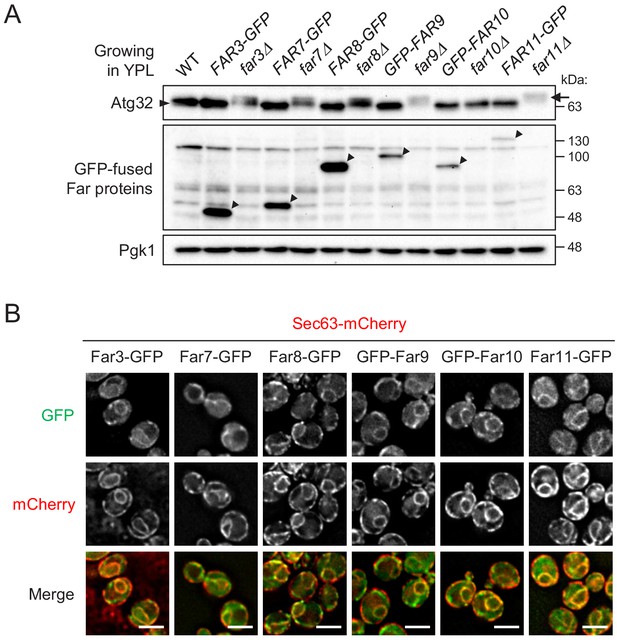
Expression, functional, and localization analyses of GFP-fused Far proteins.
(A) The indicated cells were cultured in YPL medium until the mid-log growth phase. Atg32 status was analyzed by western blot (WB) with an anti-Atg32 antibody. GFP-fused Far proteins (arrowheads) were detected with an anti-GFP antibody. Pgk1 was detected as a loading control. For Atg32 detection, arrowhead and arrow indicate the dephosphorylated and phosphorylated Atg32, respectively. WB experiments were independently replicated three times. (B) Cells expressing the indicated GFP-fused Far proteins were cultured in YPD medium until the early log growth phase and analyzed by fluorescence microscopy. Sec63-mCherry was used to visualize the ER. Representative images of at least 100 cells are shown. Scale bar, 4 µm.
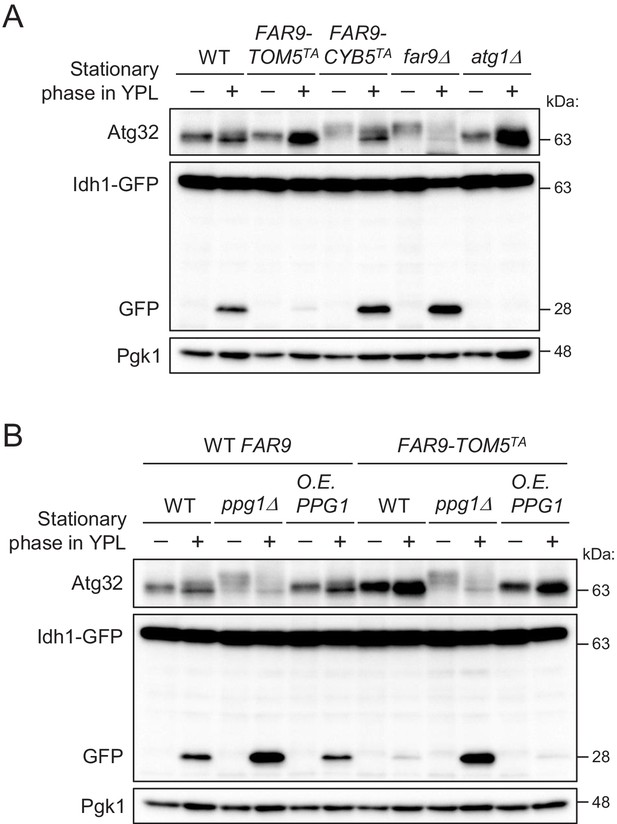
The mitochondria-localized Far complex is a limiting factor for the Ppg1-dependent inhibition of mitophagy via Atg32 dephosphorylation.
(A and B) The indicated cells expressing Idh1-GFP were continuously cultured in YPL medium and collected at 20 hr (growing phase) and 40 hr (stationary phase). Atg32 status and Idh1-GFP processing were analyzed by Western blot (WB) with anti-Atg32 and anti-GFP antibodies, respectively. WB experiments were independently replicated three times (A and B).
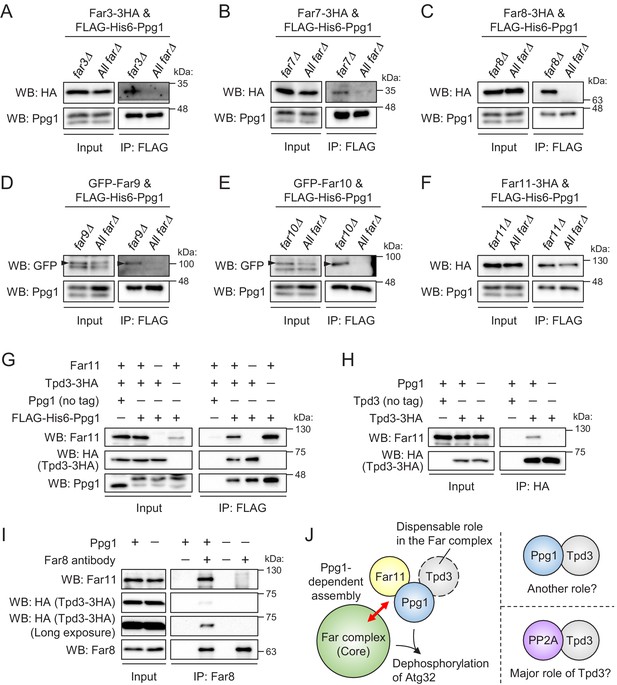
Ppg1, Far11, and Tpd3 form a subcomplex, but Tpd3 plays a limited role as a scaffold protein in the Ppg1-Far complex.
The indicated cells co-expressing FLAG-His6-Ppg1 and Far3-3HA (A), Far7-3HA (B), Far8-3HA (C), GFP-Far9 (D), GFP-Far10 (E), or Far11-3HA (F) were cultured in SMD-Trp-Ura (A–C and F) or SMD-Ura (D and E) medium until the early log growth phase. FLAG-His6-Ppg1 was precipitated from cell lysates using an anti-FLAG M2 affinity gel. Cell lysates (Input) and precipitates (IP: FLAG) were analyzed by western blot (WB) with anti-HA, anti-GFP, and anti-Ppg1 antibodies. (G) ppg1Δ TPD3-3HA, ppg1Δ far11Δ TPD3-3HA, and ppg1Δ tpd3Δ cells expressing Ppg1 (no tag) or FLAG-His6-Ppg1 were cultured in SMD-Ura medium until the early log growth phase. Cell lysates (Input) and anti-FLAG immunoprecipitates (IP: FLAG) were analyzed by WB with anti-Far11, anti-HA, and anti-Ppg1 antibodies. (H) Wild-type, TPD3-3HA, and ppg1Δ TPD3-3HA cells were cultured in YPD medium until the early log growth phase. Cell lysates (Input) and anti-HA immunoprecipitates (IP: HA) were analyzed by WB with anti-Far11 and anti-HA antibodies. (I) TPD3-3HA and ppg1Δ TPD3-3HA cells were cultured in YPD medium until the early log growth phase. Cell lysates (Input) and anti-Far8 immunoprecipitates (IP: Far8) were analyzed by WB with anti-Far11, anti-HA, and anti-Far8 antibodies. (J) Ppg1, Far11, and Tpd3 form a subcomplex, and this subcomplex binds to the core of the Far complex in a Ppg1-dependent manner. Tpd3 is dispensable for the Ppg1-Far complex, although Ppg1 and Tpd3 may play a Far complex-independent role. The major role of Tpd3 might be a scaffold protein of PP2A (Pph21 and Pph22) rather than that of Ppg1. WB experiments were independently replicated three times (A–I).
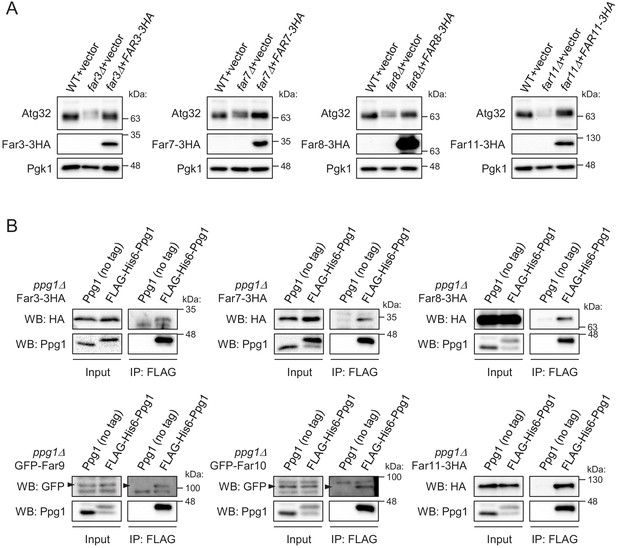
Ppg1 interacts with the Far3-7-8-9-10-11 proteins.
(A) The indicated cells were cultured in SML-Trp medium until the mid-log growth phase. Atg32 status was analyzed by western blot (WB) with an anti-Atg32 antibody. 3HA-tagged Far proteins were detected with an anti-HA antibody. (B) The indicated cells were cultured in SMD-Trp-Ura or SMD-Ura (for GFP-Far9/GFP-Far10) medium until the early log growth phase. FLAG-His6-Ppg1 was precipitated from cell lysates using an anti-FLAG M2 affinity gel. Cell lysates (Input) and the precipitates (IP: FLAG) were analyzed by WB with anti-HA, anti-GFP, and anti-Ppg1 antibodies. WB experiments were independently replicated three times (A and B).
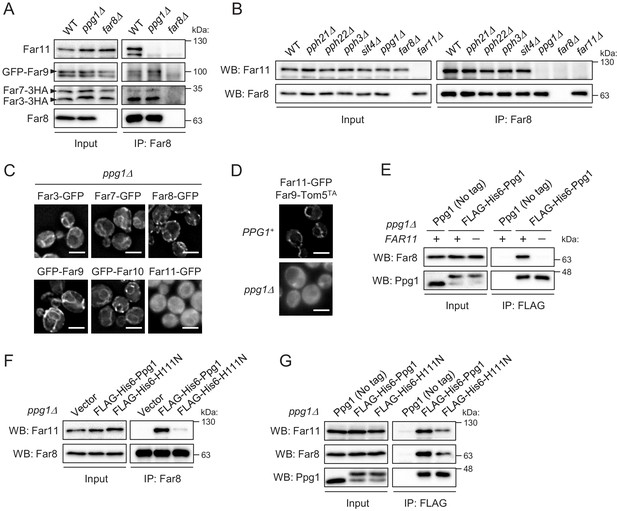
Ppg1 phosphatase activity is required for the assembling integrity of Ppg1-Far11-Far8.
(A) Wild-type, ppg1Δ, and far8Δ (negative control) cells expressing HA- or GFP-tagged Far proteins were cultured in YPD medium until the early log growth phase. Cell lysates (Input) and anti-Far8 immunoprecipitates (IP: Far8) were analyzed by western blot (WB) with anti-HA, anti-Far8, anti-Far9, and anti-Far11 antibodies. (B) The indicated cells were cultured in YPL medium until the mid-log growth phase. Atg32 status was analyzed by WB with an anti-Atg32 antibody. (C) ppg1Δ cells expressing the indicated GFP-fused Far proteins were cultured in YPD medium until the early log growth phase and analyzed by fluorescence microscopy. Representative images of at least 100 cells are shown. Scale bar, 4 µm. (D) PPG1+ and ppg1Δ cells expressing Far9-Tom5TA and Far11-GFP were cultured in YPD medium until the early log growth phase and analyzed by fluorescence microscopy. Representative images of at least 100 cells are shown. (E and G) ppg1Δ or ppg1Δ far11Δ cells expressing the indicated Ppg1 derivatives were cultured in SMD-Ura medium until the early log growth phase. FLAG-His6-Ppg1 was precipitated from cell lysates using an anti-FLAG M2 affinity gel. Cell lysates (Input) and anti-FLAG immunoprecipitates (IP: FLAG) were analyzed by WB with anti-Far8, anti-Far11, and anti-Ppg1 antibodies. (F) ppg1Δ cells expressing the indicated Ppg1 derivatives (empty vector as a negative control) were cultured in SMD-Ura medium until the early log growth phase. Cell lysates (Input) and anti-Far8 immunoprecipitates (IP: Far8) were analyzed by WB with anti-Far11 and anti-Far8 antibodies. WB experiments were independently replicated three (A, B, E, and F) or five times (G).
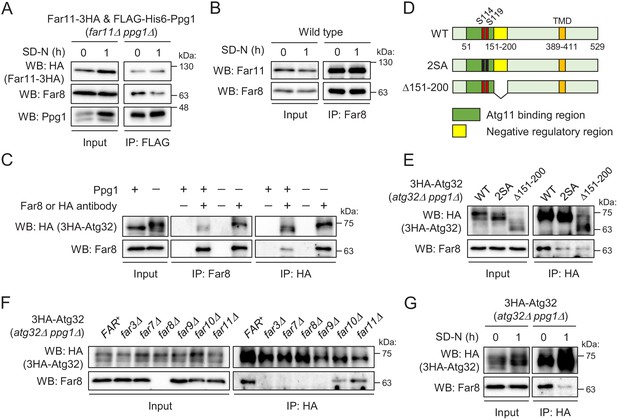
Interaction between the Far complex and phosphorylated Atg32 is impaired under mitophagy-inducing conditions.
(A) far11Δ ppg1Δ cells expressing Far11-3HA and FLAG-His6-Ppg1 were cultured in SMD-Trp-Ura medium until the early log growth phase, and the cells were then shifted to SD-N for 1 hr. Cell lysates (Input) and anti-FLAG immunoprecipitates (IP: FLAG) were analyzed by western blot (WB) with anti-HA, anti-Far8, and anti-Ppg1 antibodies. (B) Wild-type cells were cultured in YPD medium until the early log growth phase, and the cells were then shifted to SD-N medium for 1 hr. Cell lysates (Input) and anti-Far8 immunoprecipitates (IP: Far8) were analyzed by WB with anti-Far11 and anti-Far8 antibodies. (C) atg32Δ and atg32Δ ppg1Δ cells expressing 3HA-Atg32 were cultured in SMD-Ura medium until the early log growth phase. Cell lysates (Input), anti-Far8 immunoprecipitates (IP: Far8), and anti-HA immunoprecipitates (IP: HA) were analyzed by WB with anti-HA and anti-Far8 antibodies. (D) Schematic diagram of Atg32 and its derivatives. TMD, transmembrane domain; S114 and S119, serine residues phosphorylated by CK2; 2SA, S114A/S119A mutant; Δ151–200, Atg32 lacking the 151–200 amino acid region. (E–G) The indicated mutant cells expressing 3HA-Atg32 or its derivatives were cultured in SMD-Ura medium until the early log growth phase, and the cells were then shifted to SD-N medium for 1 hr (G). Cell lysates (Input) and anti-HA immunoprecipitates (IP: HA) were analyzed by WB with anti-HA and anti-Far8 antibodies. WB experiments were independently replicated three (A, B, F, and G) or four times (C and E).
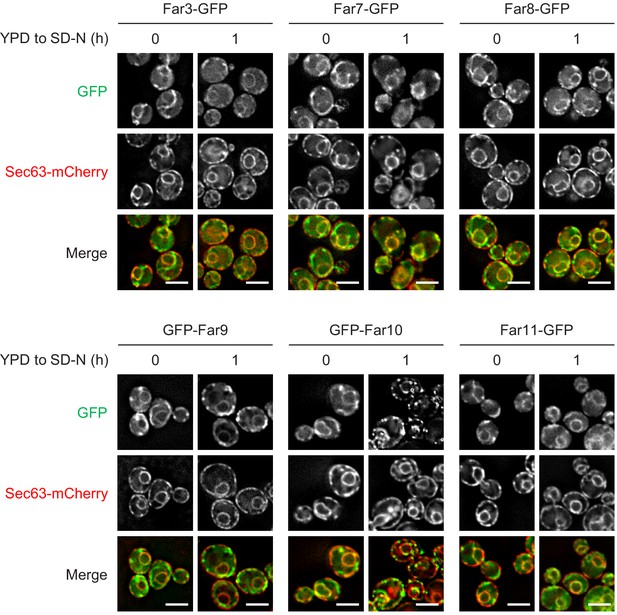
Starvation does not affect the localization of the Far proteins.
Cells expressing the indicated GFP-fused Far proteins were cultured in YPD medium until the early log growth phase, and the cells were then shifted to SD-N medium for 1 hr and analyzed by fluorescence microscopy. Sec63-mCherry was used to visualize the ER. Representative images of at least 100 cells are shown. Scale bar, 4 µm.
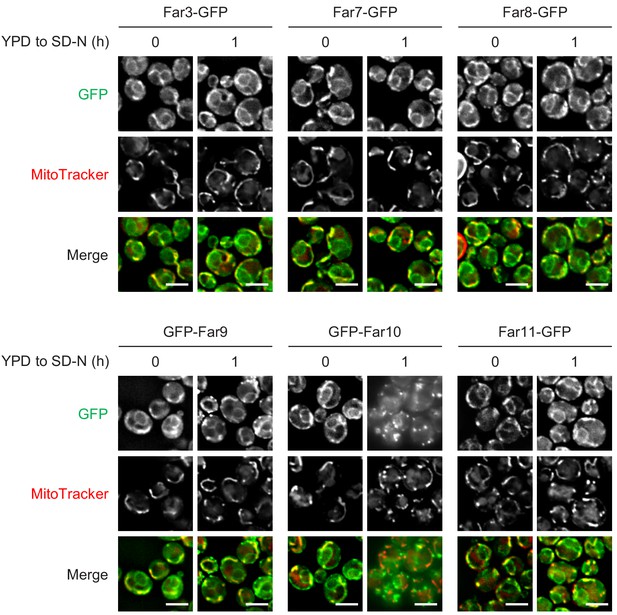
Starvation does not affect the localization of the Far proteins.
Cells expressing the indicated GFP-fused Far proteins were cultured in YPD medium until the early log growth phase, and the cells were then shifted to SD-N medium for 1 hr and analyzed by fluorescence microscopy. MitoTracker Red CMXRos was used to visualize mitochondria. Representative images of at least 100 cells are shown. Scale bar, 4 µm.
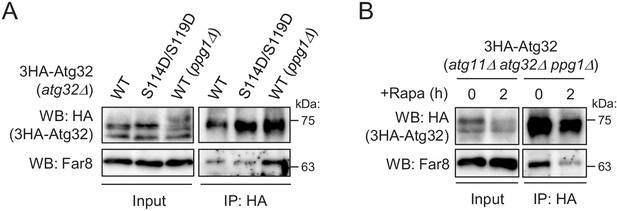
Analysis of the interaction between Atg32 and the Far complex.
(A and B) The indicated mutant cells expressing 3HA-Atg32 or 3HA-Atg32(S114D/S119D) were cultured in SMD-Ura medium until the early log growth phase, and the cells were subjected to 100 nM rapamycin treatment for 2 hr (B). Cell lysates (Input) and anti-HA immunoprecipitates (IP: HA) were analyzed by western blot (WB) with anti-HA and anti-Far8 antibodies. WB experiments were independently replicated three (A) or four times (B).
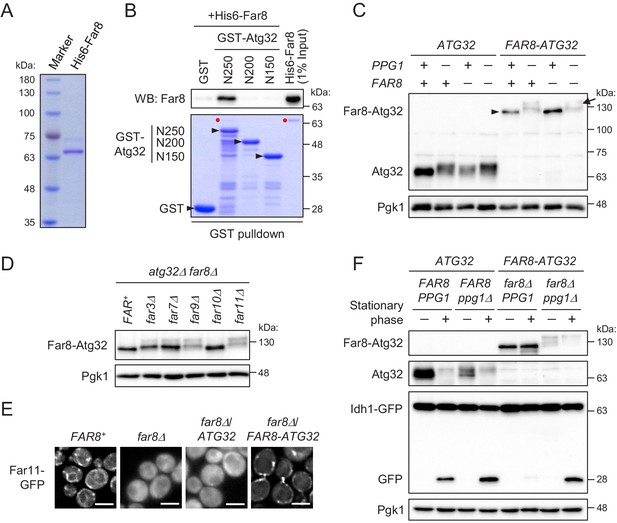
Far8 directly interacts with Atg32, and their artificial tethering prevents mitophagy.
(A) Purification of recombinant His6-Far8 protein produced in E. coli. (B) GST pull-down analysis of the interaction between Far8 and Atg32 derivatives. GST pull-down samples were loaded on an SDS-PAGE gel followed by CBB staining or western blot (WB) with an Far8 antibody. Purified His6-Far8 protein was loaded as an input sample. Pull-down experiments were replicated three times. Red dots indicate His6-Far8. (C and D) The indicated cells were cultured in SMD-Ura medium until the mid-log growth phase. Atg32/Far8-Atg32 status was analyzed by WB with an anti-Atg32 antibody. For Far8-Atg32 detection, arrowhead and arrow indicate the dephosphorylated and phosphorylated Far8-Atg32, respectively (C). (E) The indicated cells expressing Far11-GFP were cultured in YPD or SMD-Ura medium until the early log growth phase and analyzed by fluorescence microscopy. Representative images of at least 100 cells are shown. Scale bar, 4 µm. (F) The indicated cells expressing Idh1-GFP were continuously cultured in SML-Ura medium and collected at 24 hr (growing phase) and 48 hr (stationary phase). Atg32/Far8-Atg32 status and Idh1-GFP processing were analyzed by WB with anti-Atg32 and anti-GFP antibodies, respectively. WB experiments were independently replicated three (C and F) or four times (D).
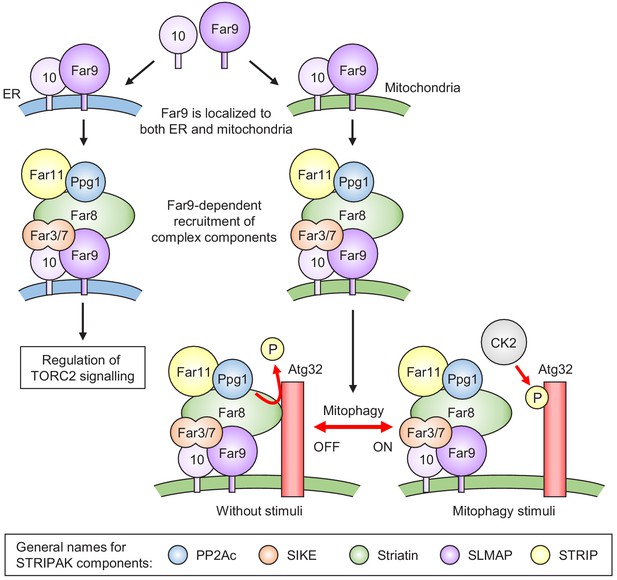
Model for the phosphoregulatory mechanism of Atg32.
Far9 is localized to both the ER and mitochondria, where the other Far complex components are assembled dependently on Far9. According to this localization pattern, the Far complex plays distinct roles in the regulation of TORC2 signaling at the ER and the regulation of mitophagy at the mitochondria. Without stimuli, the mitochondria-localized Far complex mediates the Ppg1-dependent Atg32 dephosphorylation via interaction with Atg32. Upon mitophagy stimuli, the interaction between Atg32 and the Far complex is impaired, allowing Atg32 to be phosphorylated by CK2. The general names for the common STRIPAK complex components are shown in the box.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (S. cerevisiae) | SEY6210 | DOI: 10.1128/mcb.8.11.4936 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 lys2-801 GAL | |
| Strain, strain background (S. cerevisiae) | YAI4 | This study | SEY6210 natNT::PCUP1-GFP-FAR9 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI19 | This study | SEY6210 FAR8-GFP::TRP1 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI20 | This study | SEY6210 FAR11-GFP::TRP1 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI21 | This study | SEY6210 FAR3-GFP::TRP1 SEC63-mCherry::hphNT Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI22 | This study | SEY6210 FAR7-GFP::TRP1 SEC63-mCherry::hphNT Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI23 | This study | SEY6210 FAR8-GFP::TRP1 SEC63-mCherry::hphNT Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI24 | This study | SEY6210 natNT::PCUP1-GFP-FAR9 SEC63-mCherry::hphNT Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI25 | This study | SEY6210 natNT::PCUP1-GFP-FAR10 SEC63-mCherry::hphNT Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI26 | This study | SEY6210 FAR11-GFP::TRP1 SEC63-mCherry::hphNT Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF179 | This study | SEY6210 far3::loxP-LEU2-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF180 | This study | SEY6210 far7::loxP-LEU2-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF181 | This study | SEY6210 far8::loxP-LEU2-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF182 | This study | SEY6210 far9::loxP-LEU2-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF183 | This study | SEY6210 far10::loxP-LEU2-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF184 | This study | SEY6210 far11::loxP-LEU2-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI27 | This study | SEY6210 natNT::PCUP1-GFP-FAR9ΔTA::TRP1 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI28 | This study | SEY6210 natNT::PCUP1-GFP-FAR10ΔTA::TRP1 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | TKYM307 | DOI: 10.1016/j.celrep.2018.05.064 | SEY6210 IDH1-GFP::TRP1 | |
| Strain, strain background (S. cerevisiae) | YKF170 | This study | SEY6210 IDH1-GFP::TRP1 FAR9ΔTA-3HA::HIS3 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF172 | This study | SEY6210 IDH1-GFP::TRP1 far9::kanMX far10::hphNT Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF175 | This study | SEY6210 IDH1-GFP::TRP1 FAR10ΔTA-3HA::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF176 | This study | SEY6210 IDH1-GFP::TRP1 FAR9ΔTA-3HA::HIS3 FAR10ΔTA-3HA::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF203 | This study | SEY6210 natNT::PCUP1-GFP-FAR9 SEC63-mCherry::hphNT far10::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF204 | This study | SEY6210 natNT::PCUP1-GFP- FAR9-TOM5TA::TRP1 SEC63-mCherry::hphNT Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF205 | This study | SEY6210 natNT::PCUP1-GFP- FAR9-TOM5TA::TRP1 SEC63-mCherry::hphNT far10::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF206 | This study | SEY6210 natNT::PCUP1-GFP- FAR9-CYB5TA::TRP1 SEC63-mCherry::hphNT Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF207 | This study | SEY6210 natNT::PCUP1-GFP- FAR9-CYB5TA::TRP1 SEC63-mCherry::hphNT far10::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF213 | This study | SEY6210 FAR3-GFP::TRP1 FAR9-TOM5TA::HIS3 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF214 | This study | SEY6210 FAR7-GFP::TRP1 FAR9-TOM5TA::HIS3 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF215 | This study | SEY6210 FAR8-GFP::TRP1 FAR9-TOM5TA::HIS3 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF216 | This study | SEY6210 natNT::PCUP1- GFP-FAR9-TOM5TA::TRP1 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF217 | This study | SEY6210 natNT::PCUP1-GFP- FAR10 FAR9-TOM5TA::HIS3 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF218 | This study | SEY6210 FAR11-GFP::TRP1 FAR9-TOM5TA::HIS3 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | BY4741 | DOI:https://doi.org/10.1002/(SICI)1097-0061(19980130)14:2%3C115::AID-YEA204%3E3.0.CO;2-2 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | |
| Strain, strain background (S. cerevisiae) | BY tsc11-1 | EUROSCARF (http://euroscarf.de/) | Y41093 | BY4741 tsc11-1::kanMX |
| Strain, strain background (S. cerevisiae) | YKF220 | This study | BY tsc11-1 natNT::PCUP1-GFP-FAR9 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF221 | This study | BY tsc11-1 natNT::PCUP1- GFP-FAR9-TOM5TA::TRP1 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF222 | This study | BY tsc11-1 natNT::PCUP1- GFP-FAR9-CYB5TA::TRP1 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF223 | This study | BY tsc11-1 far9::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF224 | This study | BY tsc11-1 natNT::PCUP1- GFP-FAR9ΔTA::HIS3 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | TKYM80 | DOI: 10.1016/j.devcel.2009.06.014 | SEY6210 IDH1-GFP::TRP1 atg1::HIS3 | |
| Strain, strain background (S. cerevisiae) | YKF29 | DOI: 10.1016/j.celrep.2018.05.064 | SEY6210 IDH1-GFP::TRP1 ppg1::kanMX | |
| Strain, strain background (S. cerevisiae) | YKF30 | DOI: 10.1016/j.celrep.2018.05.064 | SEY6210 IDH1-GFP::TRP1 natNT::PTEF-3HA-PPG1 | |
| Strain, strain background (S. cerevisiae) | YKF76 | DOI: 10.1016/j.celrep.2018.05.064 | SEY6210 IDH1-GFP::TRP1 far9::kanMX | |
| Strain, strain background (S. cerevisiae) | YKF225 | This study | SEY6210 IDH1-GFP::TRP1 FAR9-TOM5TA::HIS3 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF226 | This study | SEY6210 IDH1-GFP::TRP1 FAR9-TOM5TA::HIS3 ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF227 | This study | SEY6210 IDH1-GFP::TRP1 natNT::PTEF-3HA-PPG1 FAR9-TOM5TA::HIS3 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF228 | This study | SEY6210 IDH1-GFP::TRP1 FAR9-CYB5TA::HIS3 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF85 | This study | SEY6210 IDH1-GFP::TRP1 FAR3-3HA::HIS3 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF86 | This study | SEY6210 IDH1-GFP::TRP1 FAR7-3HA::HIS3 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF87 | This study | SEY6210 IDH1-GFP::TRP1 FAR8-3HA::HIS3 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF90 | This study | SEY6210 IDH1-GFP::TRP1 FAR11-3HA::HIS3 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF190 | This study | SEY6210 ppg1::kanMX far3::loxP far7::loxP far8::loxP far9::loxP-LEU2-loxP far10:: loxP-HIS3-loxP far11::loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF191 | This study | SEY6210 ppg1::kanMX far3::loxP far7::loxP far8::loxP natNT::PCUP1-GFP-FAR9 far10::loxP-HIS3-loxP far11::loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF192 | This study | SEY6210 ppg1::kanMX far3::loxP far7::loxP far8::loxP far9::loxP-LEU2-loxP natNT::PCUP1-GFP- FAR10 far11::loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF193 | This study | SEY6210 far3::loxP-LEU2:: loxP ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF194 | This study | SEY6210 far7::loxP-LEU2:: loxP ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF195 | This study | SEY6210 far8::loxP-LEU2::loxP ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF196 | This study | SEY6210 far11::loxP-LEU2::loxP ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF197 | This study | SEY6210 natNT::PCUP1-GFP-FAR9 ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF198 | This study | SEY6210 natNT::PCUP1-GFP-FAR10 ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF200 | This study | SEY6210 TPD3-3HA::hphNT Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF230 | This study | SEY6210 TPD3-3HA::hphNT ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF231 | This study | SEY6210 TPD3-3HA::hphNT ppg1::kanMX far11::loxP-LEU2-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF233 | This study | SEY6210 tpd3::natNT ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI76 | This study | SEY6210 FAR3-3HA::hphNT FAR7-3HA::HIS3 natNT::PCUP1-GFP-FAR9 Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI115 | This study | SEY6210 FAR3-3HA::hphNT FAR7-3HA::HIS3 natNT::PCUP1-GFP-FAR9 far8::loxP-LEU2-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI116 | This study | SEY6210 FAR3-3HA::hphNT FAR7-3HA::HIS3 natNT::PCUP1- GFP-FAR9 ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | BY4742 | DOI:https://doi.org/10.1002/(SICI)1097-0061(19980130)14:2%3C115::AID-YEA204%3E3.0.CO;2-2 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | |
| Strain, strain background (S. cerevisiae) | BY pph21Δ | Thermo Fisher Scientific | 13831 | BY4742 pph21::kanMX |
| Strain, strain background (S. cerevisiae) | BY pph22Δ | Thermo Fisher Scientific | 13886 | BY4742 pph22::kanMX |
| Strain, strain background (S. cerevisiae) | BY pph3Δ | Thermo Fisher Scientific | 14010 | BY4742 pph3::kanMX |
| Strain, strain background (S. cerevisiae) | BY sit4Δ | Thermo Fisher Scientific | 13744 | BY4742 sit4::kanMX |
| Strain, strain background (S. cerevisiae) | BY ppg1Δ | Thermo Fisher Scientific | 15407 | BY4742 ppg1::kanMX |
| Strain, strain background (S. cerevisiae) | BY far8Δ | Thermo Fisher Scientific | 10604 | BY4742 far8::kanMX |
| Strain, strain background (S. cerevisiae) | BY far11Δ | Thermo Fisher Scientific | 12949 | BY4742 far11::kanMX |
| Strain, strain background (S. cerevisiae) | YAI58 | This study | SEY6210 FAR3-GFP::TRP1 SEC63-mCherry::hphNT ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI59 | This study | SEY6210 FAR7-GFP::TRP1 SEC63-mCherry::hphNT ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI60 | This study | SEY6210 FAR8-GFP::TRP1 SEC63-mCherry::hphNT ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI61 | This study | SEY6210 natNT::PCUP1-GFP-FAR9 SEC63-mCherry::hphNT ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI62 | This study | SEY6210 natNT::PCUP1-GFP-FAR10 SEC63-mCherry::hphNT ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI63 | This study | SEY6210 FAR11-GFP::TRP1 SEC63-mCherry::hphNT ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF235 | This study | SEY6210 FAR11-GFP::TRP1 FAR9-TOM5TA::HIS3 SEC63-mCherry::hphNT ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF26 | DOI: 10.1016/j.celrep.2018.05.064 | SEY6210 ppg1::kanMX | |
| Strain, strain background (S. cerevisiae) | TKYM139 | DOI: 10.1016/j.devcel.2009.06.014 | SEY6210 atg32::LEU2 | |
| Strain, strain background (S. cerevisiae) | YKF57 | This study | SEY6210 atg32::LEU2 ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI70 | This study | SEY6210 atg32::LEU2 ppg1::kanMX far3::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI71 | This study | SEY6210 atg32::LEU2 ppg1::kanMX far7::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI72 | This study | SEY6210 atg32::LEU2 ppg1::kanMX far8::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI73 | This study | SEY6210 atg32::LEU2 ppg1::kanMX far9::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI74 | This study | SEY6210 atg32::LEU2 ppg1::kanMX far10::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YAI75 | This study | SEY6210 atg32::LEU2 ppg1::kanMX far11::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF35 | DOI: 10.1016/j.celrep.2018.05.064 | SEY6210 atg11::LEU2 atg32::HIS3 ppg1::kanMX | |
| Strain, strain background (S. cerevisiae) | YKF74 | DOI: 10.1016/j.celrep.2018.05.064 | SEY6210 IDH1-GFP::TRP1 far8::kanMX | |
| Strain, strain background (S. cerevisiae) | YKF160 | This study | SEY6210 IDH1-GFP::TRP1 far8::kanMX ppg1::natNT Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF260 | This study | SEY6210 FAR11-GFP::TRP1 far8:: loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | TKYM312 | DOI: 10.1016/j.celrep.2018.05.064 | SEY6210 IDH1-GFP::TRP1 atg32::LEU2 | |
| Strain, strain background (S. cerevisiae) | YKF261 | This study | SEY6210 IDH1-GFP::TRP1 atg32::LEU2 ppg1::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF262 | This study | SEY6210 IDH1-GFP::TRP1 atg32::LEU2 far8::kanMX Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF263 | This study | SEY6210 IDH1-GFP::TRP1 atg32::LEU2 far8::kanMX ppg1::natNT Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF264 | This study | SEY6210 IDH1-GFP::TRP1 atg32::LEU2 far8::kanMX far3::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF265 | This study | SEY6210 IDH1-GFP::TRP1 atg32::LEU2 far8::kanMX far7::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF266 | This study | SEY6210 IDH1-GFP::TRP1 atg32::LEU2 far8::kanMX far9::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF267 | This study | SEY6210 IDH1-GFP::TRP1 atg32::LEU2 far8::kanMX far10::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (S. cerevisiae) | YKF268 | This study | SEY6210 IDH1-GFP::TRP1 atg32::LEU2 far8::kanMX far11::loxP-HIS3-loxP Stocked in T Kanki lab. | |
| Strain, strain background (E. coli) | BL21-CodonPlus (DE3)-RIL | Agilent | Cat# 230245 | B F– ompT hsdS(rB– mB–) dcm+ Tetr gal λ(DE3) endA Hte [argU ileY leuW Camr] |
| Antibody | anti-GFP (Mouse monoclonal) | Takara Bio | Cat# 632380, RRID:AB_10013427 | WB (1:5000) |
| Antibody | anti-HA (Mouse monoclonal) | Sigma-Aldrich | Cat# H9658, RRID:AB_260092 | WB (1:2500) |
| Antibody | anti-Pgk1 (Mouse monoclonal) | Thermo Fisher Scientific | Cat# 459250, RRID:AB_ 2532235 | WB (1:5000) |
| Antibody | anti-mouse IgG (Goat polyclonal, Peroxidase conjugated) | Merck Millipore | Cat# AP124P, RRID:AB_90456 | WB (1:10000) |
| Antibody | anti-rabbit IgG (Goat polyclonal, Peroxidase conjugated) | Jackson ImmunoResearch | Cat# 111-035-003, RRID:AB_2313567 | WB (1:10000) |
| Antibody | anti-Atg32 (Rabbit polyclonal) | DOI: 10.1091/mbc.E11-02-0145 | WB (1:2500) | |
| Antibody | anti-Ppg1 (Rabbit polyclonal) | This study | WB (1:1000) Stocked in T Kanki lab. | |
| Antibody | anti-Far8 (Rabbit polyclonal) | This study | WB (1:1000) Stocked in T Kanki lab. | |
| Antibody | anti-Far9 (Rabbit polyclonal) | This study | WB (1:1000) Stocked in T Kanki lab. | |
| Antibody | anti-Far11 (Rabbit polyclonal) | This study | WB (1:1000) Stocked in T Kanki lab. | |
| Recombinant DNA reagent | pCu416 (plasmid) | DOI: 10.1016/s0076- 6879(99)06010-3 | CEN/ARS URA3 PCUP1 | |
| Recombinant DNA reagent | pCu416-PPG1 (plasmid) | DOI: 10.1016/j.celrep.2018.05.064 | CEN/ARS URA3 PCUP1-PPG1 | |
| Recombinant DNA reagent | pCu416-FLAG- His6-PPG1 (plasmid) | DOI: 10.1016/j.celrep.2018.05.064 | CEN/ARS URA3 PCUP1- FLAG-His6-PPG1 | |
| Recombinant DNA reagent | pCu416-FLAG- His6-PPG1H111N (plasmid) | This study | CEN/ARS URA3 PCUP1- FLAG-His6-PPG1H111N Stocked in T Kanki lab. | |
| Recombinant DNA reagent | pCu414-FAR3-3HA (plasmid) | This study | CEN/ARS TRP1 PCUP1-FAR3-3HA Stocked in T Kanki lab. | |
| Recombinant DNA reagent | pCu414-FAR7-3HA (plasmid) | This study | CEN/ARS TRP1 PCUP1-FAR7-3HA Stocked in T Kanki lab. | |
| Recombinant DNA reagent | pCu414- FAR8-3HA (plasmid) | This study | CEN/ARS TRP1 PCUP1-FAR8-3HA Stocked in T Kanki lab. | |
| Recombinant DNA reagent | pCu414- FAR11-3HA (plasmid) | This study | CEN/ARS TRP1 PCUP1-FAR11-3HA Stocked in T Kanki lab. | |
| Recombinant DNA reagent | pCu416- 3HA-ATG32 (plasmid) | This study | CEN/ARS URA3 PCUP1-3HA-ATG32 Stocked in T Kanki lab. | |
| Recombinant DNA reagent | pCu416-3HA- ATG32-2SA (plasmid) | This study | CEN/ARS URA3 PCUP1-3HA-ATG32S114A/S119A Stocked in T Kanki lab. | |
| Recombinant DNA reagent | pCu416-3HA- ATG32Δ151–200 (plasmid) | This study | CEN/ARS URA3 PCUP1-3HA-ATG32Δ151–200 Stocked in T Kanki lab. | |
| Recombinant DNA reagent | pCu416-ATG32 (plasmid) | DOI: 10.1091/mbc.E11-02-0145 | CEN/ARS URA3 PCUP1-ATG32 | |
| Recombinant DNA reagent | pCu416- FAR8-ATG32 (plasmid) | This study | CEN/ARS URA3 PCUP1-FAR8-ATG32 Stocked in T Kanki lab. | |
| Recombinant DNA reagent | pRS416-ATG32 (plasmid) | DOI: 10.1016/j.devcel.2009.06.014 | CEN/ARS URA3 PATG32-ATG32 | |
| Recombinant DNA reagent | pRS416- FAR8-ATG32 (plasmid) | This study | CEN/ARS URA3 PATG32-FAR8-ATG32 Stocked in T Kanki lab. | |
| Recombinant DNA reagent | pPROEX-HTb (plasmid) | Invitrogen | Cat# 10711018 | Ampr lacIq Ptrc-His6 |
| Recombinant DNA reagent | pPROEX-FAR8 (plasmid) | This study | Ampr lacIq Ptrc-His6-FAR8 Stocked in T Kanki lab. | |
| Recombinant DNA reagent | pGEX-4T-1 (plasmid) | GE Healthcare | Cat# 28954549 | Ampr lacIq Ptac-GST |
| Recombinant DNA reagent | pGEX-ATG32(N250) (plasmid) | DOI: 10.1091/mbc.E11-02-0145 | Ampr lacIq Ptac-GST-ATG32(N250) | |
| Recombinant DNA reagent | pGEX-ATG32(N200) (plasmid) | This study | Ampr lacIq Ptac-GST-ATG32(N200) Stocked in T Kanki lab. | |
| Recombinant DNA reagent | pGEX-ATG32(N150) (plasmid) | This study | Ampr lacIq Ptac-GST-ATG32(N150) Stocked in T Kanki lab. | |
| Commercial assay or kit | EzWestLumi plus | Atto | Cat# WSE-7120 | |
| Commercial assay or kit | Clarity Max Western ECL Substrate | Bio-Rad | Cat# 1705062 | |
| Commercial assay or kit | anti-FLAG M2 affinity gel | Sigma-Aldrich | Cat# A2220 | |
| Commercial assay or kit | Protein G Sepharose 4 Fast Flow | GE Healthcare | Cat# 17061801 | |
| Commercial assay or kit | Ni Sepharose 6 Fast Flow | GE Healthcare | Cat# 17531801 | |
| Commercial assay or kit | Glutathione Sepharose 4 Fast Flow | GE Healthcare | Cat# 17075601 | |
| Chemical compound, drug | MitoTracker Red CMXRos | Thermo Fisher Scientific | Cat# M7512 | (50 nM) |
| Chemical compound, drug | Rapamycin | LC Laboratories | Cat# R-5000 | (100 nM) |
| Software, algorithm | Image Lab | Bio-Rad | ||
| Software, algorithm | MetaMorph 7 | Molecular Devices |





