PRD-2 directly regulates casein kinase I and counteracts nonsense-mediated decay in the Neurospora circadian clock
Figures
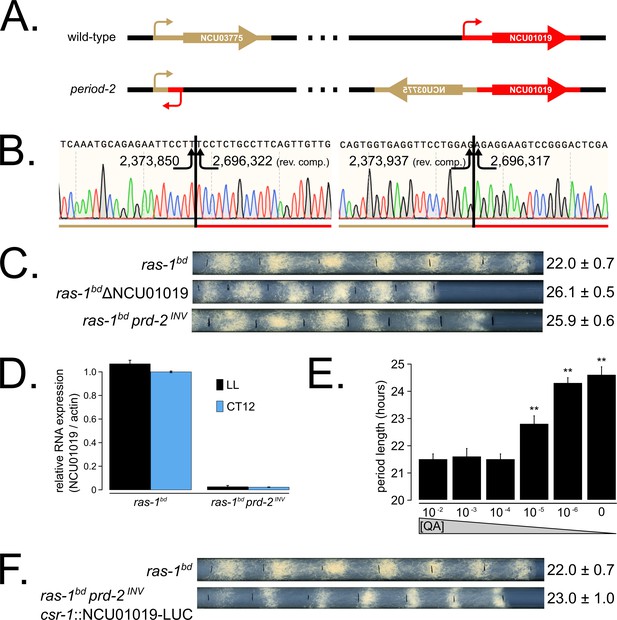
The prd-2 phenotype derives from reduced expression of NCU01019.
Whole genome sequencing identified a 322,386 bp inversion on linkage group V in the original prd-2 mutant strain (Lambreghts, 2012). The inversion breakpoints disrupt two loci, NCU03775 and NCU01019, depicted in cartoon form (A). Sanger sequencing confirms the DNA sequence of the left and right breakpoints, and the corresponding NC12 genome coordinates are shown at each arrowhead (B). Circadian period length was determined by race tube (RT) assay for ras-1bd controls, targeted deletion of the NCU01019 locus, and the classically derived prd-2INV mutant. The ΔNCU01019 mutant has a long period and slow growth defect similar to prd-2INV (C). NCU01019 RNA expression levels are detectable by RT-qPCR in the prd-2INV mutant but are drastically reduced compared to ras-1bd controls grown in constant light (LL) or at subjective dusk (CT12) during a circadian free run (D). After replacing the endogenous promoter of NCU01019 with the inducible qa-2 promoter, addition of high levels of quinic acid (10−2 to 10−3 M) led to a normal circadian period by RT assay (10−2 M τ = 21.5 ± 0.2 hr; 10−3 M τ = 21.6 ± 0.3 hr; 10−4 M τ = 21.5 ± 0.2 hr). Lower levels of QA inducer led to a long circadian period (10−5 M τ = 22.8 ± 0.3 hr; 10−6 M τ = 24.3 ± 0.2 hr; 0 QA τ = 24.6 ± 0.3 hr) due to reduced NCU01019 expression. Asterisks (**) indicate p<1 × 10−10 by Student’s t-test compared to 10−2 M QA RT results (E). The entire NCU01019 locus (plus 951 bases of its upstream promoter sequence) was fused in-frame with codon-optimized luciferase. Ectopic expression of this NCU01019-luc construct in the prd-2INV background rescues the long period phenotype by RT assay (F).
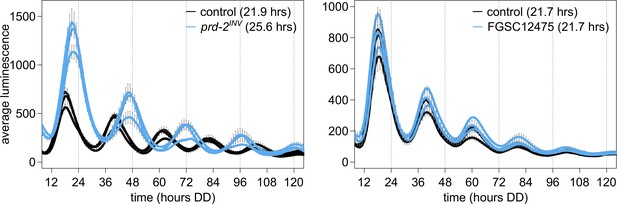
NCU03775 knockout has a normal circadian period and does not explain the prd-2INV phenotype.
96-well plate luciferase assays were used to measure the circadian period length. Traces represent the average of three technical replicates across three biological replicate experiments for: ras-1bd controls (black, τ = 21.9 ± 0.3 hr), ras-1bd prd-2INV (blue, τ = 25.6 ± 0.4 hr), FGSC2489 wild-type controls (black, τ = 21.7 ± 0.3 hr), and the ΔNCU03775 knockout strain FGSC12475 (blue, τ = 21.7 ± 0.3 hr). ΔNCU03775 has a wild-type circadian period length.
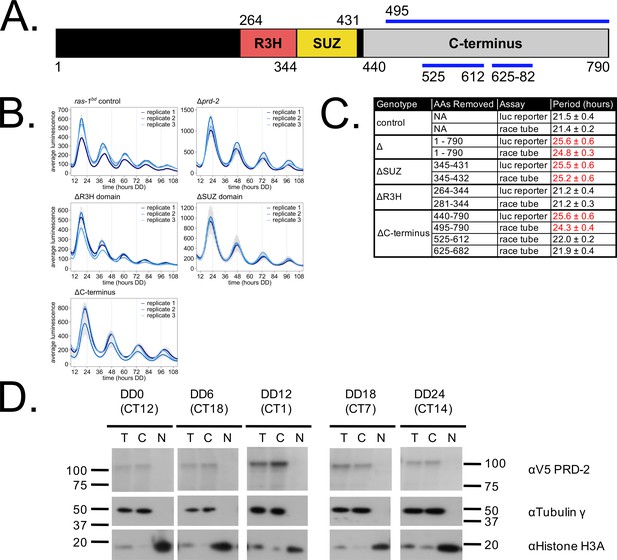
Clock-relevant protein domains and localization of PRD-2 suggest and RNA-binding function.
PRD-2 has tandemly arrayed R3H and SUZ domains associated with RNA binding proteins, and its C-terminal region is highly enriched for proline (P) and glutamine (Q). The cartoon of PRD-2 protein lists relevant amino acid coordinates (A). The native NCU01019 locus was replaced with single domain deletion mutants, and 96-well plate luciferase assays were used to measure the circadian period length in triplicate wells per biological replicate experiment. A wild-type clock period was recovered in ras-1bd controls and the prd-2ΔR3H mutant, while Δprd-2, prd-2ΔSUZ, and prd-2ΔC-terminus had long period phenotypes (B). Independently constructed strains targeted domain deletion mutants to the csr-1 locus in a Δprd-2 background (Supplementary file 1), and mutant period lengths were determined by race tube assay. Period lengths (±1 SD) show that the clock-relevant domains of PRD-2 are the SUZ domain and the C-terminus (C). Total (T), Nuclear (N), and Cytosolic (C) fractions were prepared over a circadian time course (N = 1 per time point). γ-Tubulin (NCU03954) was used as a control for cytoplasmic localization and histone H3 (NCU01635) for nuclear localization. PRD-2 tagged with a C-terminal V5 epitope tag is localized to the cytoplasm throughout the circadian cycle (D).
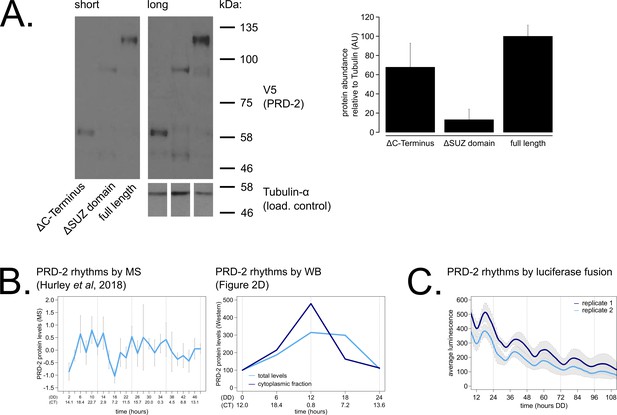
PRD-2 protein levels are slightly rhythmic and are detectable in protein domain deletion mutants.
PRD-2 protein levels were measured from at least three biological replicates using strains where the endogenous NCU01019 locus was replaced with V5-tagged domain deletion constructs: prd-2ΔC-Terminus(Δ440–790), prd-2ΔSUZ(Δ345–431), and full length. Long and short exposures of a representative immunoblot are shown with quantification relative to tubulin loading controls (A). The C-terminal deletion strain has ~68% PRD-2 levels compared to the full length control, and the SUZ deletion has ~13% levels. Both are above the low levels of qa-driven NCU01019 needed to induce the long period phenotype (Figure 1E). MS data from a previous study (Hurley et al., 2018) revealed low amplitude rhythms in PRD-2 abundance with a broad peak in the subjective circadian night and early morning (~CT18 – 2). Circadian Time (CT) was calculated as described previously (Kelliher et al., 2020). PRD-2 abundance was also quantified from the localization time course (Figure 2D) relative to tubulin and relative to the first time point. Peak PRD-2 protein abundance was observed in the subjective morning from both MS and immunoblot data (B), corresponding with the rise in frq transcript levels (Aronson et al., 1994). To confirm rhythms in PRD-2 protein expression, the complete NCU01019 5’-UTR and coding sequence, including 951 bp of upstream promoter sequence but lacking its endogenous 3’-UTR sequence, was fused in-frame with codon-optimized luciferase (Gooch et al., 2008). This construct was transformed into the prd-2WT background at the csr-1 locus, and PRD-2 protein cycles in abundance (τ = 21.7 ± 0.8 hr). PRD-2 protein peaks during the circadian day (CT7.5±1) by luciferase fusion (C), slightly delayed relative to its morning peak by western blot and MS.
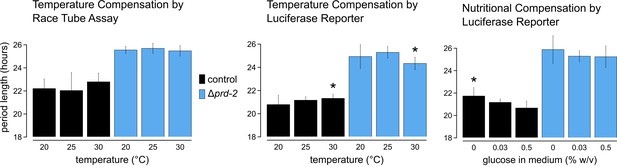
Temperature and nutritional compensation are normal in ΔNCU01019.
Temperature and nutritional compensation were assessed in ras-1bd controls compared to ras-1bd Δprd-2. Race tubes (RTs) were incubated at 20°, 25°, or 30°C to determine free running period length. Temperature did not significantly affect period length for controls (ANOVA p=0.598) or for the Δprd-2 mutant (ANOVA p=0.756) RTs. 96-well plates were incubated at 20°, 25°, or 30°C to determine free running period length. Period was significantly different at 30°C for both genotypes (Asterisks [*]: control 20°C vs 30°C, Tukey test p=0.023; Δprd-2 20°C vs 30°C, Tukey test p=0.037; Δprd-2 25°C vs 30°C, Tukey test p=0.0002). 96-well plates were run with 0%, 0.03%, or 0.5% glucose w/v to test nutritional compensation. Period length was significantly different at 0% glucose for controls only (Asterisk [*]: control 0% vs 0.5%, Tukey test p=0.00005; control 0% vs 0.03%, Tukey test p=0.034; Δprd-2 ANOVA p=0.183).
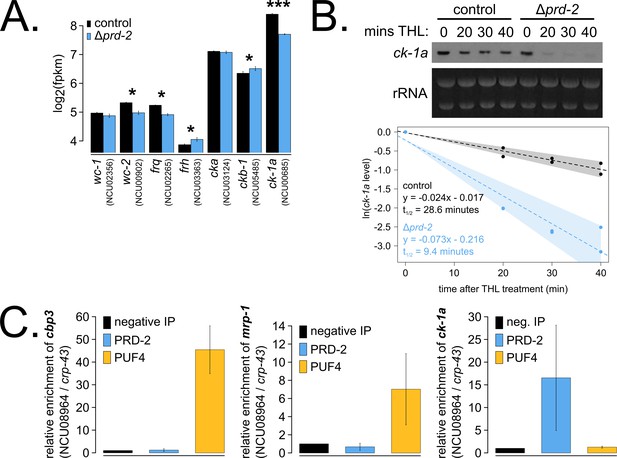
The core clock target of PRD-2 is the casein kinase I transcript.
Control and Δprd-2 cultures were grown in the light at 25°C in Bird medium for 48 hr prior to RNA isolation. Expression levels for core clock genes were measured by RNA-sequencing (N = 3 biological replicates per strain), and log2-transformed FPKM values are shown. Asterisks indicate p<0.05 (*) or p<5 × 10−5 (***) by Student’s t-test compared to control levels. The ck-1a transcript is >1.5× less abundant in Δprd-2 (A). ck-1a mRNA degradation kinetics were examined by Northern blot in a time course after treatment with thiolutin (THL) at approximately CT1 (N = 2 biological replicates). RNA levels were quantified using ImageJ, natural log transformed, fit with a linear model (glm in R, Gaussian family defaults), and half-life was calculated assuming first order decay kinetics (ln(2)/slope). Shaded areas around the linear fit represent 95% confidence intervals on the slope. The ck-1a transcript is 3× less stable in Δprd-2 (B). The PUF4 (NCU16560) RNA-binding protein pulls down known target transcripts cbp3 (NCU00057) and mrp-1 (NCU07386) by RT-qPCR (N = 3 biological replicates). PRD-2 CLIP samples were processed in parallel with PUF4 positive controls, and PRD-2 binds the ck-1a transcript in vivo (C).
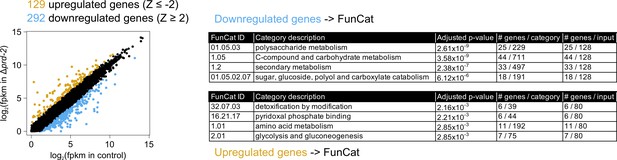
Hundreds of genes have altered expression levels in the Δprd-2 mutant but a common pathway or sequence motif was not detected.
RNA-seq data were first filtered for low expression. Out of 9730 annotated N. crassa genes, 8622 were expressed in four of six samples (>0 FPKM units in triplicate control and Δprd-2). FPKM units for 8622 expressed genes were log2-transformed, averaged, subtracted from control, and Z-scores computed. In all, 129 genes (gold) were upregulated in Δprd-2 (Z-score < −2) and 292 genes (blue) were downregulated in Δprd-2 (Z-score >2). Hypothesizing that PRD-2 is an RNA-binding protein that stabilizes its target transcripts (Figure 3B), we searched for enriched sequence motifs in the untranslated regions of the 292 downregulated genes using Weeder2 (212 annotated 5’-UTRs and 226 annotated 3’-UTRs searched). Zero motifs scored better than 1.5 from Weeder2 output compared to background Neurospora nucleotide frequencies (data not shown). Up- and downregulated gene categories were then run through FunCat to determine functionally enriched categories of genes in the putative PRD-2 regulon. Of the 292 downregulated genes, 128 were input to FunCat, and the top scoring functional categories indicated that carbohydrate and secondary metabolism were decreased in Δprd-2. Out of 128 upregulated genes, 80 were also input to FunCat, and other metabolism categories were identified, which could indicate altered central carbon metabolism in the Δprd-2 mutant, correlating with its slow growth phenotype (Figure 1C).
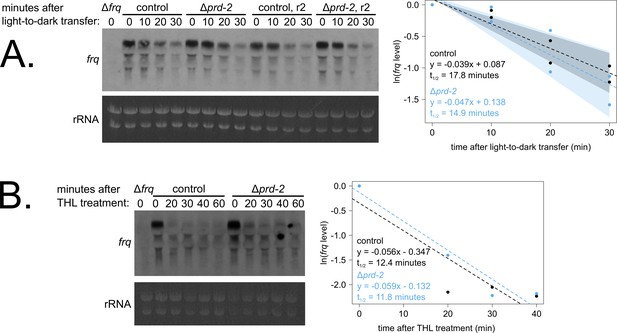
Loss of prd-2 has little effect on stability of the frq transcript.
frq mRNA degradation kinetics were examined by northern blot in a time course after light-to-dark transfer (N = 2 biological replicates). RNA levels were quantified using ImageJ, natural log transformed, fit with a linear model (glm in R, Gaussian family defaults), and half-life was calculated assuming first order decay kinetics (ln(2)/slope). Shaded areas around the linear fit represent 95% confidence intervals on the slope. The frq half-life is approximately 3 min shorter in Δprd-2 but is not statistically different from the control (A). Using the same total RNA samples as shown in Figure 3B, frq degradation was examined by northern blot in a time course after treatment with thiolutin (THL) at approximately CT1 (N = 1 biological replicate). The stability of the frq transcript is not significantly altered in Δprd-2 after THL treatment (B) or light-to-dark transfer (A).
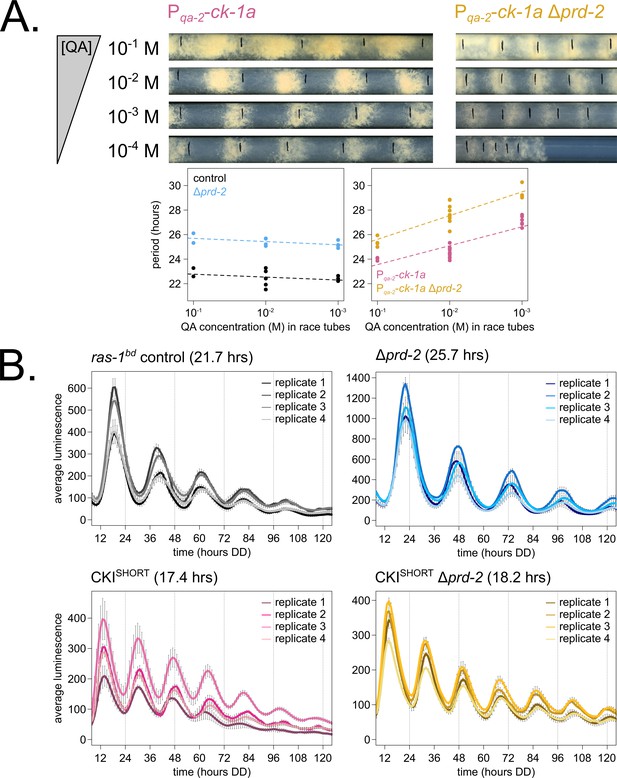
Genetically increasing casein kinase I (CKI) levels or activity rescues the Δprd-2 long period phenotype.
Representative race tubes (RTs) from ras-1bd Pqa-2-ck-1a single (pink) and ras-1bd Pqa-2-ck-1a Δprd-2 double (yellow) mutants are shown with growth using the indicated concentrations of quinic acid (QA) to drive expression of ck-1a. All results are shown in a scatterplot, where each dot represents one RT’s free running period length. ras-1bd controls (black) had an average period of 22.5 ± 0.5 hr (N = 12), and period length was not significantly affected by QA concentration (ANOVA p=0.297). ras-1bd Δprd-2 controls (blue) had an average period of 25.4 ± 0.4 hr (N = 10), and period length was not significantly affected by QA concentration (ANOVA p=0.093). Period length of ras-1bd Pqa-2-ck-1a single mutants (pink) was significantly altered across QA levels (ANOVA p=3.6 × 10−6), and the average period at 10−1 M QA was 24.3 ± 0.5 hr (N = 4). Period length of ras-1bd Pqa-2-ck-1a Δprd-2 double mutants (yellow) was also significantly affected by QA levels (ANOVA p=8.1 × 10−8), and the average period at 10−1 M QA was 25.4 ± 0.4 hr (N = 4). The double mutant period length was not genetically additive at high levels of QA induction (A). A hyperactive CKI allele was constructed by expressing the shortest isoform only (CKISHORT). 96-well plate luciferase assays were used to measure the circadian period length. Traces represent the average of three technical replicates across four biological replicate experiments for: ras-1bd controls (gray, τ = 21.7 ± 0.3 hr), ras-1bd Δprd-2 (blue, τ = 25.7 ± 0.6 hr), ras-1bd CKISHORT (pink, τ = 17.4 ± 0.3 hr), and ras-1bd CKISHORT Δprd-2 double mutants (yellow, τ = 18.2 ± 0.3). CKISHORT is completely epistatic to Δprd-2 in double mutants (B).
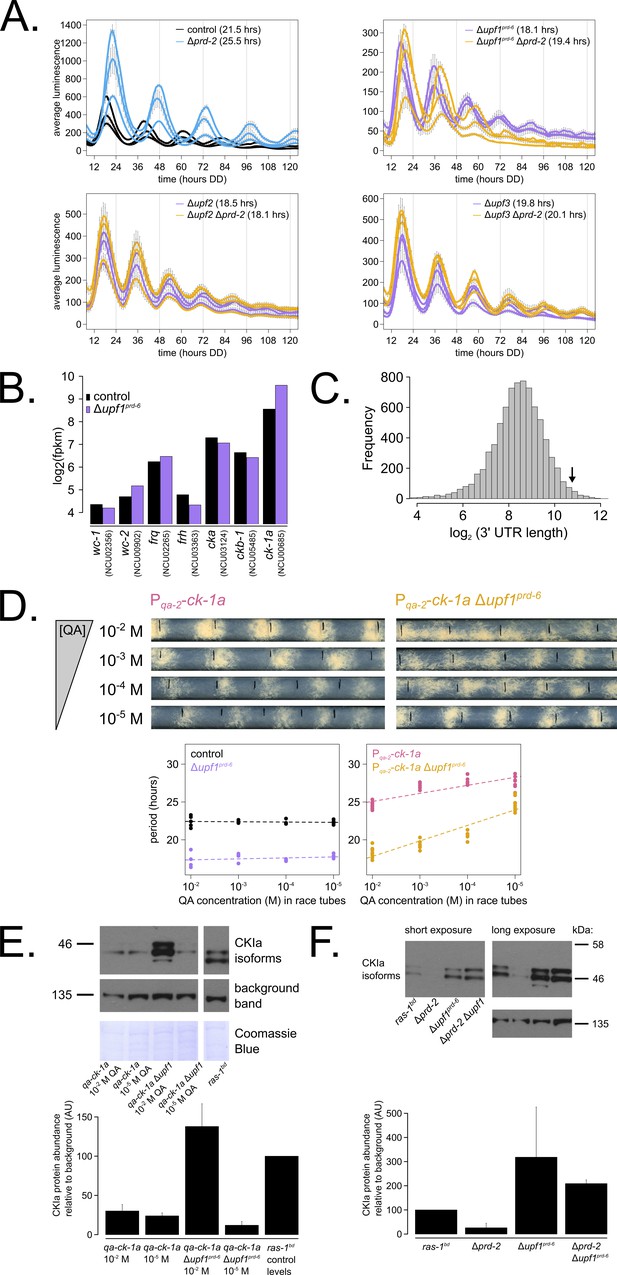
Nonsense-mediated decay (NMD) negatively regulates casein kinase I (CKI) levels via UPF1PRD-6, establishing a basis for the upf1prd-6 prd-2 genetic epistasis on circadian period length.
96-well plate luciferase assays were used to measure the circadian period length in triplicate wells per three biological replicate experiments for: ras-1bd controls (black, τ = 21.5 ± 0.3 hr), ras-1bd Δprd-2 (blue, τ = 25.5 ± 0.4 hr); ras-1bd Δupf1prd-6 (purple, τ = 18.1 ± 0.2 hr), ras-1bd Δupf1prd-6Δprd-2 double mutants (yellow, τ = 19.4 ± 0.7 hr); ras-1bd Δupf2 (purple, τ = 18.5 ± 0.5 hr), ras-1bd Δupf2 Δprd-2 double mutants (yellow, τ = 18.1 ± 0.3 hr); ras-1bd Δupf3 (purple, τ = 19.8 ± 0.3 hr), ras-1bd Δupf3 Δprd-2 double mutants (yellow, τ = 20.1 ± 0.2 hr). Each individual NMD subunit knockout is epistatic to the Δprd-2 long period phenotype (A). Raw RNA-seq data from a previous study (Wu et al., 2017) were analyzed using the same pipeline as data from Figure 3A (see Materials and methods). Control and Δupf1prd-6 gene expression levels (log2-transformed) are shown for core clock genes. The ck-1a transcript is >2× more abundant in Δupf1prd-6 (B). 3’-UTR lengths from 7793 genes were mined from the N. crassa OR74A genome annotation (FungiDB version 45, accessed on 10/25/2019), and plotted as a histogram. The arrow marks the 3’-UTR of ck-1a, which is 1739 bp and within the top 100 longest annotated UTRs in the entire genome (C). Representative race tubes (RTs) from ras-1bd Pqa-2-ck-1a single (pink) and ras-1bd Pqa-2-ck-1a Δupf1prd-6 double (yellow) mutants are shown at the indicated concentrations of quinic acid to drive expression of ck-1a. All results are shown in a scatterplot, where each dot represents one RT’s free running period length. ras-1bd controls (black) had an average period of 22.4 ± 0.4 hr (N = 20), and period length was not significantly affected by QA concentration (ANOVA p=0.605). ras-1bd Δupf1prd-6 controls (purple) had an average period of 17.5 ± 0.6 hr (N = 16), and period length was not significantly affected by QA concentration (ANOVA p=0.362). Period length of ras-1bd Pqa-2-ck-1a single mutants (pink) was significantly altered across QA levels (ANOVA p=2.9×10−8), and the average period at 10−5 M QA was 27.6 ± 0.8 hr (N = 8). Period length of ras-1bd Pqa-2-ck-1a Δupf1prd-6 double mutants (yellow) was also significantly affected by QA levels (ANOVA p=9.4×10−12), and the average period at 10−5 M QA was 24.7 ± 0.9 hr (N = 8). Thus, the double mutant period length was not genetically additive at low levels of QA induction, and the short period phenotype of Δupf1prd-6 is rescued (D). CKI protein levels were measured from the indicated genotypes grown in 0.1% glucose liquid culture medium (LCM) with QA supplemented at the indicated concentrations for 48 hr in constant light. A representative immunoblot of three biological replicates is shown, and replicates are quantified in the bar graph relative to ras-1bd control CKI levels from a 2% glucose LCM culture (E). CKI protein levels were measured from the indicated genotypes grown in 2% glucose LCM for 48 hr in constant light. A representative immunoblot of three biological replicates is shown, and replicates are quantified in the bar graph relative to ras-1bd control CKI levels (F). CKI protein levels are increased in Δupf1prd-6, decreased in the Δprd-2 mutant, and Δupf1prd-6 is epistatic to Δprd-2 with respect to CKI levels and circadian period length.
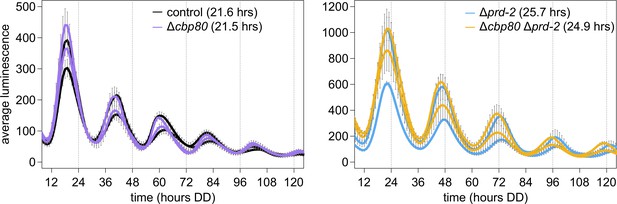
The cap-binding protein CBP80 (NCU04187) is not required for a normal clock, and does not alter the Δprd-2 long period phenotype, suggesting that ck-1a degradation is controlled by NMD machinery without the Exon Junction Complex and nuclear cap-binding complex.
96-well plate luciferase assays were used to measure the circadian period length. Traces represent the average of three technical replicates across two biological replicate experiments for: ras-1bd controls (black, τ = 21.6 ± 0.4 hr), ras-1bd Δcbp80 (purple, FGSC22441, τ = 21.5 ± 0.3 hr), ras-1bd Δprd-2 (blue, τ = 25.7 ± 0.2 hr), and ras-1bd Δcbp80 Δprd-2 (yellow, τ = 24.9 ± 0.2 hr). ΔNCU04187 has a wild-type circadian period length and does not genetically interact with Δprd-2, suggesting that ck-1a regulation does not require CBP80.
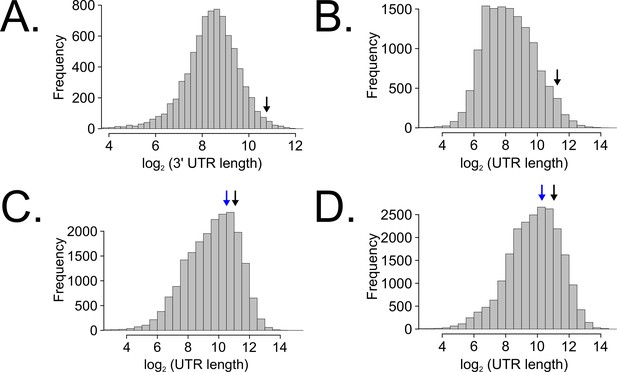
Long untranslated regions (UTRs) are characteristic of casein kinase I gene orthologs across species.
Neurospora 3’-UTR lengths were mined from 7793 annotated genes (as described in Figure 5C) and plotted as a histogram. The black arrow marks the 3’-UTR of ck-1a (NCU00685) at 1739 bp in length (A). UTR lengths from Drosophila melanogaster were mined from 13,552 uniquely annotated genes (Ensembl GTF version BDGP6, accessed on 8/5/2020 from Illumina iGenomes) and plotted as a histogram. The black arrow marks the UTR of dbt (FBgn0002413) at 2443 bp in length (B). UTR lengths from Mus musculus were mined from 20,477 uniquely annotated genes (Ensembl GTF version GRCm38, accessed on 8/5/2020 from Illumina iGenomes) and plotted as a histogram. The black arrow marks the UTR of CSNK1D (ENSMUSG00000025162) at 2157 bp in length, and the blue arrow corresponds to CSNK1E (ENSMUSG00000022433) at 1456 bp (C). UTR lengths from Homo sapiens were mined from 22,401 uniquely annotated genes (Ensembl GTF version GRCh37, accessed on 8/5/2020 from Illumina iGenomes) and plotted as a histogram. The black arrow marks the UTR of CSNK1D (ENSG00000141551) at 2113 bp in length, and the blue arrow corresponds to CSNK1E (ENSG00000213923) at 1247 bp (D).
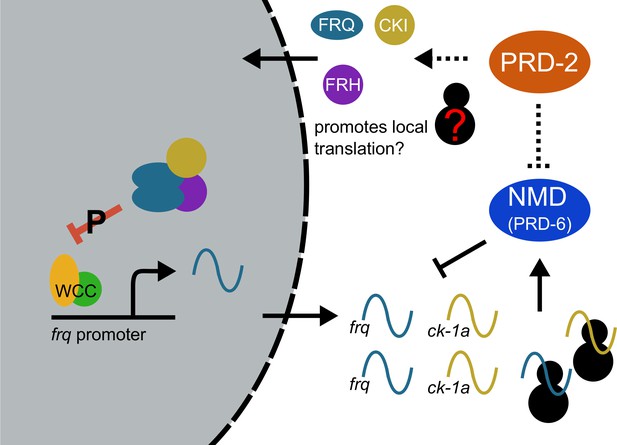
Counterbalancing regulation of casein kinase I (CKI) provides a unifying genetic model for the action of PRD-2 and UPF1PRD-6 in the circadian oscillator.
The NMD complex (UPF1PRD-6, UPF2, and UPF3) targets the frq and ck-1a transcripts for degradation (upstream uORFs in frq; long 3’-UTR in ck-1a). PRD-2 binds to and stabilizes ck-1a transcripts (dashed lines), which could also promote local translation and complex formation for the negative arm of the clock. In the absence of PRD-2, the long period phenotype is due to low CKI levels, and in the absence of NMD, the short period phenotype is due to high CKI levels.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Neurospora crassa) | prd-2 | FungiDB | NCU01019 | |
| Gene (Neurospora crassa) | upf1prd-6 | FungiDB | NCU04242 | |
| Gene (Neurospora crassa) | ck-1a | FungiDB | NCU00685 | |
| Strain, strain background (Neurospora crassa) | Supplementary file 1 | This study; Fungal Genetics Stock Center (FGSC) | ||
| Antibody | Anti-V5 (mouse monoclonal) | ThermoFisher | Cat. # R960-25 | (1:3000) |
| Antibody | Anti-tubulin alpha (mouse monoclonal) | Fitzgerald | Cat. # 10R-T130a | (1:10,000) |
| Antibody | Anti-CKI (rabbit polyclonal) | Generous gift from Michael Brunner (University of Heidelberg) | (1:1000) | |
| Antibody | Anti-FLAG M2 magnetic beads (mouse monoclonal) | Sigma | Cat. # M8823 | 30 μl beads incubated with 10 mg total protein for UV-CLIP |
| Recombinant DNA reagent | c box-luc (plasmid-derived construct) | As described, PMID:25635104 | <500 bp of the frq promoter driving codon-optimized luciferase; targeted to the csr-1 locus for selection | |
| Chemical compound, drug | D-quinic acid | Sigma | Cat. # 138622 | 1 M stock solution, pH adjusted to 5.8 with NaOH |
| Chemical compound, drug | Allele-In-One Mouse Tail Direct Lysis Buffer | Allele Biotechnology | Cat. # ABP-PP-MT01500 | 50 μl reagent mixed withNeurospora asexual spores for gDNA isolation |
| Chemical compound, drug | Thiolutin | Cayman Chemical | Cat. # 11350 | Stock solution prepared in DMSO |
| Software, algorithm | Custom R software | https://github.com/cmk35 | UTR length analyses from Figure 5—figure supplement 2 |
Additional files
-
Supplementary file 1
Neurospora crassa strains used in this study.
- https://cdn.elifesciences.org/articles/64007/elife-64007-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64007/elife-64007-transrepform-v2.docx





