A KDM5–Prospero transcriptional axis functions during early neurodevelopment to regulate mushroom body formation
Figures
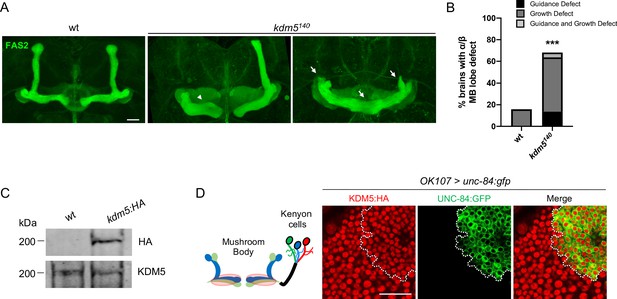
kdm5140 pharate adults have neuromorphological defects of the mushroom body (MB).
(A) Representative α/β lobe Z projections of pharate wild-type (NP4707rev2 revertant) and kdm5140 strains. Arrows indicate growth defects, and arrowheads indicate guidance defects. The α/β lobes are revealed with anti-fasciclin 2. (B) Quantification of α/β lobe defects in wild-type and kdm5140 strains. n = 19–22 (mean n = 21). ***p<0.001 (chi-square test with Yates’ correction). (C) Western blot of w1118 and kdm5:HA adult heads confirming wild-type expression levels of lysine demethylase 5 (KDM5):HA within our endogenously tagged kdm5:HA strain. Anti-HA (top) and anti-KDM5 (bottom) loading control. (D) Schematic of an adult MB with its associated Kenyon cells (left). OK107-Gal4 is used to drive expression of UNC-84:GFP, an inner nuclear membrane GFP reporter, within MB neuroblasts, mushroom body-ganglion mother cells, and Kenyon cells (right). Brains are counterstained with anti-HA to demonstrate the presence of endogenously tagged KDM5:HA within Kenyon cell nuclei. Scale bars represent 20 μm.
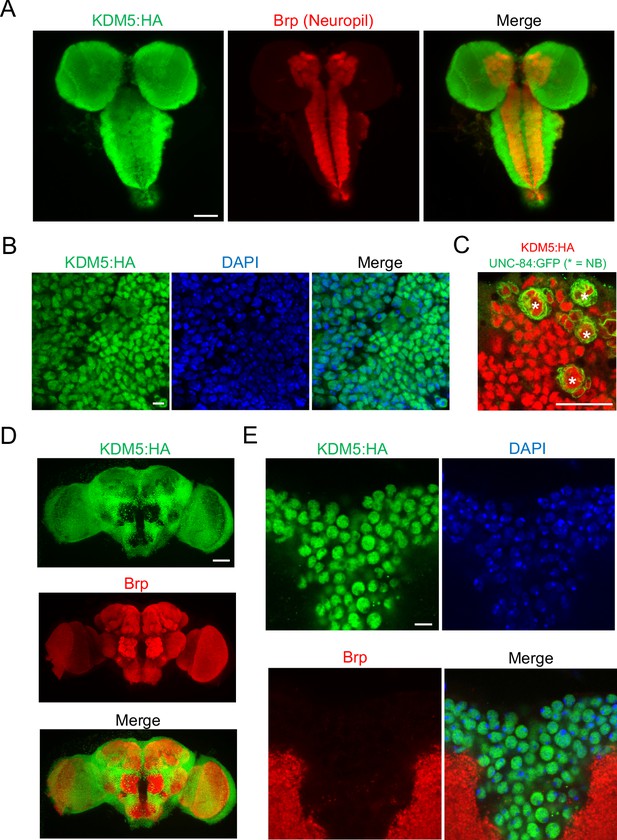
lysine demethylase 5 (KDM5) is broadly expressed in nuclei of the Drosophila larval central nervous system (CNS) and adult brain.
(A) Maximal Z projection of third-instar larval CNS revealing broad expression of endogenously tagged KDM5:HA and stained with anti-HA and anti-Bruchpilot (anti-Brp). (B) Cortical region of a larval (WL3) brain lobe stained with anti-HA and DAPI, showing nuclear localization of endogenously tagged KDM5:HA. (C) Neuroblast (NB)-specific wor-Gal4 driving expression of UNC-84:GFP to demonstrate endogenously tagged KDM5:HA expression within nuclei of WL3 central brain NBs (marked by *) and presumptive ganglion mother cells. UNC-84:GFP perdures for approximately 2–3 cell divisions. (D) Maximal Z projection of an adult brain revealing broad expression of endogenously tagged KDM5:HA with anti-HA and counterstained for neuropil with anti-Brp. (E) Dorsoanterior cortical region of an adult brain with endogenously tagged KDM5:HA. The nuclear localization of KDM5:HA is revealed by anti-HA, DAPI, and anti-Brp staining. Scale bars represent 50 μm in (A) and (D), 5 μM in (B) and (E), and 20 μM in (C).
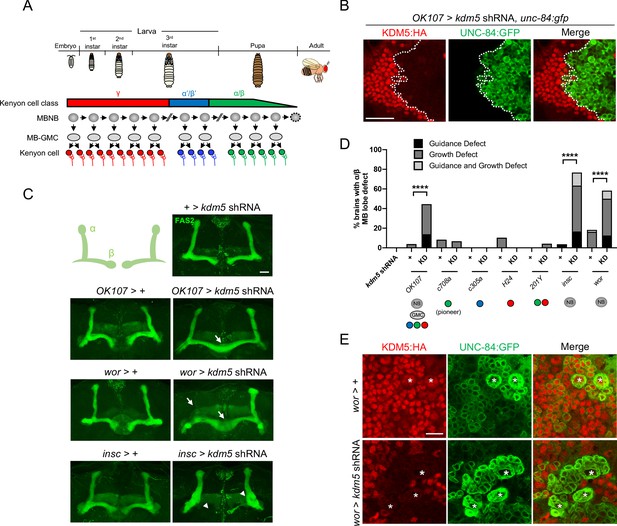
Depletion of lysine demethylase 5 (KDM5) within neural precursors results in neuroanatomical defects of the mushroom body (MB).
(A) Schematic showing the sequential generation of distinct subclasses of Kenyon cells throughout development. MB neuroblasts (MBNBs) are eliminated via apoptosis (dotted line) immediately prior to eclosion. (B) OK107-Gal4 driving expression of UNC-84:GFP to reveal shRNA-mediated KDM5:HA depletion within adult Kenyon cell nuclei. (C) Representative Z projections of adult kdm5 knockdown animals exhibiting significant α/β lobe defects and their respective kdm5 shRNA and GAL4 controls. The antibody anti-fasciclin 2 is used to visualize α/β lobes. Arrows indicate growth defects, and arrowheads indicate guidance defects. (D) Quantification of α/β MB lobe defects in flies expressing kdm5 shRNA driven by neural progenitor cell- and Kenyon cell-specific drivers. ‘KD’ indicates shRNA-mediated knockdown of kdm5. n = 16–49 (mean n = 29). ****p<0.0001 (chi-square test with Bonferroni correction). (E) Z projection of larval cortex revealing wor-Gal4-driven expression of kdm5 shRNA and unc-84:gfp transgenes. KDM5:HA depletion is observed in presumptive ganglion mother cells and post-mitotic cells surrounding NBs (marked by asterisks). Scale bars represent 20 μm in (B) and (C) and 10 μm in (E).
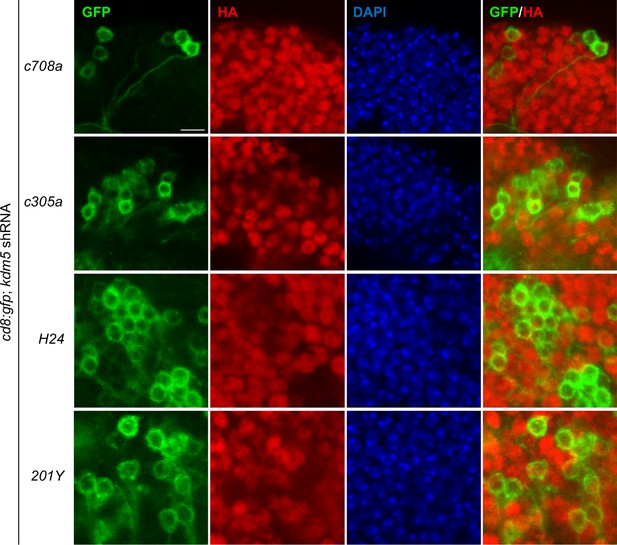
Validation of kdm5 knockdown within subpopulations of mature mushroom body (MB) neurons.
MB Gal4 drivers promoting the expression of CD8:GFP to reveal shRNA-mediated lysine demethylase 5 (KDM5):HA depletion within subpopulations of mature Kenyon cells within the adult brain. Scale bar represents 5 μM.
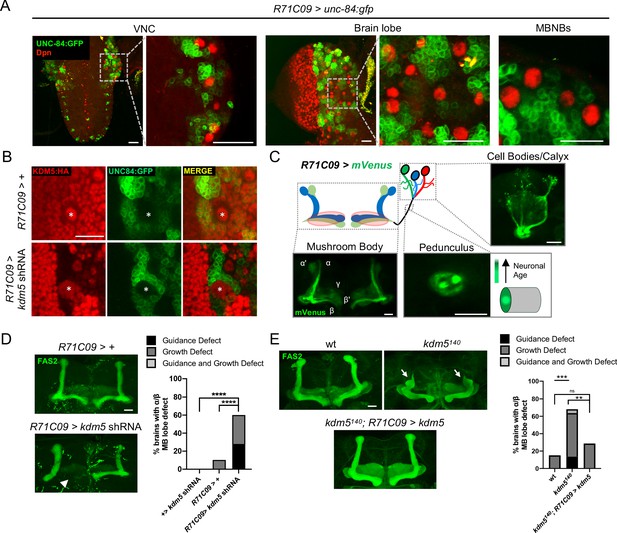
Expression pattern of R71C09-Gal4 within ganglion mother cells and immature neurons of the Drosophila central nervous system.
(A) Whole-mount Z projections of a larval ventral nerve cord (left), brain lobe (middle), and brain cortical region (right). Z projections show R71C09-Gal4-driven expression of UNC-84:GFP, counterstained with neuroblast (NB)-specific anti-Dpn. (B) Z projection of larval cortex revealing R71C09-Gal4-driven expression of UNC-84:GFP with and without kdm5 shRNA. NBs are marked by an asterisk. (C) Optical sections of an adult mushroom body (MB) with its associated pedunculus, Kenyon cell bodies, and calyces expressing an R71C09-Gal4-driven mVenus reporter. R71C09-Gal4 strongly drives mVenus expression in newly born neurons located within core fibers of the pedunculus. (D) Representative Z projections (left) and quantification (right) of adult α/β MB lobe defects in flies expressing kdm5 shRNA driven by R71C09-Gal4. The antibody anti-fasciclin 2 (anti-Fas2) is used to visualize α/β lobes. n = 15–39 (mean n = 26). Arrowhead indicates a guidance defect. ****p<0.0001 (chi-square test with Bonferroni correction). (E) Representative α/β lobe Z projections and quantification of pharate wild-type (NP4707rev2 revertant), kdm5140, and kdm5140; R71C09 > kdm5 rescue strains. Arrows indicate growth defects. The α/β lobes are revealed with anti-Fas2. n = 20–28 (mean n = 24). **p<0.01; ***p<0.001 (chi-square test with Bonferroni correction). Scale bars represent 20 μm.
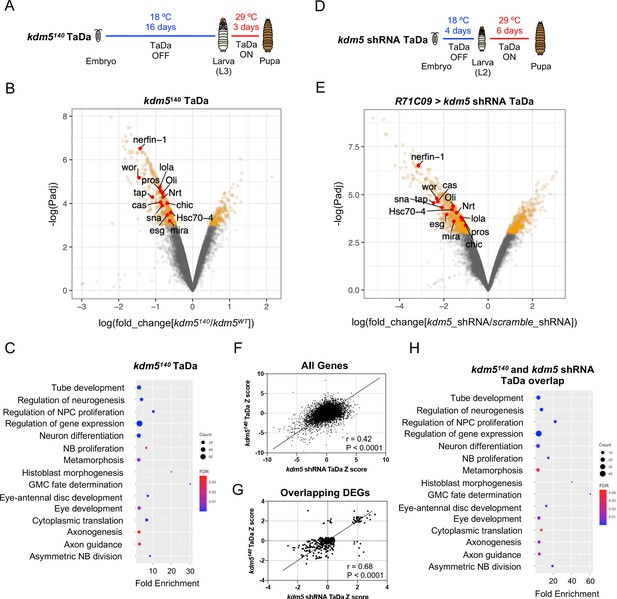
Transcriptome profiling of lysine demethylase 5 (KDM5)-depleted ganglion mother cells (GMCs) and immature neurons by targeted DamID (TaDa) reveals KDM5-regulatory networks critical for ganglion mother cell (GMC) proliferation and neurodevelopment.
(A) Timeline of TaDa induction within GMCs and immature neurons of kdm5140 WL3 animals and pupae. (B) Volcano plot of differentially expressed genes (DEGs) within GMCs and immature neurons of kdm5140 animals compared to wild type. Genes with a false discovery rate (FDR) < 0.05 are in red, with those labeled involved in GMC proliferation and neurodevelopment. TaDa analyses were performed in quintuplicate. (C) Representative distribution of ontology terms for DEGs in kdm5140 GMCs and immature neurons using a PANTHER Overrepresentation Test (Fisher’s exact test with FDR < 0.05). (D) Timeline of TaDa and kdm5 shRNA induction within GMCs and immature neurons of WL3 animals and pharate adults. (E) Volcano plot of DEGs within kdm5 shRNA GMCs and immature neurons compared to those expressing a scrambled shRNA. Genes with an FDR < 0.05 are in red, with those labeled involved in GMC proliferation and neurodevelopment. TaDa analyses were performed in triplicate. (F) Correlation of Z scores between DEGs of kdm5140 and kdm5 shRNA TaDa datasets (Deming regression; p<0.0001). (G) Correlation of Z scores between overlapping DEGs of kdm5140 and kdm5 shRNA TaDa datasets (Deming regression; p<0.0001). (H) Representative distribution of ontology terms for DEGs in overlapping kdm5140 and kdm5 shRNA TaDa datasets utilizing a PANTHER Overrepresentation Test (Fisher’s exact test with FDR < 0.05).
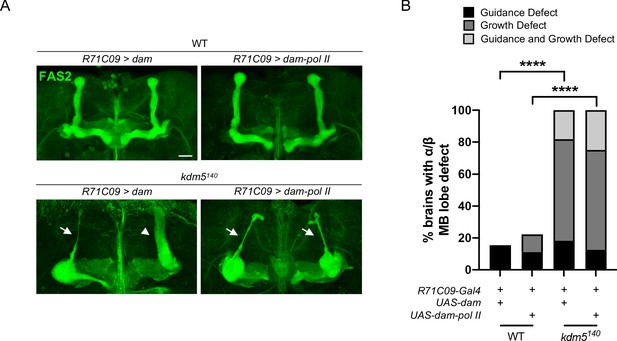
kdm5140 pharate adults expressing targeted DamID (TaDa) genetic elements present with significant mushroom body (MB) morphological defects.
(A) Representative α/β lobe Z projections of pharate kdm5140 animals expressing TaDa genetic elements and their respective controls. The antibody anti-fasciclin 2 is used to visualize α/β lobes. Arrows indicate growth defects. Scale bar represents 20 μm. (B) Quantification of α/β lobe defects in pharate kdm5140 animals expressing TaDa genetic elements and their respective controls. n = 8–22 (mean n = 13). ****p<0.0001 (chi-square test with Bonferroni correction).
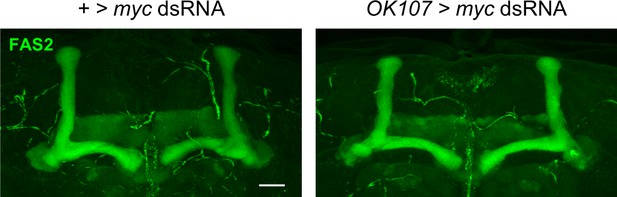
Knockdown of myc within Kenyon cells does not result in morphological defects of the mushroom body.
Representative α/β lobe Z projections of adults expressing myc dsRNA driven by OK107-Gal4. No gross morphological defects were detected in the KD or control conditions n = 29–30.
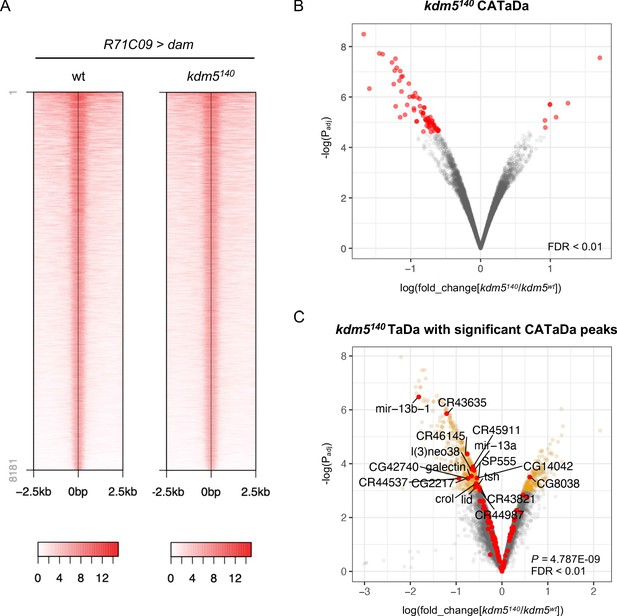
Chromatin accessibility profiling using targeted DamID (CATaDa) analyses reveal minimal changes to chromatin accessibility within ganglion mother cells (GMCs) and immature neurons upon loss of kdm5.
(A) Representative CATaDa profiles of R71C09-Gal4 expressing cells for kdm5140 and wild type. Heat maps show Dam binding profiles for the greatest 8181 peaks for each genotype. (B) Volcano plot showing changes to chromatin accessibility within GMCs and immature neurons of kdm5140 animals compared to wild type. Genes with an FDR < 0.01 are in yellow. CATaDa analyses were performed in quintuplicate. (C) Volcano plot showing changes to chromatin accessibility for differentially expressed genes (DEGs) from kdm5140 TaDa. In yellow are kdm5140 TaDa DEGs with an FDR < 0.05. In red are kdm5140 TaDa DEGs with associated changes in chromatin accessibility for FDR < 0.01 (Fisher’s exact test, p=4.79E-09).
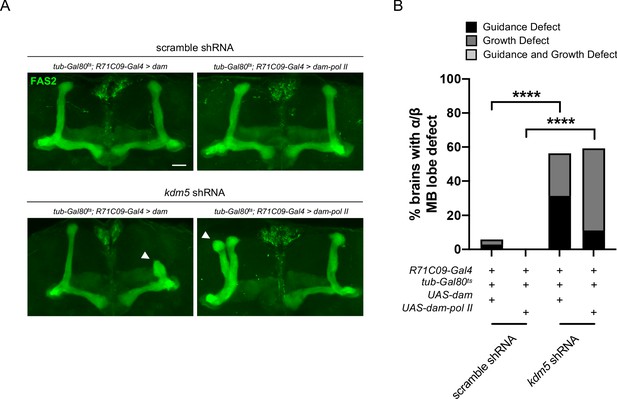
Adults expressing R71C09-Gal4-driven kdm5 shRNA in tandem with targeted DamID (TaDa) genetic elements present with significant mushroom body morphological defects.
(A) Representative α/β lobe Z projections of adult R71C09 > kdm5 shRNA animals expressing TaDa genetic elements and their respective controls. The antibody anti-fasciclin 2 is used to visualize α/β lobes. Arrowheads indicate guidance defects. Scale bar represents 20 μm. (B) Quantification of α/β lobe Z projections of adult R71C09 > kdm5 shRNA animals expressing TaDa genetic elements and their respective controls. n = 16–34 (mean n = 28). ****p<0.0001 (chi-square test with Bonferroni correction).
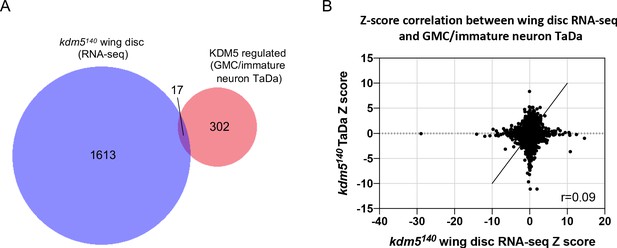
Transcriptome analyses reveal that lysine demethylase 5 (KDM5)-regulated gene expression is tissue-specific.
(A) Venn diagram illustrating intersection of kdm5140 and kdm5 shRNA overlapping targeted DamID (TaDa) data with that from a previously published kdm5140 mRNA-seq wing disc dataset (Drelon et al., 2018). (B) Correlation of Z scores between kdm5140 TaDa and kdm5140 wing disc mRNA-seq (Drelon et al., 2018) datasets. Pearson’s R = 0.09.
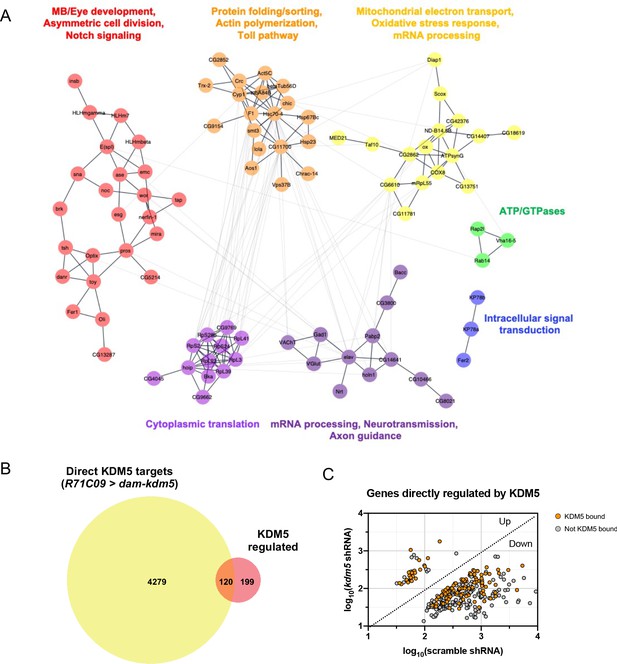
Network of known and predicted interactions between lysine demethylase 5 (KDM5)-regulated aenes and analysis of direct KDM5 targets.
(A) Gene network analysis and community clustering were performed using Cytoscape with a minimum confidence score of 0.4. Networks with greater than two nodes are shown and color-coded based on cluster. Labels indicate general categories of overlapping kdm5140 and kdm5 shRNA targeted DamID (TaDa) differentially expressed genes (DEGs) within each cluster. (B) Venn diagram illustrating intersection of similarly dysregulated kdm5140 and kdm5 shRNA overlapping DEGs with direct KDM5 targets from R71C09 > dam-kdm5 TaDa. (C) Analysis of the 319 similarly dysregulated kdm5140 and kdm5 shRNA overlapping DEGs (with values plotted from the kdm5 shRNA TaDa). Direct KDM5 targets are labeled in orange. Venn diagram created with BioVenn (Hulsen et al., 2008).
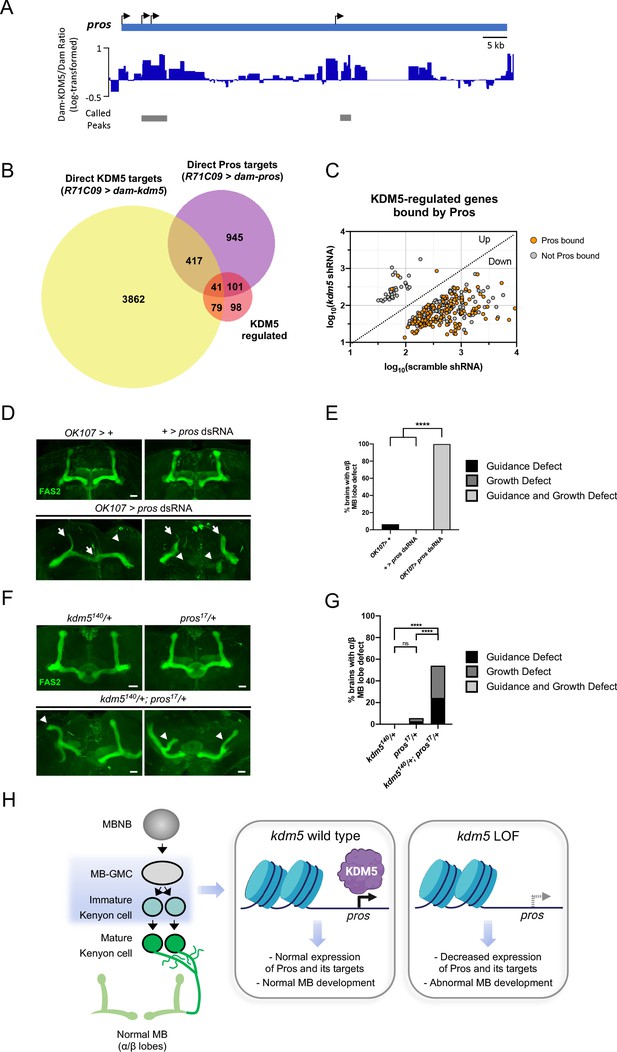
Neuromorphological and transcriptomic analyses reveal a genetic interaction between prospero and lysine demethylase 5 (KDM5).
(A) Integrative genomics viewer (IGV) plot showing average KDM5 occupancy at pros transcriptional start sites across six replicates. Scale bars represent the log2 ratio change between Dam-KDM5 and Dam samples. Called peaks are indicated by gray bars. (B) Venn diagram illustrating intersection of similarly dysregulated kdm5140 and kdm5 shRNA overlapping differentially expressed genes (DEGs) with Dam-KDM5 direct binding data and a previously published Dam-Pros targeted DamID (TaDa) binding dataset (Fisher’s exact test, p=2.20E-16 for KDM5-dysregulated and Pros gene overlap) (Liu et al., 2020). (C) Analysis of the 319 similarly dysregulated kdm5140 and kdm5 shRNA overlapping DEGs (with values plotted from the kdm5 shRNA TaDa). Direct Pros targets from the previously published Dam-Pros TaDa dataset (Liu et al., 2020) are labeled in orange. (D) Representative Z projections of OK107 > pros RNAi adults exhibiting significant α/β lobe defects. The α/β lobes are revealed by anti-fasciclin 2 (Fas2). (E) Quantification of α/β mushroom body (MB) lobe defects in flies expressing pros shRNA driven by OK107-Gal4. n = 16–19 (mean n = 17). ****p<0.0001 (chi-square test with Bonferroni correction). (F) Representative Z projections of representative kdm5140/+ and pros17/+ heterozygous adult α/β MB lobes (top) and kdm5140/+; pros17/+ transheterozygous adult α/β MB lobes (bottom). The α/β lobes are revealed by anti-Fas2. (G) Quantification of α/β MB lobe defects in kdm5140/+; pros17/+ transheterozygous adults and heterozygous controls. n = 28–37 (mean n = 34). ****p<0.0001 (chi-square test with Bonferroni correction). (H) Model proposing a genetic interaction between pros and kdm5 within ganglion mother cells (GMCs) and immature α/β Kenyon cells. KDM5 binds to the pros locus and positively regulates its transcription. Loss of kdm5 leads to downregulation of pros and its targets, resulting in defects to MB neurodevelopment and cognitive function. Image was created with BioRender and Venn diagrams with BioVenn (Hulsen et al., 2008). Scale bars represent 20 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Mouse monoclonal anti-brp | DSHB | Cat# nc82; RRID:AB_2314866 | IF (1:50) |
| Antibody | Rabbit monoclonal anti-HA-Tag | Cell Signaling Technology | Cat# 3724; RRID:AB_1549585 | IF (1:100) |
| Antibody | Mouse monoclonal anti-HA-Tag | Cell Signaling Technology | Cat# 2367; RRID:AB_10691311 | IF (1:100) WB (1:1000) |
| Antibody | Rat monoclonal anti-Deadpan | Abcam | Cat# ab195173; RRID:AB_2687586 | IF (1:100) |
| Antibody | Goat polyclonal anti-mouse Alexa-488 | Thermo Fisher Scientific | Cat# A32723; RRID:AB_2633275 | IF (1:500) |
| Antibody | Goat polyclonal anti-mouse Alexa-568 | Thermo Fisher Scientific | Cat# A11004; RRID:AB_2534072 | IF (1:500) |
| Antibody | Goat polyclonal anti-rabbit Alexa-488 | Thermo Fisher Scientific | Cat# A11034; RRID:AB_2576217 | IF (1:500) |
| Antibody | Goat polyclonal anti-rabbit Alexa-568 | Thermo Fisher Scientific | Cat# A11004, RRID:AB_2534072 | IF (1:500) |
| Antibody | Goat polyclonal anti-rat Alexa-568 | Thermo Fisher Scientific | Cat# A11077; RRID:AB_2534121 | IF (1:500) |
| Antibody | Mouse monoclonal anti-Fas2 | DSHB | Cat# 1D4; RRID:AB_528235 | IF (1:25) |
| Antibody | Rabbit polyclonal anti-KDM5 | Secombe Lab; Secombe et al., 2007 | RRID:AB_2569502 | WB (1:1000) |
| Antibody | IRDye 680RD donkey monoclonal anti-mouse IgG secondary antibody | LI-COR Biosciences | LI-COR Biosciences Cat# 925-68072, RRID:AB_2814912 | WB (1:8000) |
| Antibody | IRDye 800CW donkey monoclonal anti-rabbit IgG secondary antibody | LI-COR Biosciences | LI-COR Biosciences Cat# 926-32213, RRID:AB_621848 | WB (1:8000) |
| Cell line (Escherichia coli) | NEB 5-alpha competent E. coli | New England BioLabs | Cat# C2987 | |
| Commercial assay or kit | Clontech CloneAmp HiFi PCR Premix | Clontech | Cat# 639298 | |
| Commercial assay or kit | Advantage 2 cDNA polymerase | Clontech | Cat# 639201 | |
| Commercial assay or kit | Agencourt AMPure XP Beads | Beckman Coulter | Cat# A63880 | |
| Commercial assay or kit | Takara In-Fusion HD Cloning Plus | Takara | Cat# 638909 | |
| Commercial assay or kit | Quick Ligation Kit | New England BioLabs | Cat# M2200S | |
| Commercial assay or kit | Zymo Quick-DNA miniprep plus | Zymo Research | Cat# D4069 | |
| Commercial assay or kit | Macherey-Nagel NucleoSpin Gel and PCR Clean-up Kit | Takara | Cat# 740609.250 | |
| Commercial assay or kit | Qubit dsDNA HS Assay Kit | Invitrogen | Cat# Q32851 | |
| Genetic reagent (Drosophila melanogaster) | Drosophila: kdm5:3xHA | This study | N/A | Endogenous kdm5:HA strain (Figures 2–4). Available from lead contact. |
| Genetic reagent (D. melanogaster) | Drosophila: OK107-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_854 | |
| Genetic reagent (D. melanogaster) | Drosophila: 5XUAS-unc84::2XGFP | Janelia Research Campus; Henry et al., 2012 | N/A | |
| Genetic reagent (D. melanogaster) | Drosophila: wor-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_56553 | |
| Genetic reagent (D. melanogaster) | Drosophila: UAS-kdm5RNAI | Bloomington Drosophila Stock Center | RRID:BDSC_35706 | |
| Genetic reagent (D. melanogaster) | Drosophila: insc-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_8751 | |
| Genetic reagent (D. melanogaster) | Drosophila: c708a-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_50743 | |
| Genetic reagent (D. melanogaster) | Drosophila: c305a-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_30829 | |
| Genetic reagent (D. melanogaster) | Drosophila: H24-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_51632 | |
| Genetic reagent (D. melanogaster) | Drosophila: 201Y-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_4440 | |
| Genetic reagent (D. melanogaster) | Drosophila: UAS-Dcr-2 | Bloomington Drosophila Stock Center | RRID:BDSC_24650 | |
| Genetic reagent (D. melanogaster) | Drosophila: GMR71C09-GAL4 | Bloomington Drosophila Stock Center | RRID:BDSC_39575 | |
| Genetic reagent (D. melanogaster) | Drosophila: 20XUAS-IVS-CsChrimson.mVenus | Bloomington Drosophila Stock Center | RRID:BDSC_55136 | |
| Genetic reagent (D. melanogaster) | Drosophila: kdm5NP4707 | Kyoto Stock Center; Hayashi et al., 2002 | ; stock# 104754 | |
| Genetic reagent (D. melanogaster) | Drosophila: kdm5140 | Secombe Lab; Drelon et al., 2018 | ||
| Genetic reagent (D. melanogaster) | Drosophila: UASt-kdm5 | Secombe Lab; Secombe et al., 2007 | ||
| Genetic reagent (D. melanogaster) | Drosophila: UAS-dMycRNAi | Vienna Drosophila Resource Center | Stock# KK106066 | |
| Genetic reagent (D. melanogaster) | Drosophila: UAS-prosRNAi | Bloomington Drosophila Stock Center | RRID:BDSC_42538 | |
| Genetic reagent (D. melanogaster) | Drosophila: tubP-Gal80ts | Bloomington Drosophila Stock Center | RRID:BDSC_7019 | |
| Genetic reagent (D. melanogaster) | Drosophila: UAS-LT3-NDam | Brand Lab; Southall et al., 2013 | ||
| Genetic reagent (D. melanogaster) | Drosophila: UAS-LT3-NDam-RpII215 | Brand Lab; Southall et al., 2013 | ||
| Genetic reagent (D. melanogaster) | Drosophila: w1118 | Bloomington Drosophila Stock Center | RRID:BDSC_5905 | |
| Genetic reagent (D. melanogaster) | Drosophila: UAS-LT3-NDam-kdm5 | This study | N/A | Used for KDM5 TaDa (Figures 6 and 7). Available from lead contact. |
| Sequence-based reagent | AdRt | Vogel et al., 2007 | PCR primers | CTAATACGACTCACTATAGGGCAGCGTGGTCGCGGCCGAGGA |
| Sequence-based reagent | AdRb | Vogel et al., 2007 | PCR primers | TCCTCGGCCG |
| Sequence-based reagent | DamID_PCR | Vogel et al., 2007 | PCR primers | GGTCGCGGCCGAGGATC |
| Sequence-based reagent | scram_shRNA | This study | PCR primers | GGATAATAGAATAGTTATATTCAAGCATATTCTATTATCC |
| Sequence-based reagent | Fw DsRed_KDM5_AarI | This study | PCR primers | tatagtgtcttcggggccgaCAGGAGCTGTGGCGCATTCTAGAAAC |
| Sequence-based reagent | PAM Rv | This study | PCR primers | AATCTGGAACATCGTATGGGTACTGCGGCCGCGCTCGCGC |
| Sequence-based reagent | PAM Fw | This study | PCR primers | AGCAGCGGGCGGTGCAATCGGCGCGAGCGCGGCCGCAGTA |
| Sequence-based reagent | Rv DsRed_KDM5_AarI | This study | PCR primers | gattatctttctagggttaaAGGAAAAAGTCAAATAAAACGTAAGAAAACTTTGC |
| Sequence-based reagent | Fw DsRed_KDM5_SapI | This study | PCR primers | gactatctttctagggttaaTCAAAGGCGAAGGCGACTCT |
| Sequence-based reagent | Rv DsRed_KDM5_SapI | This study | PCR primers | atatggtcttcttttcccggAACATGTTCCTCTTTTAAGGTGCTCTTT |
| Sequence-based reagent | Dam-kdm5_NotI-Fw | This study | PCR primers | cgcagatctgcggccgATGTCCGCCAAAACTGAGG |
| Sequence-based reagent | Dam-kdm5_XbaI-Rv | This study | PCR primers | acaaagatcctctagCTACCGCGCCGATTGCAC |
| Recombinant DNA reagent | pU6-BbsI-chiRNA | Addgene; Gratz et al., 2013 | RRID:Addgene_45946 | |
| Recombinant DNA reagent | pValium20 | Drosophila Genomics Resource Center; Ni et al., 2009 | DGRC# 1467 | |
| Recombinant DNA reagent | pHD-ScarlessDsRed | Drosophila Genomics Resource Center | DGRC# 1364 | |
| Software, algorithm | Fiji | https://fiji.sc/ | RRID:SCR_002285 | |
| Software, algorithm | Prism 6 | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | Cytoscape | https://cytoscape.org/ | RRID:SCR_003032 | |
| Software, algorithm | Gene Ontology | http://www.geneontology.org | RRID:SCR_002811 | |
| Software, algorithm | R 3.5.1 | The R Foundation | RRID:SCR_001905 | |
| Software, algorithm | ggplot2 (R package) | CRAN | RRID:SCR_014601 | |
| Software, algorithm | damidseq_pipeline | Marshall and Brand, 2015a | http://owenjm.github.io/damidseq_pipeline/ | |
| Software, algorithm | find_peaks | Wolfram et al., 2012 | http://github.com/owenjm/find_peaks | |
| Software, algorithm | bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | |
| Software, algorithm | BioVenn | Hulsen et al., 2008 | https://www.biovenn.nl/ | |
| Other | Vectashield Mounting Medium | Vector Labs | Cat# H-1000 | |
| Other | Normal donkey serum | Fisher Scientific | Cat# 50-413-367 | |
| Other | DAPI-Fluoromount-G | SouthernBiotech | Cat# OB010020 | |
| Other | T4 DNA ligase | New England BioLabs | Cat# M0202 | |
| Other | T4 polynucleotide kinase | New England BioLabs | Cat# M0201 | |
| Other | AarI | Thermo Scientific | Cat# ER1581 | |
| Other | SapI | New England BioLabs | Cat# R0569 | |
| Other | BpiI | Thermo Scientific | Cat# ER1011 | |
| Other | NotI | New England BioLabs | Cat# R0189 | |
| Other | PstI | New England BioLabs | Cat# R0140 | |
| Other | NheI | New England BioLabs | Cat# R0131 | |
| Other | EcoRI | New England BioLabs | Cat# R0101 | |
| Other | DpnI | New England BioLabs | Cat# R0176 | |
| Other | DpnII | New England BioLabs | Cat# R0543 | |
| Other | AlwI | New England BioLabs | Cat# R0513 |
Additional files
-
Supplementary file 1
DEGs from kdm5140 TaDa.
Related to Figure 5.
- https://cdn.elifesciences.org/articles/63886/elife-63886-supp1-v2.xlsx
-
Supplementary file 2
GO categories from kdm5140 TaDa.
Related to Figure 5.
- https://cdn.elifesciences.org/articles/63886/elife-63886-supp2-v2.xlsx
-
Supplementary file 3
DEGs from kdm5 shRNA TaDa.
Related to Figure 5.
- https://cdn.elifesciences.org/articles/63886/elife-63886-supp3-v2.xlsx
-
Supplementary file 4
GO categories from kdm5 shRNA TaDa.
Related to Figure 5.
- https://cdn.elifesciences.org/articles/63886/elife-63886-supp4-v2.xlsx
-
Supplementary file 5
KDM5-regulated genes that are direct Dam-KDM5 targets.
- https://cdn.elifesciences.org/articles/63886/elife-63886-supp5-v2.xlsx
-
Supplementary file 6
KDM5-regulated genes that are direct Dam-Pros targets.
Related to Figure 7.
- https://cdn.elifesciences.org/articles/63886/elife-63886-supp6-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63886/elife-63886-transrepform-v2.docx





