Inducible mechanisms of disease tolerance provide an alternative strategy of acquired immunity to malaria
Figures
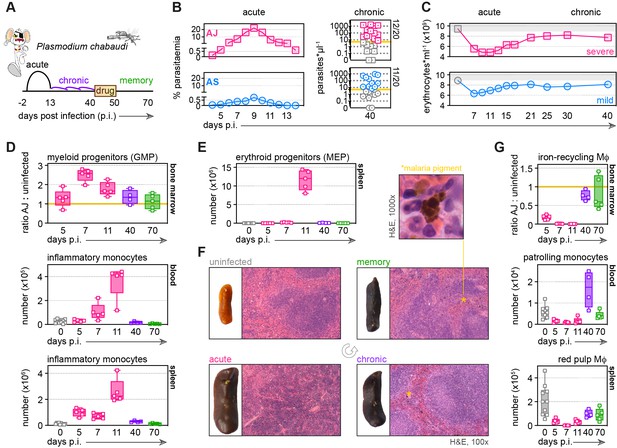
Malaria triggers emergency myelopoiesis and obliterates tissue-resident macrophages.
(A) C57Bl/6 mice were infected with Plasmodium chabaudi AJ or AS sporozoites; the blood-stage of infection started 2 days later after the release of merozoites from the liver. Mice were chronically infected for 40 days, at which point we administered the antimalarial drug chloroquine. Memory responses were assessed 30 days thereafter. Note that we exclusively use days post infection (p.i.) to refer to the blood-stage of malaria. (B) Acute parasitaemia was monitored daily using Giemsa stained thin blood films and chronic infection was verified 40 days p.i. by qPCR (n = 20 per group). Symbols below the limit of detection (5 parasites*μl−1) are coloured grey and these mice were excluded from the study. (C) The mean number of erythrocytes*ml−1 is shown before (grey symbols) and during infection (n = 10 for AJ and n = 14 for AS). Severe anaemia is defined as >50% loss of red cells. (D and E) Inflammatory monocytes and progenitor cells (granulocyte monocyte progenitors [GMP] and megakaryocyte erythroid progenitors [MEP]) from uninfected mice (0 days p.i.), AJ-infected mice (5, 7, 11, and 40 days p.i.) and once-infected mice (memory, 70 days p.i.) were analysed by flow cytometry (n = 4–5 per time point, box-plots show median and IQR). Uninfected age-matched controls were analysed at each time point and pooled for graphing (n = 10); absolute counts are shown for blood and spleen. In (D), GMP are shown as a ratio of infected:uninfected at each time point because bone marrow cellularity increases with age. (F) Paraffin-embedded spleen sections were H&E stained (11 days p.i. for acute AJ infection) – examples of malaria pigment in chronically infected and once-infected mice are marked with an asterisk. (G) Tissue-resident macrophages (Mɸ) and patrolling monocytes from uninfected mice, AJ-infected mice, and once-infected mice were analysed by flow cytometry (n = 4–5 per time point, box-plots show median and IQR). Absolute counts (blood and spleen) and cell ratios (bone marrow) are shown exactly as described for (D and E). See Supplementary file 1 for all antibody panels and gating strategies.
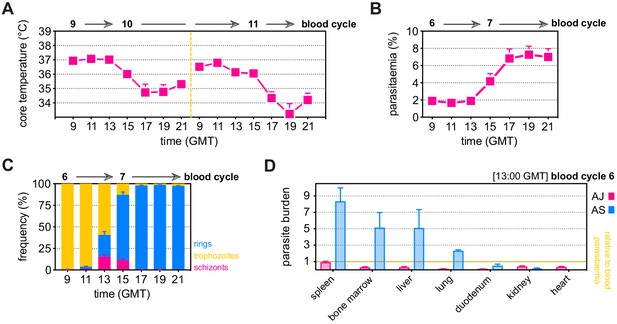
P. chabaudi AJ causes severe disease without sequestering in host tissues.
(A–C) C57Bl/6 mice were infected with P. chabaudi AJ sporozoites; the blood-stage of infection started 2 days later after the release of merozoites from the liver. We refer to the emergence of parasites from the liver as the start of blood cycle 1, after which parasites undergo schizogony approximately every 24 hr to start the next blood cycle. Mice were housed under reverse light conditions (lights OFF 07:00, lights ON 19:00 GMT) so that schizogony would peak at 13:00 GMT. (A) Core body temperature was measured every 2 hr (09:00–21:00 GMT) as parasites transitioned from blood cycle 9 to 10 and again from blood cycle 10 to 11 (n = 20, mean + SEM). (B and C) Parasitaemia was monitored every 2 hr by Giemsa stained thin blood films (09:00–21:00 GMT) as parasites transitioned from blood cycle 6 to 7 (n = 9, mean + SEM). The percentage of infected red cells is shown in (B), whereas the proportion of parasites at the ring, trophozoite, and schizont stages is shown in (C). Note that hypothermia is most severe after the peak of schizogony when all schizonts have ruptured. (D) C57Bl/6 mice were infected with mosquito-transmitted blood-stage parasites (P. chabaudi AS or AJ). Circulating parasitaemia was measured at the peak of schizogony (13:00 GMT) as parasites transitioned from blood cycle 6 to 7 (n = 3 per group, mean + SEM). At the same time, mice were euthanised and organs were fixed in neutral buffered formalin for histology. Sequestration rates in each organ were assessed by counting the percentage of infected red cells inside blood vessels and normalising this number to circulating parasitaemia (see Materials and methods). For high-resolution images and a guide to identifying infected red cells on tissue sections, you can refer to our publication on the sequestration and histopathology of P. chabaudi (Brugat et al., 2014).
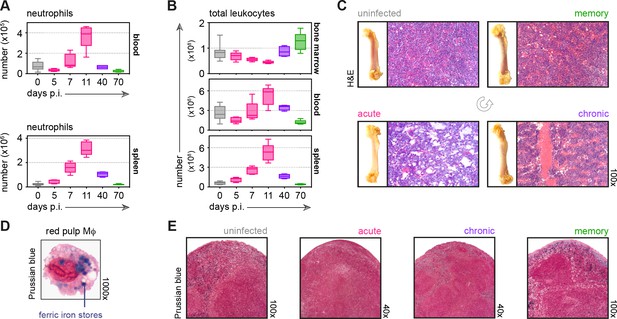
Malaria causes major disturbances in tissue structure and integrity.
(A and B) Neutrophils (A) and total leukocytes (B) from uninfected mice (0 days p.i.), AJ-infected mice (5, 7, 11, and 40 days p.i.) and once-infected mice (memory, 70 days p.i.) were analysed by flow cytometry (n = 4–5 per time point, box-plots show median and IQR). Uninfected age-matched controls were analysed at each time point and pooled for graphing (n = 10). (C) Paraffin-embedded femur sections were H&E stained (11 days p.i. for acute AJ infection) – note that during acute infection bones appear translucent, as cellularity drops. (D) Red pulp macrophages (Mɸ) were flow-sorted from uninfected control mice and stained with Prussian Blue (intracellular ferric iron stores) and Neutral Red (nuclei). (E) Paraffin-embedded spleen sections were stained with Prussian Blue and Neutral Red (11 days p.i. for acute AJ infection). See Supplementary file 1 for all antibody panels and gating strategies.
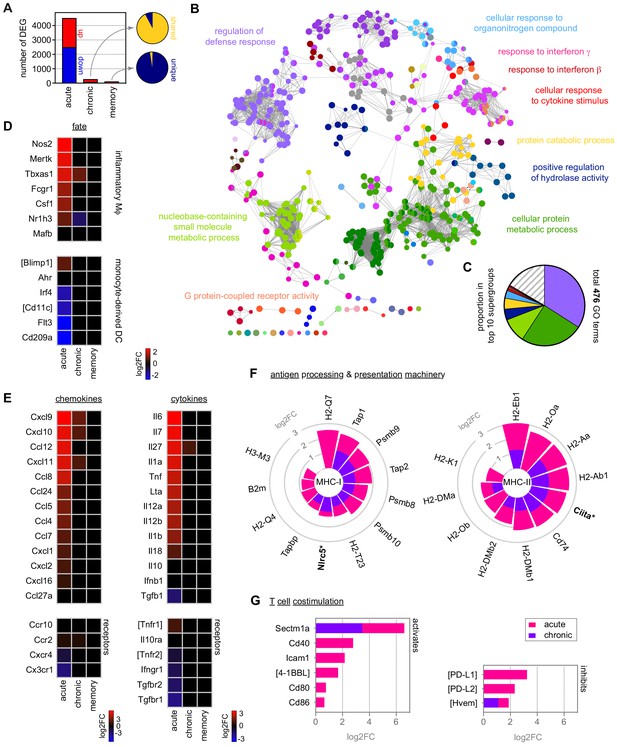
Monocytes differentiate into inflammatory macrophages in naive hosts but become quiescent in chronic infection.
(A) RNA sequencing of spleen monocytes flow-sorted from AJ-infected mice (7 and 40 days p.i.for acute and chronic, respectively) and once-infected mice (memory, 70 days p.i.). At each time point, the number of differentially expressed genes (DEG, padj <0.01, >1.5-fold change) was assessed relative to uninfected controls. Pies show the proportion of shared or unique DEG between chronic and acute infection (top) or memory and chronic infection (bottom). (B and C) ClueGO network of DEG in spleen monocytes during acute infection. Each node represents a significantly enriched gene ontology (GO) term and node size is determined by padj. Related GO terms that share >40% of genes are connected by a line and organised into functional groups (each given a unique colour). Supergroups are formed when GO terms are shared between more than one group. The names of the top 10 (super)groups (lowest padj) are displayed and (C) shows their proportion of total GO terms. (D–G) Log2FC of (D) signature genes used to predict monocyte differentiation into inflammatory macrophages (Mɸ) or monocyte-derived dendritic cells (DC); (E) chemokines, cytokines and their receptors; (F) class I and class II antigen processing and presentation pathways (inc MHC transactivators*); and (G) T cell co-stimulation and inhibitory ligands. At each time point, log2FC is shown relative to uninfected controls. Square brackets indicate that common gene names were used. At each time point in (A–G), n = 5–6 for infected mice and n = 6–7 for uninfected controls.
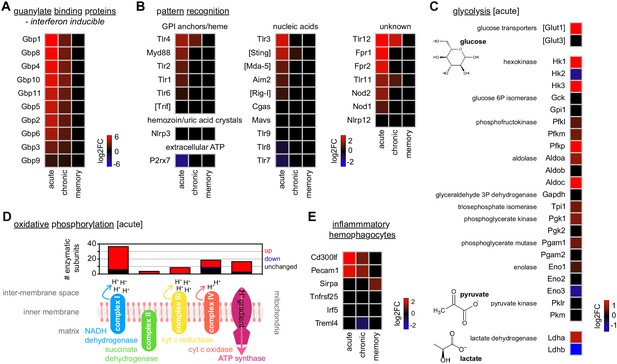
Monocytes upregulate glycolysis and oxidative phosphorylation as they differentiate into inflammatory macrophages.
(A and B) RNA sequencing of spleen monocytes flow-sorted from AJ-infected mice (7 and 40 days p.i.for acute and chronic, respectively) and once-infected mice (memory, 70 days p.i.). Shown is the log2 fold change (FC) of (A) interferon-inducible guanylate binding proteins and (B) pattern recognition receptors (PRR). At each time point, log2FC is shown relative to uninfected controls. If parasite- or host-derived ligands have been identified for PRR in malaria these ligands are labelled. (C) Log2FC of the major glucose transporters and glycolytic enzymes in spleen monocytes flow-sorted from AJ-infected mice (acute, 7 days p.i.) – data are shown relative to uninfected controls. Note that under anaerobic conditions pyruvate can be further converted to lactate by lactate dehydrogenase, which is also shown. (D) Transcriptional regulation of oxidative phosphorylation in spleen monocytes flow-sorted from AJ-infected mice (acute, 7 days p.i.). Data show the number of enzymatic subunits that are transcriptionally up- or downregulated compared to uninfected controls (padj <0.01, >1.5-fold change) – all subunits that are required to form complex I to IV in the electron transport chain and ATP synthase are shown. (E) Log2FC of signature genes used to predict monocyte differentiation into inflammatory hemophagocytes; data are shown relative to uninfected controls. At each time point in (A–E), n = 5–6 for infected mice and n = 6–7 for uninfected controls. Square brackets indicate that common gene names were used.
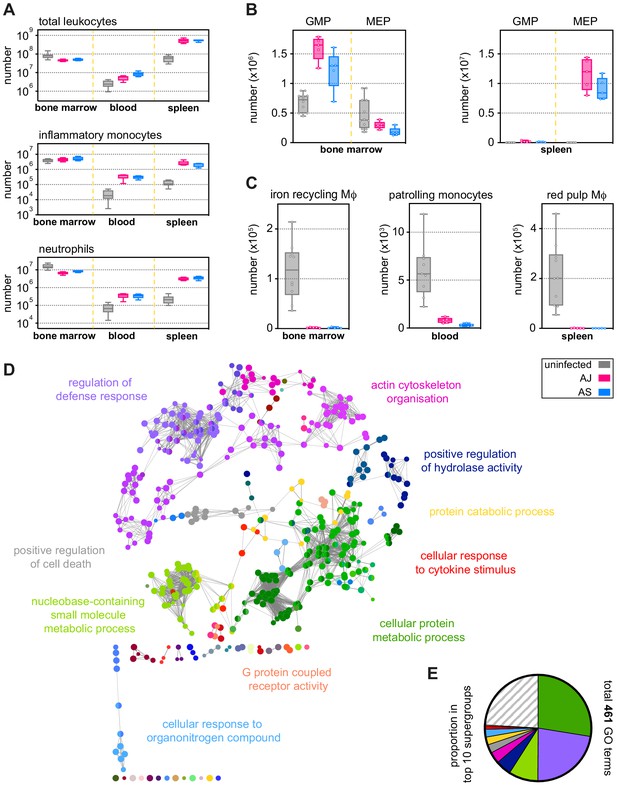
Parasite genotype does not influence the emergency myeloid response to malaria.
(A–C) Inflammatory and patrolling monocytes, neutrophils, progenitors, and tissue-resident macrophages (Mɸ) from uninfected mice (n = 10), AJ-infected mice (n = 5), and AS-infected mice (n = 5) were analysed by flow cytometry (box-plots show median and IQR). Data shown represent the peak of the acute phase response, which is (A) 11 days p.i. for inflammatory monocytes and neutrophils in bone marrow, blood, and spleen; (B) 7 days p.i. for granulocyte monocyte progenitors (GMP) and megakaryocyte erythroid progenitors (MEP) in bone marrow and 11 days p.i. in the spleen; and (C) 7 days p.i. for tissue-resident Mɸ and patrolling monocytes. (D and E) ClueGO network of differentially expressed genes (DEG) in spleen monocytes flow-sorted from AS-infected mice (acute, 7 days p.i., n = 5). DEG were called relative to uninfected controls (n = 7). Each node represents a significantly enriched gene ontology (GO) term and node size is determined by padj. Related GO terms that share >40% of genes are connected by a line and organised into functional groups (each given a unique colour). Supergroups are formed when GO terms are shared between more than one group. The names of the top 10 (super)groups (lowest padj) are displayed and (E) shows their proportion of total GO terms. See Supplementary file 1 for all antibody panels and gating strategies.
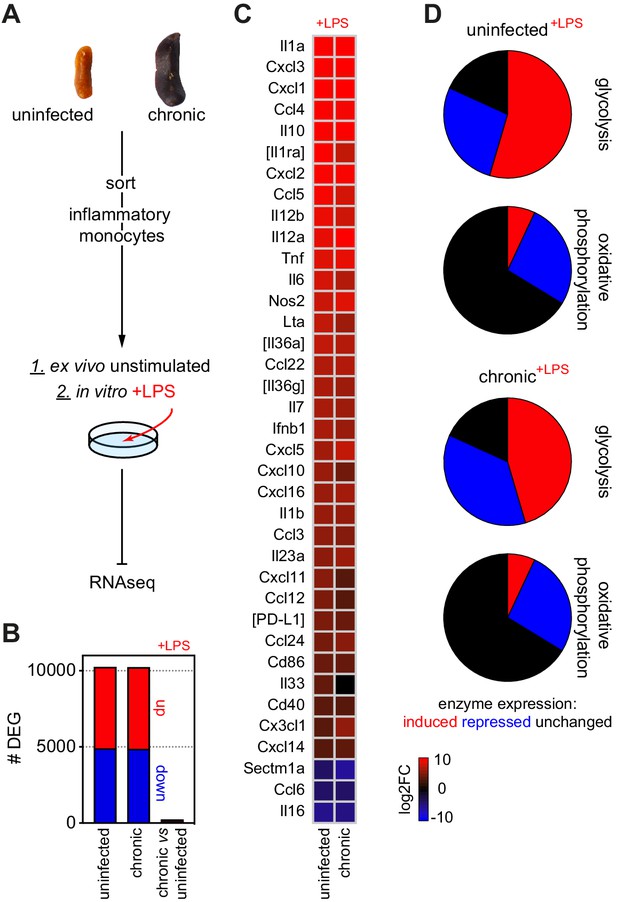
Quiescent monocytes can differentiate into inflammatory macrophages when removed from the spleen.
(A) Spleen monocytes were flow-sorted from uninfected mice (n = 4) or mice chronically infected with P. chabaudi AJ (40 days p.i., n = 5). Sorted cells were then prepared immediately for RNA sequencing (ex vivo) or stimulated in vitro with LPS for 4 hr before RNA sequencing. (B) The number of differentially expressed genes (DEG, padj <0.01, >1.5-fold change) between LPS-stimulated and unstimulated (ex vivo) monocytes is shown for uninfected mice (left bar) and chronically infected mice (centre bar). Note that a direct comparison between LPS-stimulated monocytes from chronically infected mice and uninfected controls yielded only 229 DEG (right bar). (C) Log2 fold change (FC) of inflammatory markers showing the differentiation of monocytes in vitro after LPS stimulation (data shown relative to ex vivo samples). Square brackets indicate that common gene names were used. (D) Transcriptional regulation of glycolysis and oxidative phosphorylation after LPS stimulation (relative to ex vivo samples). Data include the major glucose transporters and glycolytic enzymes, and all enzymatic subunits that form complex I to IV in the electron transport chain and ATP synthase.
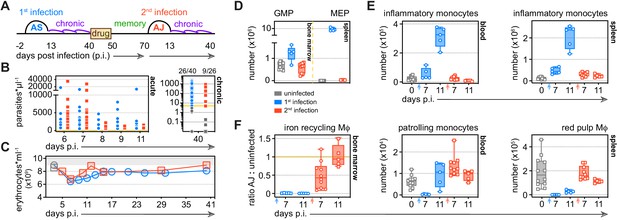
Tolerance persists to protect host tissues during reinfection.
(A) Malaria reinfection model: C57Bl/6 mice were first infected with P. chabaudi AS sporozoites. Chronic infection was confirmed by qPCR at 40 days p.i. and drug-treated with the antimalarial drug chloroquine. Thirty days thereafter mice were infected for a second time with P. chabaudi AJ. (B) Circulating parasite density was calculated using percentage parasitaemia (limit of detection 0.01%) and red cell counts, and is presented as the number of parasites*μl−1 throughout the acute phase of first and second infection. No statistically significant difference was detected at any timepoint (padj <0.05, Mann-Whitney test corrected for multiple comparisons using Holm-Šidák method). Chronic infection was verified 40 days p.i. by qPCR (n = 40 in 1st infection and n = 26 in 2nd infection) and symbols below the limit of detection (5 parasites*μl−1) are coloured grey; these mice were excluded from the study. (C) The mean number of erythrocytes*ml−1 is shown before (grey symbols) and during first and second infection (n = 14 per group). (D–F) Inflammatory and patrolling monocytes, progenitors and tissue-resident macrophages (Mɸ) from mice experiencing their first (n = 5 per time point) or second (n = 5–10 per time point) infection were analysed by flow cytometry (box-plots show median and IQR). Uninfected age-matched controls were analysed at each time point and pooled for graphing (n = 12); absolute counts are shown except for tissue-resident Mɸ in the bone marrow. In this case, data are presented as a ratio of infected:uninfected at each time point. In (D), granulocyte monocyte progenitors (GMP) and megakaryocyte erythroid progenitors (MEP) are shown 11 days p.i. See Supplementary file 1 for all antibody panels and gating strategies.
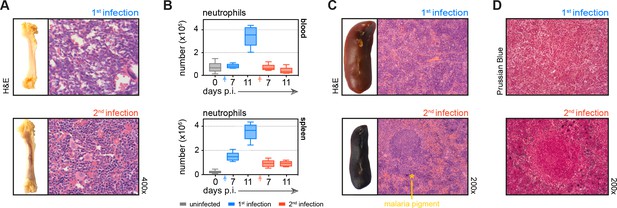
Tissue architecture is preserved and key homeostatic processes are maintained in tolerised hosts.
(A) Paraffin-embedded femur sections were H&E stained during the acute phase of first or second infection (11 days p.i.). (B) Neutrophils from mice experiencing their first (n = 5 per time point) or second (n = 5–10 per time point) infection were analysed by flow cytometry (box-plots show median and IQR). Uninfected age-matched controls were analysed at each time point and pooled for graphing (n = 12). (C and D) Paraffin-embedded spleen sections were H&E (C) or Prussian Blue/Neutral Red (D) stained during the acute phase of first or second infection (11 days p.i.). Note that malaria pigment deposited during first infection persists and can still be observed during reinfection (marked with an asterisk). See Supplementary file 1 for all antibody panels and gating strategies.
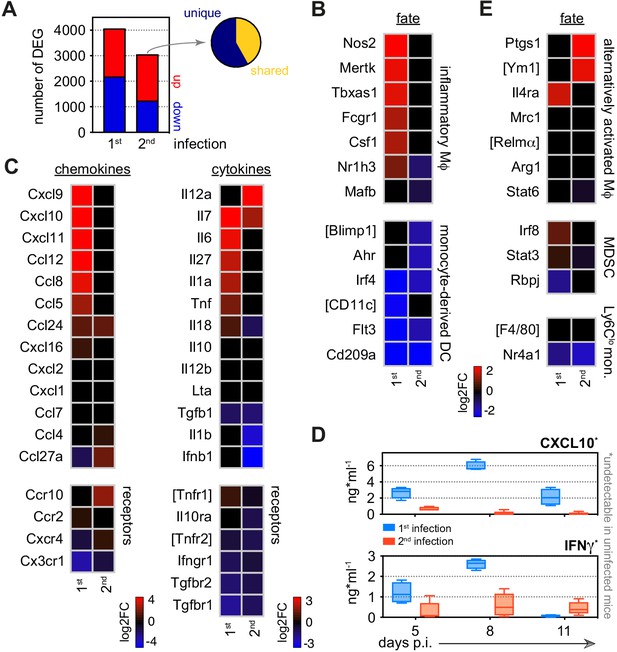
Monocytes minimise inflammation in tolerised hosts.
(A) RNA sequencing of spleen monocytes flow-sorted during the acute phase of first or second infection (7 days p.i.). In each case, the number of differentially expressed genes (DEG, padj <0.01, >1.5-fold change) was assessed relative to uninfected controls. Pie shows the proportion of DEG unique to second infection (i.e. not shared with first infection). (B–C) Log2 fold change (FC) of (B) signature genes used to predict monocyte differentiation into inflammatory macrophages (Mɸ) or monocyte-derived dendritic cells (DC); and (C) chemokines, cytokines, and their receptors. Log2FC is shown relative to uninfected controls. (D) Plasma concentration of CXCL10 and IFNɣ during the acute phase of first (n = 4) or second (n = 5) infection (box-plots show median and IQR). Note that both plasma proteins were below the limit of detection in uninfected controls. (E) Log2FC of signature genes used to predict monocyte differentiation towards alternative fates (log2FC is shown relative to uninfected controls). In (A–C and E), n = 5 for infected mice and n = 6–7 for uninfected controls. Square brackets indicate that common gene names were used.
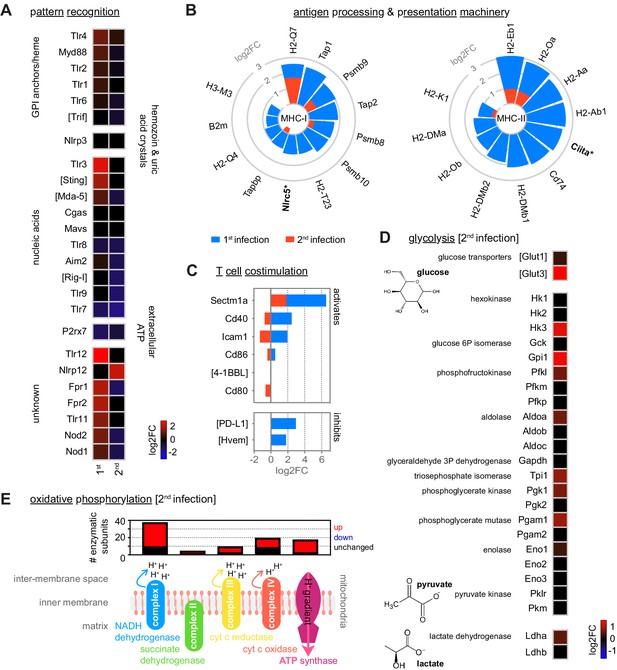
Monocytes minimise inflammation in tolerised hosts.
(A–C) RNA sequencing of spleen monocytes flow-sorted from mice experiencing their first or second infection (acute, 7 days p.i.). Shown is the log2 fold change (FC) of (A) pattern recognition receptors (PRR); (B) class I and class II antigen processing and presentation pathways (inc MHC transactivators*); and (C) T cell co-stimulation and inhibitory ligands. Data are shown relative to uninfected controls. If parasite- or host-derived ligands have been identified for PRR in malaria these ligands are labelled. (D) Log2FC of the major glucose transporters and glycolytic enzymes in spleen monocytes during the acute phase of second infection (7 days p.i.) – data are shown relative to uninfected controls. Note that under anaerobic conditions pyruvate can be further converted to lactate by lactate dehydrogenase, which is also shown. (E) Transcriptional regulation of oxidative phosphorylation in spleen monocytes during the acute phase of second infection (7 days p.i.). Data show the number of enzymatic subunits that are transcriptionally up- or downregulated compared to uninfected controls (padj <0.01, >1.5-fold change) – all subunits that are required to form complex I to IV in the electron transport chain and ATP synthase are shown. In (A–E), n = 5 for infected mice and n = 6–7 for uninfected controls. Square brackets indicate that common gene names were used.
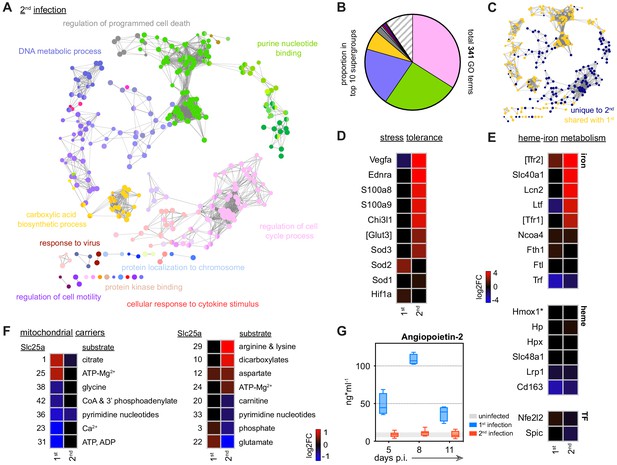
Disease tolerance is established after one malaria episode.
(A and B) ClueGO network of DEG in spleen monocytes during the acute phase of second infection. Each node represents a significantly enriched gene ontology (GO) term and node size is determined by padj. Related GO terms that share >40% of genes are connected by a line and organised into functional groups (each given a unique colour). Supergroups are formed when GO terms are shared between more than one group. The names of the top 10 (super)groups (lowest padj) are displayed and (B) shows their proportion of total GO terms. (C) Recoloured clueGO network indicating the GO terms that are unique to second infection (or shared with first infection). (D–F) Log2 fold change (FC) of genes that (D) promote stress tolerance; (E) regulate heme-iron metabolism; and (F) encode mitochondrial carrier proteins. Log2FC is shown relative to uninfected controls. (G) Plasma concentration of Angiopoietin-2 during the acute phase of first (n = 4) or second (n = 8) infection (box-plots show median and IQR). The grey shaded area represents min. to max. measurements in uninfected controls (n = 3). In (A–F), n = 5 for infected mice and n = 6–7 for uninfected controls. Square brackets indicate that common gene names were used.
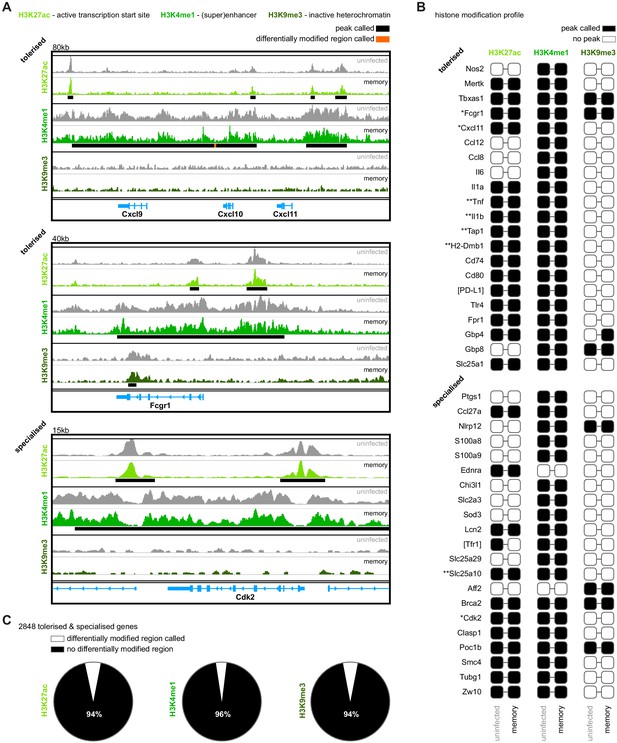
Malaria does not induce epigenetic reprogramming of bone marrow monocytes.
(A) Chromatin immunoprecipitation (ChIP)seq of bone marrow monocytes flow-sorted from once-infected mice (AJ, memory, 70 days p.i.) and uninfected controls. Shown are the Integrative Genomics Viewer (IGV) traces (autoscaled) of three loci encoding genes that are transcriptionally tolerised (upregulated in first but not second infection) or specialised (upregulated only in second infection). Peaks were called relative to non-immunoprecipitated input DNA and are shown for uninfected controls (black line). Regions of the genome that were differentially marked between once-infected mice and uninfected controls are underlined in orange (differentially modified regions). (B) Histone modification profiles of bone marrow monocytes from once-infected mice and uninfected controls. Black squares indicate that a peak was called within 10 kb (H3K27ac and H3K9me3) or 100 kb (H3K4me1) of the transcription start site. Square brackets indicate that common gene names were used. IGV traces are shown in (A) and Figure 6—figure supplement 1 for genes marked with one or two asterisks, respectively. (C) Pies show the proportion of tolerised/specialised genes (n = 2848) annotated with or without a differentially modified region (annotated to the nearest gene). In (A–C), the data shown are pooled from independent biological replicates (see Materials and methods).
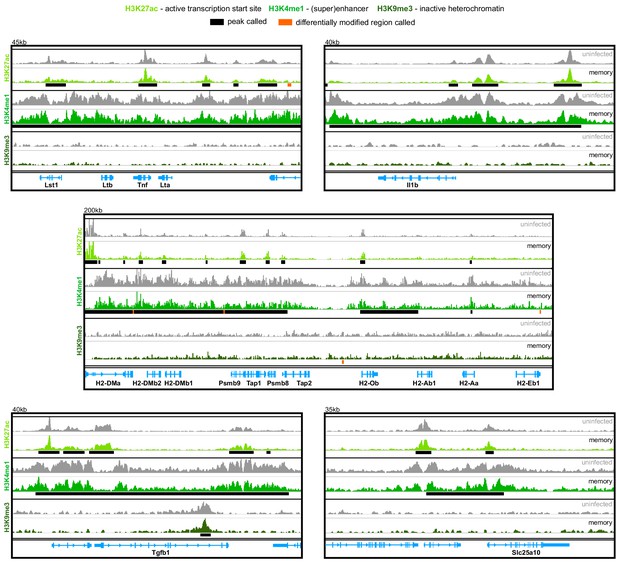
Malaria does not induce epigenetic reprogramming of bone marrow monocytes.
Chromatin immunoprecipitation (ChIP)seq of bone marrow monocytes flow-sorted from once-infected mice (AJ, memory, 70 days p.i.) and uninfected controls. Shown are the Integrative Genomics Viewer (IGV) traces (autoscaled) of five loci encoding immune and metabolic genes. Peaks were called relative to non-immunoprecipitated input DNA and are shown for uninfected controls (black line). Regions of the genome that were differentially marked between once-infected mice and uninfected controls are underlined in orange (differentially modified regions). The data shown are pooled from independent biological replicates (see Materials and methods).
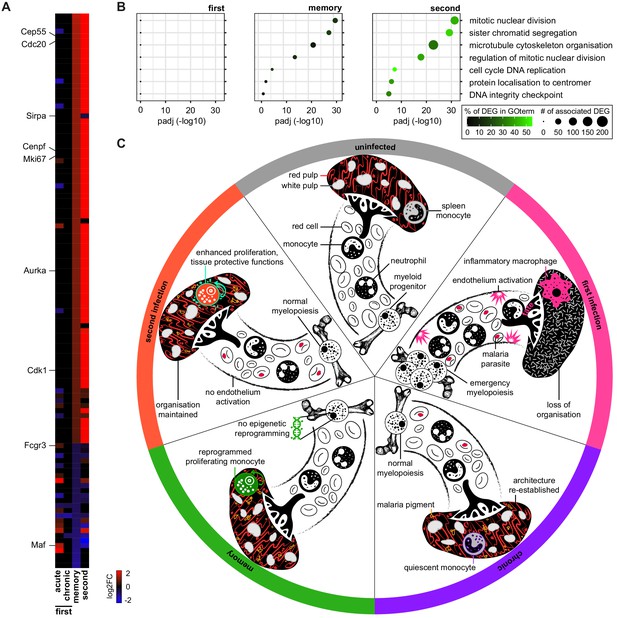
Monocytes are transcriptionally reprogrammed in the remodelled spleen.
(A) RNA sequencing of spleen monocytes flow-sorted from AJ-infected mice (7 and 40 days p.i. for acute and chronic, respectively), once-infected mice (memory, 70 days p.i.), and reinfected mice (acute, 7 days p.i.). The heatmap shows all 111 differentially expressed genes in once-infected mice (DEG, relative to uninfected controls, padj <0.01, >1.5-fold change). (B) GO analysis of DEG in spleen monocytes during first infection (7 days p.i.), memory phase (70 days p.i.) and second infection (7 days p.i.). Mice were infected with P. chabaudi AJ and the top GO terms in once-infected mice are shown. (C) Working model of disease tolerance in malaria, showing the major changes in the myeloid compartment throughout chronic infection, convalescence and reinfection. Note that in the memory phase (one month after drug cure) there is no evidence that monocytes are epigenetically reprogrammed in the bone marrow but they are transcriptionally reprogrammed in the spleen. We therefore propose that the remodelled spleen imprints monocytes with tissue protective functions. In (A and B) n = 5–6 for infected mice and n = 6–7 for uninfected controls. (C) Icons credit: https://thenounproject.com/.
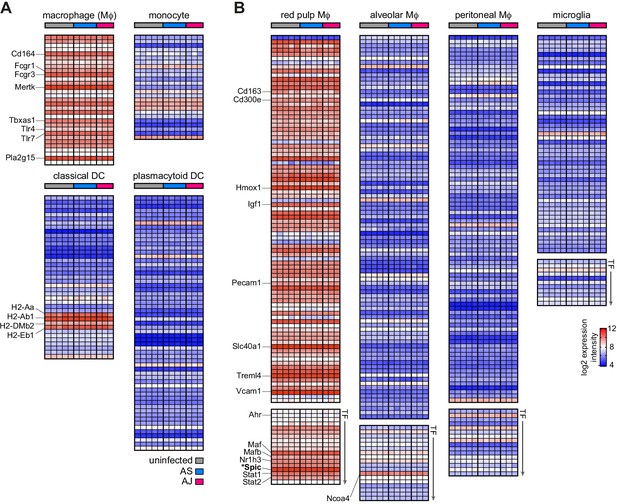
Tissue printing shapes the transcriptional programme of recruited monocytes to meet the needs of the niche.
(A and B) Microarray of red pulp macrophages (Mɸ) flow-sorted from the spleens of once-infected mice (memory, 100 days p.i.) and age-matched uninfected controls. Data are displayed as the RMA (robust multi-array average) normalised log2 expression intensity for each gene and each column represents one mouse. Details of how we curated signature genelists for (A) mononuclear phagocytes (inc dendritic cells, DC) and (B) tissue resident Mɸ are provided in the Materials and methods section. Spic is the master transcription factor (TF) for red pulp Mɸ fate (Kohyama et al., 2009) and is marked with an asterisk. Note that pairwise comparisons between each of the three groups (uninfected, AS, and AJ) revealed zero differentially expressed genes (padj <0.05) (n = 5 for uninfected mice and n = 3–4 for once-infected mice).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus C57Bl/6J female) | C57Bl/6 | The Jackson Laboratory | RRID:IMSR_JAX:000664 | Bred and housed in individually ventilated cages (SPF conditions) at the University of Edinburgh |
| Strain, strain background (Plasmodium chabaudi chabaudi AS) | P. chabaudi AS | The European malaria reagent repository http://www.malariaresearch.eu | Clone 28AS11 | |
| Strain, strain background (Plasmodium chabaudi chabaudi AJ) | P. chabaudi AJ | Clone 96AJ15 | ||
| Strain, strain background (Anopheles stephensi SD500) | Mosquitoes | Reared in-house at the University of Edinburgh | ||
| Antibody | Anti-mouse B220 (rat monoclonal) | Clone RA3-6B2 eBioscience - sold by ThermoFisher | RRID:AB_10717389 | (0.2 μl) per test = 2 million cells in 100 μl volume |
| Antibody | Anti-mouse CD3ε (Armenian hamster monoclonal) | Clone 145–2 C11 BioLegend | RRID:AB_312676 | (0.3 μl) per test |
| Antibody | Anti-mouse CD4 (rat monoclonal) | Clone RM4-5 BioLegend | RRID:AB_312718 | (0.3 μl) per test |
| Antibody | Anti-mouse CD8a (rat monoclonal) | Clone 53–6.7 BioLegend | RRID:AB_312750 | (0.3 μl) per test |
| Antibody | Anti-mouse CD11b (rat monoclonal) | Clone M1/70 BioLegend | RRID:AB_312798 | (0.1 μl) per test |
| Antibody | Anti-mouse CD11c (Armenian hamster monoclonal) | Clone N418 BioLegend | RRID:AB_313776 | (0.15 μl) per test |
| Antibody | Anti-mouse CD16/32 (rat monoclonal) | Clone 93 eBioscience - sold by ThermoFisher | RRID:AB_469598 | (0.5 μl) per test |
| Antibody | TruStain FcX anti-mouse CD16/32 (rat monoclonal) | Clone 93 BioLegend | RRID:AB_1574973 | (2 μl) per test blocks FcɣR II/III prior to antibody staining |
| Antibody | Anti-mouse CD19 (rat monoclonal) | Clone 6D5 BioLegend | RRID:AB_313646 | (0.1 μl) per test |
| Antibody | Anti-mouse CD27 (Armenian hamster monoclonal) | Clone LG.7F9 eBioscience - sold by ThermoFisher | RRID:AB_465614 | (0.3 μl) per test |
| Antibody | Anti-mouse CD34 (rat monoclonal) | Clone RAM34 eBioscience - sold by ThermoFisher | RRID:AB_465021 | (0.4 μl) per test |
| Antibody | Anti-mouse CD71 (rat monoclonal) | Clone RI7217 BioLegend | RRID:AB_10899739 | (0.3 μl) per test |
| Antibody | Anti-mouse CD115/Csf1r (rat monoclonal) | Clone AFS98 BioLegend | RRID:AB_2562760 | (0.3 μl) per test |
| Antibody | Anti-mouse CD135/Flt3 (rat monoclonal) | Clone A2F10 eBioscience - sold by ThermoFisher | RRID:AB_465859 | (2.5 μl) per test |
| Antibody | Anti-mouse CD169 (rat monoclonal) | Clone 3D6.112 BioLegend | RRID:AB_2563910 | (1 μl) per test |
| Antibody | Anti-mouse cKit/CD117 (rat monoclonal) | Clone 2B8 eBioscience - sold by ThermoFisher | RRID:AB_1834421 | (0.3 μl) per test |
| Antibody | Anti-mouse CX3CR1 (mouse monoclonal) | Clone SA011F11 BioLegend | RRID:AB_2564493 | (0.3 μl) per test |
| Antibody | Anti-mouse F4/80 (rat monoclonal) | Clone BM8 BioLegend | RRID:AB_10901171 | (0.8 μl) per test |
| Antibody | Anti-mouse IAb (mouse monoclonal) | Clone AF6-120.1 BioLegend | RRID:AB_313724 | (0.5 μl) per test |
| Antibody | Anti-mouse Ly6C (rat monoclonal) | Clone HK1.4 BioLegend | RRID:AB_2562177 | (0.1 μl) per test |
| Antibody | Anti-mouse Ly6G (rat monoclonal) | Clone 1A8-Ly6g eBioscience - sold by ThermoFisher | RRID:AB_2573893 | (0.2 μl) per test |
| Antibody | Anti-mouse NK1.1 (mouse monoclonal) | Clone PK136 BioLegend | RRID:AB_313396 | (0.3 μl) per test |
| Antibody | Anti-mouse Nr4a1/Nur77 (mouse monoclonal) | Clone 12.14 eBioscience - sold by ThermoFisher | RRID:AB_1257209 | (0.3 μl) per test intracellular stain |
| Antibody | Anti-mouse Sca1/Ly6a (rat monoclonal) | Clone D7 BioLegend | RRID:AB_2562275 | (2 μl) per test |
| Antibody | Anti-mouse Ter119 (rat monoclonal) | Clone Ter119 BioLegend | RRID:AB_313712 | (0.3 μl) per test |
| Antibody | Anti-mouse VCAM-1 (rat monoclonal) | Clone 429 BioLegend | RRID:AB_1595594 | (0.5 μl) per test |
| Antibody | Anti-H3K27ac ChIPseq grade (rabbit polyclonal) | Diagenode #C15410196 see our optimised ChIPseq protocol dx.doi.org/10.17504/protocols.io.bja3kign | RRID:AB_2637079 | (2 μg) per ChIP |
| Antibody | Anti-H3K4me1 ChIPseq grade (rabbit polyclonal) | Diagenode #C15410037 see our optimised ChIPseq protocol dx.doi.org/10.17504/protocols.io.bja3kign | RRID:AB_2561054 | (5 μg) per ChIP |
| Antibody | Anti-H3K9me3 ChIPseq grade (rabbit polyclonal) | Diagenode #C15410193 see our optimised ChIPseq protocol dx.doi.org/10.17504/protocols.io.bja3kign | RRID:AB_2616044 | (1 μg) per ChIP |
| Sequence-based reagent | Forward primer | 5-GCGAGAAAGTTAAAAGAATTGA-3 | For measuring P. chabaudi blood-stage parasitaemia by quantitative PCR | |
| Sequence-based reagent | Reverse primer | 5-CTAGTGAGTTTCCCCGTGTT-3 | ||
| Sequence-based reagent | Probe | [6FAM] - AAATTAAGCCGCAAGCTCCACG - [TAM] | ||
| Commercial assay or kit | Quick DNA Universal Microprep Kit | Zymo Research | D4074 | |
| Commercial assay or kit | IFNɣ mouse ELISA kit, extra sensitive | Invitrogen - sold by ThermoFisher | BMS609 | |
| Commercial assay or kit | IP-10 (CXCL10) mouse ELISA kit | Invitrogen - sold by ThermoFisher | BMS6018 | |
| Commercial assay or kit | Mouse/rat Angiopoietin-2 quantine ELISA kit | R&D Systems | MANG20 | |
| Commercial assay or kit | Foxp3 / Transcription Factor Staining Buffer Set | eBioscience - sold by ThermoFisher | 00-5523-00 | |
| Commercial assay or kit | SMART-Seq v4 Ultra Low Input RNA Kit | Takara Bio | 634891 | |
| Commercial assay or kit | Nextera XT DNA Library Preparation Kit | Illumina | FC-131-1024 | |
| Commercial assay or kit | True MicroChIP kit | Diagenode | C01010130 | |
| Commercial assay or kit | MicroPlex Library Preparation Kit v2 | Diagenode | C05010012 | |
| Commercial assay or kit | RNA Clean and Concentrator-5 Kit | Zymo Research | R1013 | |
| Commercial assay or kit | GeneChip WT Pico Kit | Affymetrix - sold by ThermoFisher | 902622 | |
| Commercial assay or kit | GeneChip Mouse Gene 1.0 ST Array | Affymetrix - sold by ThermoFisher | 901168 | |
| Chemical compound, drug | 4-Aminobenzoic acid | Sigma-Aldrich | A9878 | |
| Chemical compound, drug | Chloroquine diphosphate salt | Sigma-Aldrich | C6628 | Dissolve in water, dosage: 100 mg/kg by oral gavage |
| Chemical compound, drug | Lipopolysaccharide from Escherichia coli 0111:B4 | Sigma-Aldrich | L4391 | |
| Software, algorithm | bowtie2 v2.2.7 | (Langmead and Salzberg, 2012) http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | RRID:SCR_016368 | |
| Software, algorithm | DESeq2 | (Love et al., 2014) https://bioconductor.org/packages/release/bioc/html/DESeq2.html | RRID:SCR_015687 | |
| Software, algorithm | Cytoscape v3.8.0 | (Shannon et al., 2003) https://cytoscape.org/ | RRID:SCR_003032 | |
| Software, algorithm | clueGO v2.5.4 | (Bindea et al., 2009; Mlecnik et al., 2014) http://apps.cytoscape.org/apps/cluego | RRID:SCR_005748 | |
| Software, algorithm | HOMER v4.10 | (Heinz et al., 2010) http://homer.ucsd.edu/ | RRID:SCR_010881 | |
| Software, algorithm | Integrative genomics viewer (IGV) | (Thorvaldsdóttir et al., 2013) http://www.broadinstitute.org/igv/ | RRID:SCR_011793 |
Additional files
-
Supplementary file 1
Gating strategies for flow cytometry and cell sorting.
Flow cytometry was performed using the listed antibodies and panels. We gated myeloid cells and their progenitors in bone marrow, blood, and spleen using FlowJo v9; flow profiles of uninfected mice are displayed alongside the acute phase of a first malaria episode (P. chabaudi AJ). In every case, gating was performed identically between uninfected and infected mice with one exception (marked with an asterisk); to identify myeloid and erythroid progenitors in the bone marrow of infected mice we had to adjust our first gate (on lineage negative live singlets) due to the well-known upregulation of Sca-1 during acute infection (Belyaev et al., 2010). Note that CD115 (Csf1r) was replaced with CD11c when sorting monocytes as engagement of the Csf1 receptor has been shown to induce transcriptional changes (Jung et al., 2000).
- https://cdn.elifesciences.org/articles/63838/elife-63838-supp1-v1.pdf
-
Supplementary file 2
Quantitative changes in the histone modification profiles of once-infected mice.
Bone marrow monocytes were flow-sorted from once-infected mice (AJ, memory, 70 days p.i.) and uninfected controls for chromatin immunoprecipitation (ChIP)seq; differences in their histone modification profiles were then quantified by calling differentially modified regions (DMR, annotated to the nearest gene). Shown is a list of all tolerised/specialised genes annotated with a DMR, ordered by peak score.
- https://cdn.elifesciences.org/articles/63838/elife-63838-supp2-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63838/elife-63838-transrepform-v1.docx





