Cerebrospinal fluid proteome maps detect pathogen-specific host response patterns in meningitis
Figures
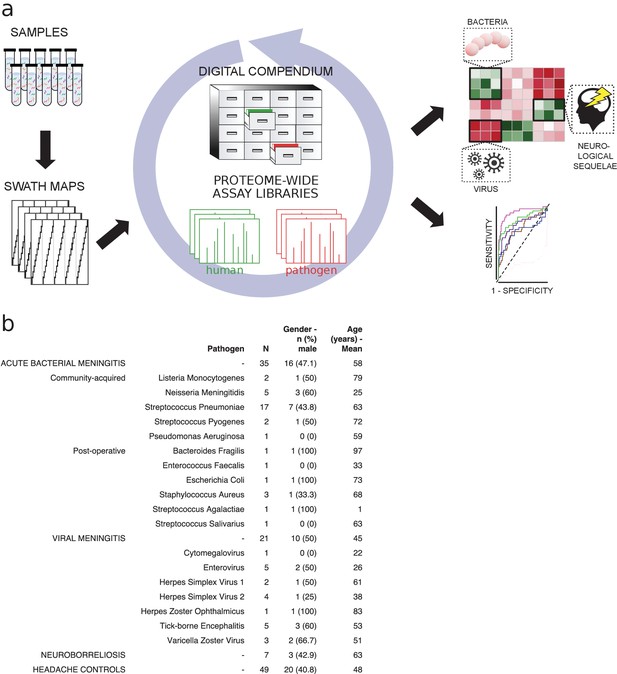
Method flow chart and patient cerebrospinal fluid (CSF) samples.
(a) Fifty microliters of CSF from each sample were prepared and analyzed using both shotgun mass spectrometry (MS) and sequential window acquisition of all theoretical fragment ion spectra (SWATH-MS) for the construction of CSF proteome assay library and production of a digital compendium of SWATH-MS proteome maps. These SWATH-MS maps were post-acquisition interrogated with previously established assay library from 28 healthy human organs or primary cells to enable the quantification of proteins enriched in relevant tissues such as brain, plasma, or immune cells. The SWATH-MS maps were further interrogated with the CSF assay library for determining protein profiles correlating with acute bacterial meningitis, viral meningitis, neuroborreliosis and control samples. (b) A summary showing the number of CSF samples in each sample group, as well as the bacterial or viral strains that caused meningitis. The distribution of gender (as percentage in male) and average age for each group is also presented.
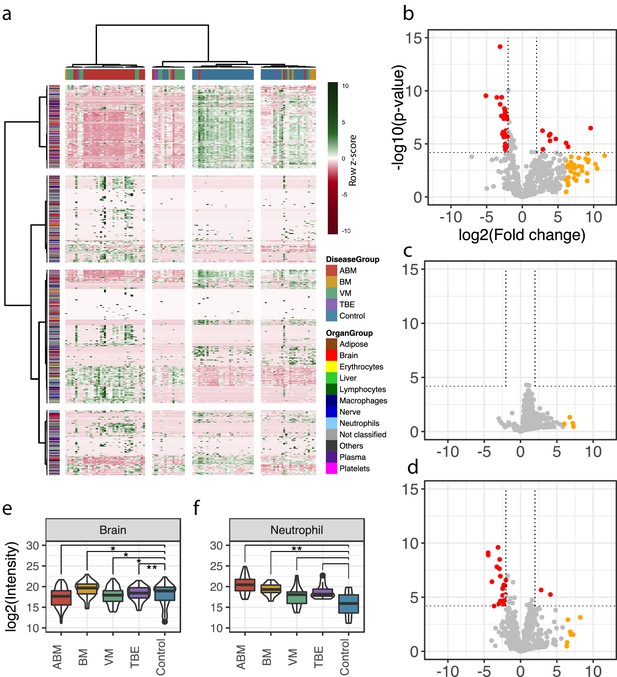
Cerebrospinal fluid proteome analysis.
(a) The quantified proteins (rows) and samples (columns) were clustered and visualized in a heat map. Top horizontal color bar classified sample groups, and left vertical color bar classified the human tissue assignments for each protein. The data was row-wise normalized by Z-score transformation, and column and row clustering was performed with ward.D and canberra clustering criterions. (b–d) Differentially expressed proteins between control and acute bacterial meningitis (ABM) (b), neuroborreliosis (BM) (c), and viral meningitis (VM) (d) are shown in volcano plots. Statistically significant proteins (Hochberg-corrected p-values≤0.05 and log2 fold change ≤ −2 and ≥2) are labeled in red. Statistically non-significant proteins with a high fold change of ≥64 are labeled in yellow. (e, f) Log2 scaled average intensities of brain and neutrophil-associated proteins for ABM, BM, VM, tick-borne encephalitis (TBE), and control are presented. The significance of the changes was calculated with a standard Student's t-test and marked with asterisk (one or two star significance).
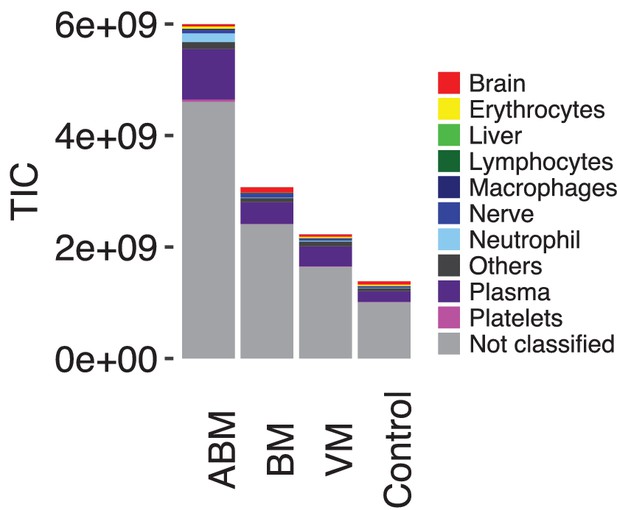
Overview of average protein content and intensities.
Average total ion currents (TIC) for acute bacterial meningitis (ABM), neuroborreliosis (BM), viral meningitis (VM), and control groups are presented, and the contribution of each tissue group is shown with color.
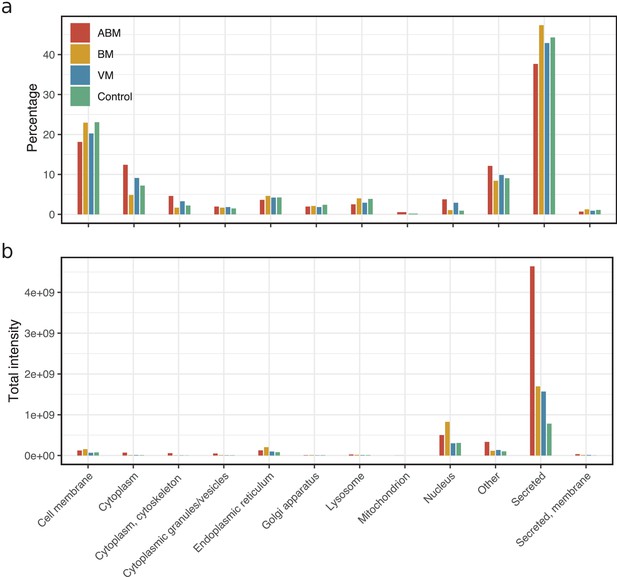
Predicted subcellular compartments of detected proteins.
The detected proteins in this dataset were matched to the human reference proteome from UniProt (Proteome ID: UP000005640, March 2021), and the annotation for subcellular location was obtained. The primary subcellular compartment was selected, and these are visualized as grouped bar plots for each sample group; acute bacterial meningitis (ABM; red), neuroborreliosis (BM; yellow), viral meningitis (VM; blue), and control (green) bars. (a) Percentage on y-axis depicts the proportion of a particular subcellular compartment within the sample group. (b) The total average intensity of each protein belonging to corresponding subcellular compartment was summed and shown. Separate compartment groups were made in case two primary subcellular compartments were indicated for a protein (e.g., ‘Secreted, membrane’). Proteins that were annotated to multiple subcellular compartments are classified as ‘Other.’
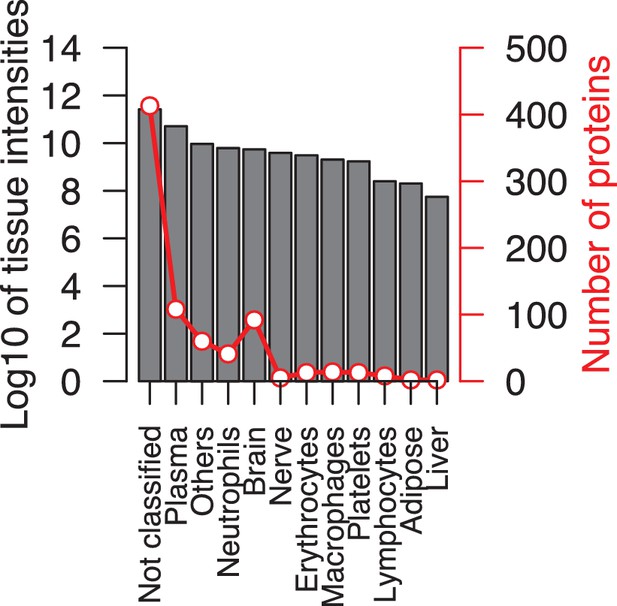
Overview of average protein content and intensities.
A dual-axis graph showing the average intensities for each 12 tissues (left y-axis) and the number of proteins associated to each tissue (right y-axis).
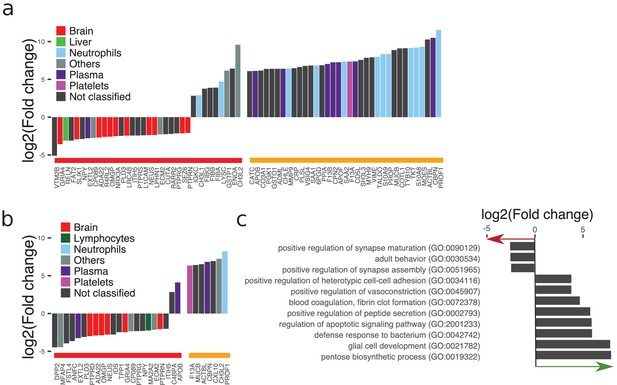
Analysis of differences in the cerebrospinal fluid (CSF) of patients with meningitis.
(a, b) The tissue assignment, protein abundances, and identities of proteins with statistical significance (Benjamini–Hochberg-corrected p-value≤0.05 and log2 fold change ≤ −2 and ≥2) are shown and labeled with red horizontal line. Statistically non-significant proteins with high fold change of ≥64 were considered of biological interest and labeled with yellow horizontal line. The data is shown for acute bacterial meningitis (ABM) in (a) and for viral meningitis (VM) in (b). (c) Proteins selected for ABM were annotated by gene ontology terms. The average fold change for all proteins included in each term is plotted to show the direction of change compared to controls.
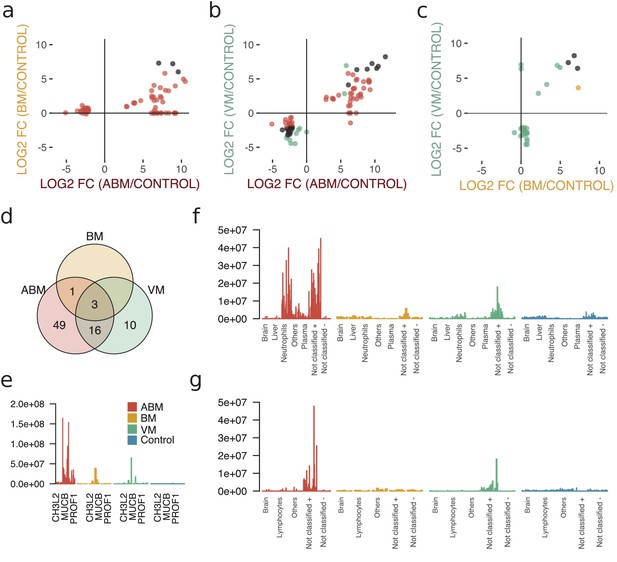
Cross-comparison of cerebrospinal fluid (CSF) protein composition between acute bacterial meningitis (ABM), neuroborreliosis (BM), and viral meningitis (VM).
(a–c) Significantly regulated proteins (Benjamini–Hochberg-corrected p-value≤0.05 and log2 fold change of ≤ −2 and ≥2) together with proteins with high fold change of ≥64 for all three groups were selected, and their fold changes of two disease group at a time were plotted against each other: ABM versus BM (a), ABM versus VM (b), and BM versus VM (c). The points are colored based on their significance in either one group only (red: ABM; orange: BM; green: VM) or in both (black). Proteins absent in one group were plotted at the null line for that group for visualization purposes. (d) Venn diagram summarizes the overlap of the proteins shown in (a–c) for the different groups. The intensities of the three proteins shared in all three groups (e), 49 proteins specific to ABM (f), and 10 proteins specific to VM (g) are shown as bar plots for every CSF sample. Proteins not classified to a tissue were further divided into two groups depending on if they were downregulated (-) or upregulated (+) compared to control.
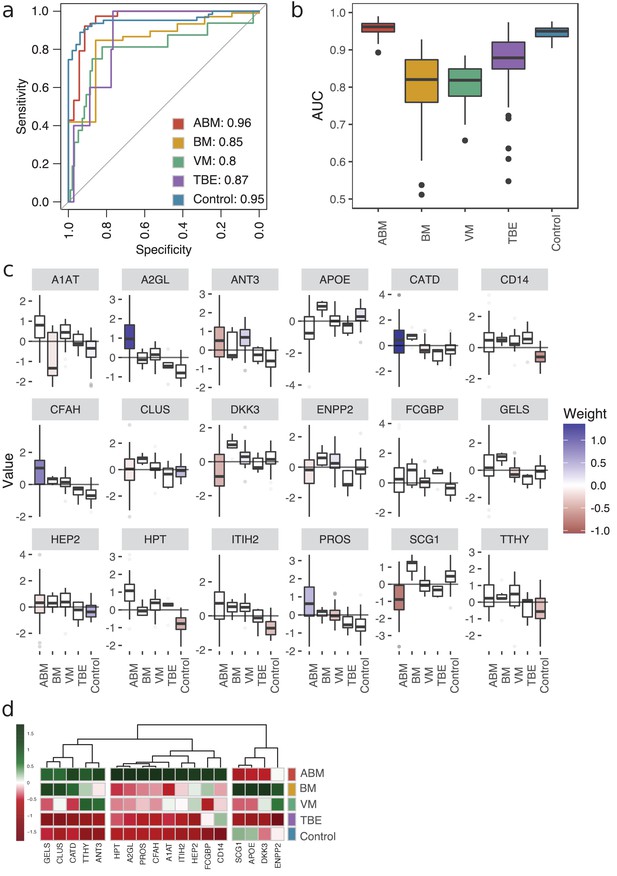
Predictive proteomic profiling using a LASSO regression model.
(a) Predictive proteomic profiling was performed on all detected and manually curated proteins (n = 771) by LASSO regression model to assign predictive scores to discriminate acute bacterial meningitis (ABM), neuroborreliosis (BM), viral meningitis (VM), tick-borne encephalitis (TBE), and control samples from each other. The samples were split into four randomly selected folds, and the modeling was repeated 100 times. The average area under the curve (AUC) of the sensitivity and specificity of the predicted model is presented in a receiver operating characteristic (ROC) curve. (b) The variation of the AUC across the 100 repetitions is shown as box plots. (c) All proteins that were detected in each fold and in ≥90% of the 100 repetitions (18 proteins) were selected as important proteins for the construction of the LASSO prediction model, and their value (e.g., log-transformed, centered, and scaled protein abundances) plotted as box plots. The fill color of the box plot represents the weight coefficient assigned to each individual protein, where blue and red represent positive or negative effect of that protein for prediction of that specific sample group. (d) The selected 18 proteins are presented in a heat map, with sample groups as rows and sample group averages for each protein as columns.
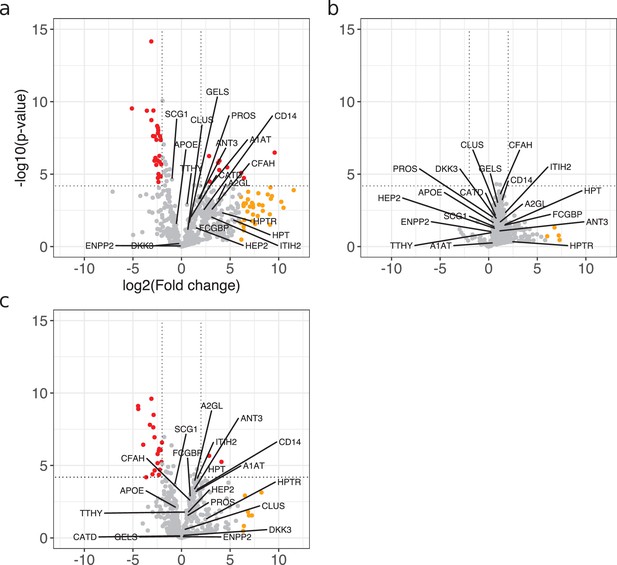
Visualization of the 18 predictive proteins in volcano plots.
(a–c) Differentially expressed proteins between control and acute bacterial meningitis (ABM) (a), neuroborreliosis (BM) (b), and viral meningitis (VM) (c) are shown in volcano plots. Statistically significant proteins (Hochberg-corrected p-values≤0.05 and log2 fold change ≤ −2 and ≥2) are labeled in red. Statistically non-significant proteins with a high fold change of ≥64 are labeled in yellow. These volcano plots are identical to the volcano plots in Figure 2b–d. The 18 predictive proteins selected as important proteins for the construction of the LASSO prediction model are labeled in each volcano plot. The color of the protein labels complies with the volcano plot colors.
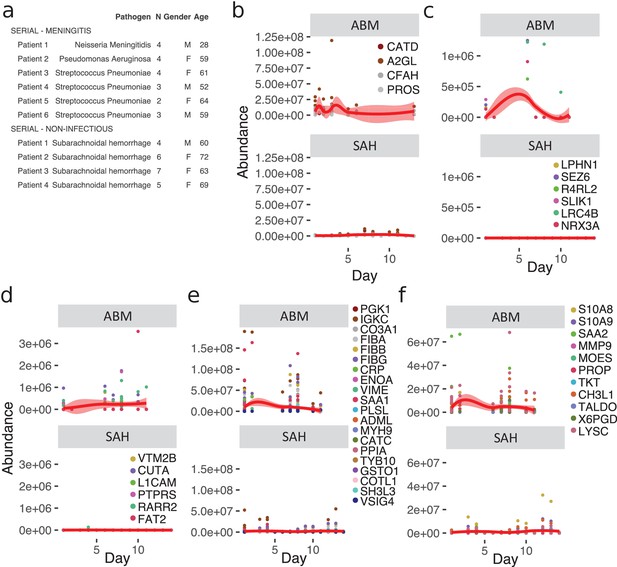
Longitudinal investigation of protein abundances in acute bacterial meningitis (ABM) and subarachnoidal hemorrhage (SAH).
(a) Summary of the cerebrospinal fluid (CSF) samples collected from patients with ABM (n = 6) or SAH (n = 4) for longitudinal analysis of CSF proteome is presented. (b–f) Based on previous results, five groups of proteins were selected for longitudinal analysis in ABM up to 10 days and in SAH up to 13 days after admittance to hospital: four proteins selected by LASSO as predictive and indicative of ABM (b), brain-associated proteins downregulated in AMB (c), downregulated proteins not classified to a tissue and specific to ABM (d), upregulated proteins not classified to a tissue and specific to ABM (e), and neutrophil proteins upregulated in ABM (f).
Additional files
-
Supplementary file 1
Cross-referencing the tissue assignments based on Malmström et al. (unpublished work).
The tissue assignments based on Malmström et al. (unpublished work) for proteins in Figures 3f, g, 4c, and 5b–f and Figure 2—figure supplement 4a, b matched and compared to publicly available and published protein tissue assignment repositories.
- https://cdn.elifesciences.org/articles/64159/elife-64159-supp1-v2.xlsx
-
Supplementary file 2
The full data generated from 112 data-independent acquisition MS-runs (acute bacterial meningitis [ABM]: n = 25, neuroborreliosis [BM]: n = 7, viral meningitis [VM]: n = 21 of which tick-borne encephalitis [TBE]: n = 5 and controls: n = 49).
The data was manually curated to remove immunoglobulin variable chain proteins.
- https://cdn.elifesciences.org/articles/64159/elife-64159-supp2-v2.xlsx
-
Supplementary file 3
The full data generated from the data-independent acquisition MS-runs from the longitudinal study.
The cohort consists of longitudinal samples collected from six acute bacterial meningitis (ABM) patients (6 original samples used in this study and additional 14 longitudinal samples) and from four patients with subarachnoidal hemorrhage (SAH, 22 longitudinal samples). The data was manually curated to remove immunoglobulin variable chain proteins.
- https://cdn.elifesciences.org/articles/64159/elife-64159-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64159/elife-64159-transrepform-v2.docx





