Stretch-activated ion channels identified in the touch-sensitive structures of carnivorous Droseraceae plants
Figures
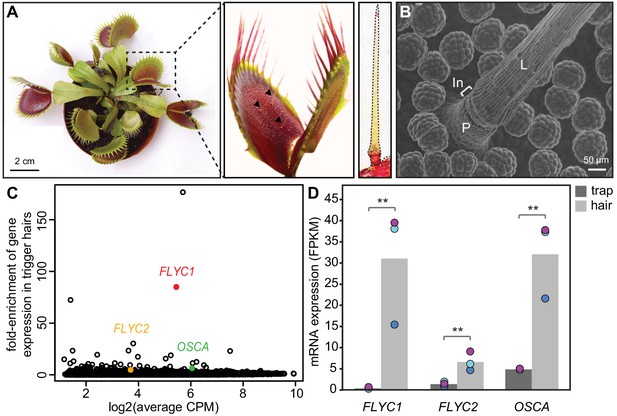
Identification of putative mechanosensory channels in the Venus flytrap trigger hair.
(A) Representative image of a soil-grown Venus flytrap clone (left), Venus flytrap leaf (center), and single trigger hair (right). Black arrowheads in the center picture indicate trigger hairs on leaf. (B) Scanning electron micrograph of a trigger hair. Cells of the lever (L), indentation zone (In) and podium (P) are indicated. Also seen are the digestive glands on the floor of the lobe. (C) Fold enrichment of protein-coding genes of >100 amino acids in length (black circles) in the trigger hair relative to the trap. FLYC1, FLYC2, and OSCA are shown in red, orange, and green, respectively. CPM, counts per million of mapped sequencing reads. (D) Average Fragments Per Kilobase of transcript per Million mapped reads (FPKM) for FLYC1, FLYC2, and OSCA in traps and trigger hairs. Dots of the same color indicate paired biological replicates. **FDR < 0.005.
-
Figure 1—source data 1
Size estimates of two Arabidopsis (Col-0) samples compared to our Venus flytrap strain (CP01).
- https://cdn.elifesciences.org/articles/64250/elife-64250-fig1-data1-v1.docx
-
Figure 1—source data 2
Summary of sequencing reads used to build the de novo transcriptome (NCBI Transcriptome Shotgun Assembly Sequence Database accession # GHJF00000000).
- https://cdn.elifesciences.org/articles/64250/elife-64250-fig1-data2-v1.docx
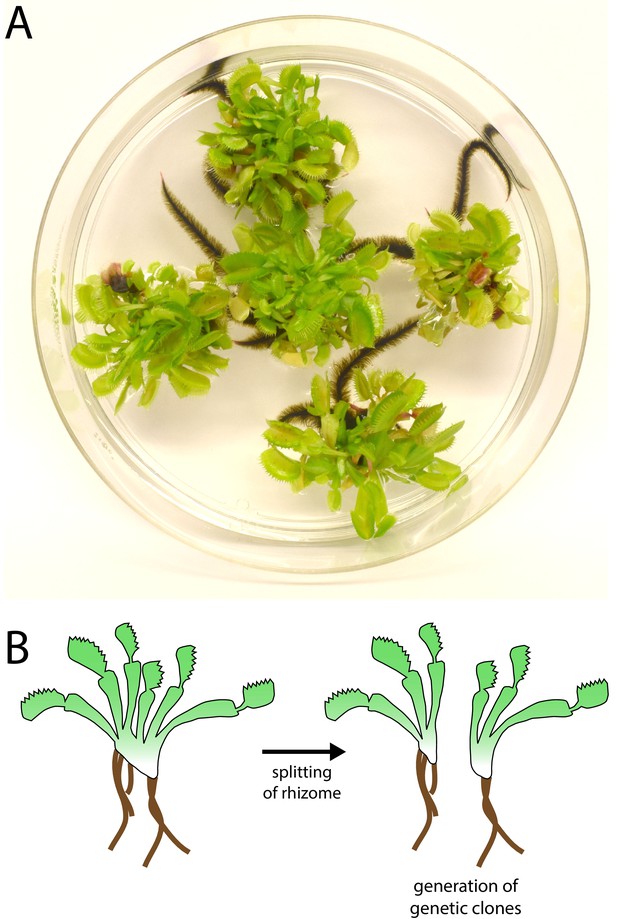
Venus flytrap clonal propagation system.
(A) Example of clonal Venus flytraps growing in tissue culture (10 cm plate) using methods adapted from Jang et al., 2003). (B) Diagram depicting the method of propagation. Rosettes were separated by splitting the rhizome. Plants were then transferred to fresh sterile growth medium for further propagation in culture or transferred to soil and ‘hardened’ for at least 2–3 months prior to experiments.
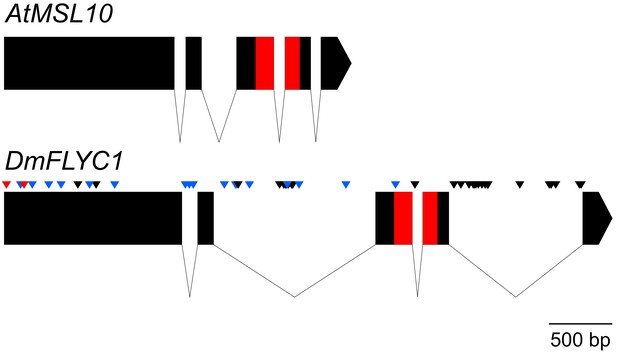
Gene structure of Venus flytrap FLYC1.
A comparison of Arabidopsis MSL10 and Venus flytrap FLYC1 genes. Boxes represent exons; lines, introns. The sequence between the start and stop codons only is shown. SNPs (allelic differences) in FLYC1 are indicated for our strain (CP01) by black arrowheads. Thirty-two SNPs in total were detected, of which only two were found in the coding region and were both silent. Additional SNPs and short sequence variations found in the cultivar 'Creeping Death' are indicated by blue arrowheads (silent variations) and red (causing amino acid changes P8S and A51G relative to strain CP01).

Gene sequence of FLYC1.
Exons, uppercase and bold; introns, lower case. SNPS and sequence variations identified in the strain CP01 and "Creeping Death" cultivar are shown in square brackets and are highlighted in red.
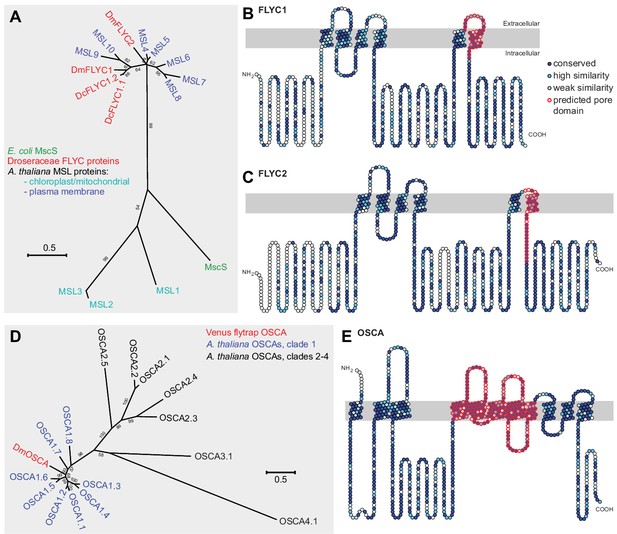
Molecular phylogenetic relationship of FLYCATCHER and OSCA proteins.
(A) Phylogenetic analysis by maximum likelihood method to show the relationship between the conserved MscS domain (Haswell and Meyerowitz, 2006) of Escherichia coli MscS protein, Arabidopsis thaliana MSL proteins (MSL1–MSL10), and Dionaea muscipula/Venus flytrap (DmFLYC1 and DmFLYC2) and Drosera capensis/Cape sundew (DcFLYC1.1 and DcFLYC1.2) FLYCATCHER proteins. (B, C) Predicted topology for DmFLYC1 (B) and DmFLYC2 (C) proteins. (D) Phylogenetic analysis by maximum likelihood method showing the relationship between A. thaliana OSCA family proteins and Venus flytrap DmOSCA. (E) Predicted topology for DmOSCA protein. In (A) and (D), bootstrap values >50 are shown; scale, substitutions per site. In (B), (C), and (E), amino acid residues that are conserved with Arabidopsis MSL10, MSL5, and OSCA1.5, respectively, are indicated in dark blue circles, whereas residues similar in identity are indicated in lighter blue circles. The predicted pore domain for each protein is indicated in red circles.
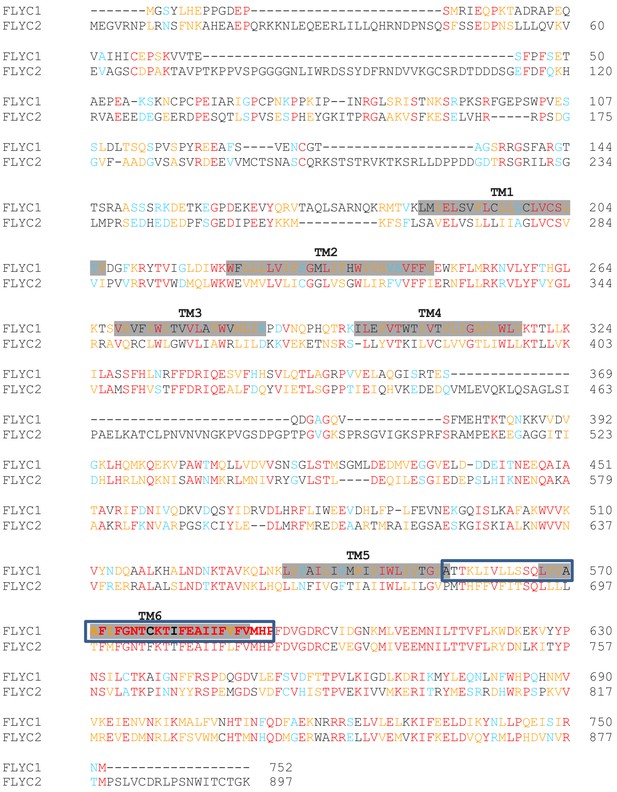
Sequence alignment of DmFLYC1 and DmFLYC2 proteins.
Conserved amino acids are shown in red. Similar and highly similar amino acids are in blue and orange, respectively. For FLYC1, also shown are the transmembrane (TM) domains shadowed in gray as predicted by Protter (Omasits et al., 2014), and the residues forming the putative pore domain modeled in Figure 4—figure supplement 3 are boxed.
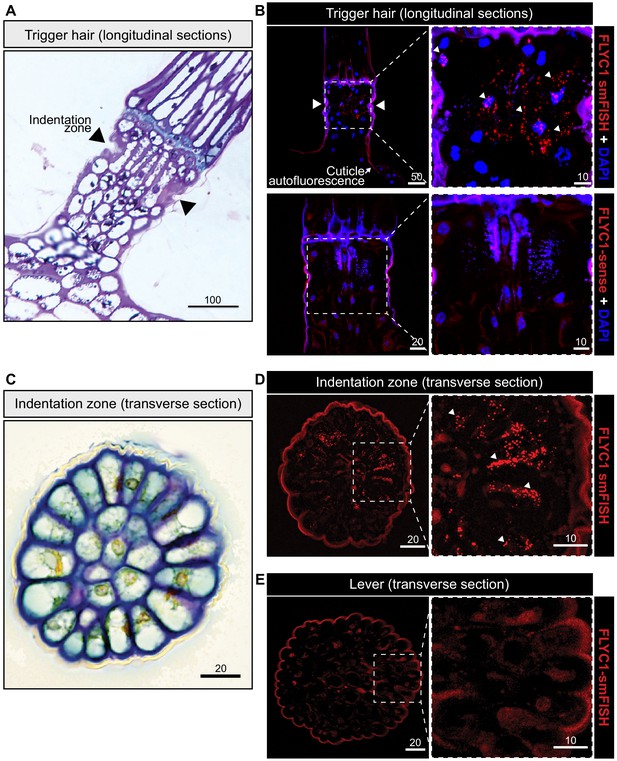
FLYC1 mRNA localization in Venus flytrap trigger hairs.
(A) Toluidine blue-stained longitudinal section through the base of a trigger hair. Elongated sensory cells are visible at the indentation zone (arrowheads). (B) Max projection through a longitudinal section after fluorescent in situ hybridization. (Top) FLYC1 transcript (red) and DAPI (blue) at low (left) and high (right) magnification of the indentation zone. Note localization of the FLYC1 transcript puncta in sensory cells. (Bottom) No signal was observed when using FLYC1-sense probes. High background fluorescence is observed in all channels. (C) Toluidine blue-stained transverse section through the indentation zone. The cells forming the outer ring are presumed to be the mechanosensors (Benolken and Jacobson, 1970). (D) Max projection through a transverse section after fluorescent in situ hybridization. FLYC1 transcript (red) at low (left) and high (right) magnification. Note localization of the FLYC1 transcript puncta predominantly in the sensory cells of the indentation zone. (E) Max projection through a transverse section of the trigger hair lever. No transcript was observed. Scale bars, μm.
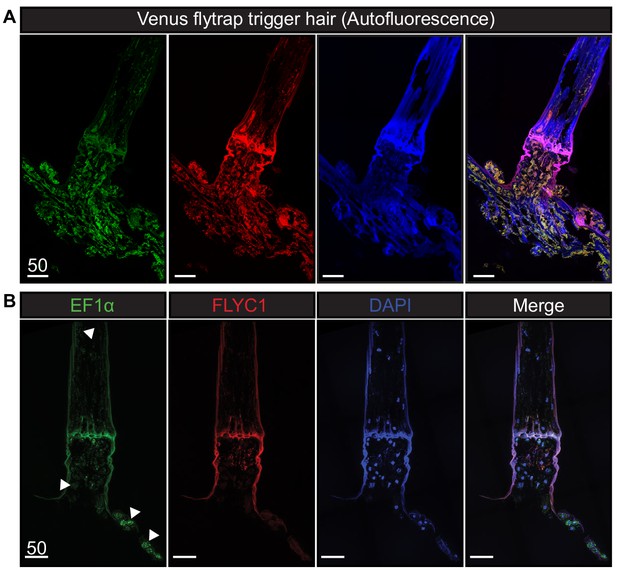
Autofluorescence and EF1α transcript expression in Venus flytrap.
(A) Unprocessed Venus flytrap trigger hair in different channels to depict autofluorescence from the plant cuticle. Images were taken with the same settings as tissue subjected to in situ hybridization. (B) A longitudinal section of a trigger hair and trap with smFISH against the housekeeping gene EF1α (green), FLYC1 (red), and DAPI (blue). White arrowheads indicate examples of EF1α expression in various cells throughout the tissue. Scale bar, µm.
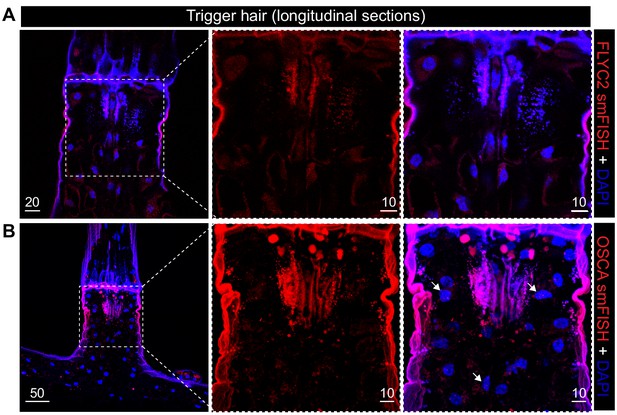
FLYC2 and DmOSCA transcript expression in trigger hair.
Max projection through longitudinal sections after fluorescent in situ hybridization of (A) FLYC2 transcript and (B) DmOSCA transcript at low (left) and high resolution (center and right). FLYC2 and DmOSCA transcript is depicted in red channel and DAPI in blue. Note no transcript was observed for FLYC2, whereas DmOSCA was observed mostly in the sensory cells. Scale bars, µm.
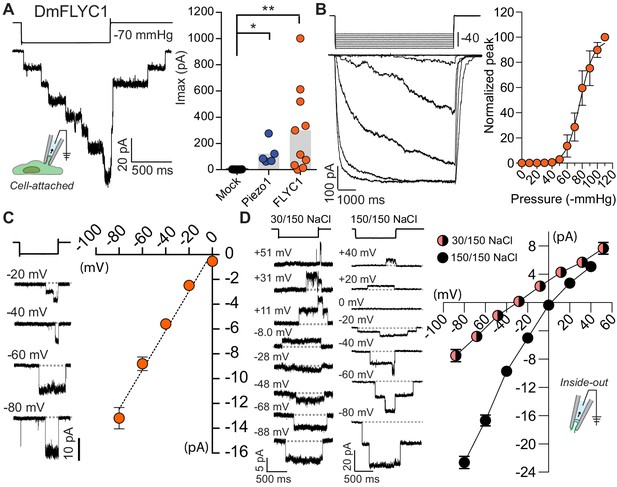
FLYC1 induces stretch-activated currents.
(A) Left, representative trace of stretch-activated current recorded from FLYC1-expressing HEK-P1KO cells in the cell-attached patch clamp configuration at −80 mV membrane potential in response to −70 mmHg pipette pressure. Stimulus trace illustrated above the current trace. Right, quantification of maximal current response from cells transfected with mock (N = 7), mouse PIEZO1 (N = 5), or FLYC1 plasmid (N = 10). p=0.0251 (mock vs. PIEZO1); p=0.0070 (mock vs. FLYC1); Dunn’s multiple comparison test. (B) Left, currents in response to graded negative pressure steps from 50 to 90 mmHg (Δ5 mmHg) at −60 mV membrane potential. Right, average pressure–response curve normalized to peak current across cells (pressure–response curve for absolute peak values is plotted in Figure 4—figure supplement 2). A fit with Boltzmann equation revealed P50 value of 77 mmHg (N = 9). (C) Left, representative single-channel traces in response to stretch at the indicated membrane potential. Right, average I–V relationship of stretch-activated single-channel currents from FLYC1-transfected cells (N = 6). (D) Left, representative stretch-activated single-channel currents recorded from excised inside-out patch configuration in asymmetrical or symmetrical NaCl solution at the indicated membrane potential. Right, average I–V of stretch-activated single-channel currents in asymmetrical NaCl solution (N = 7, red/black circles) or symmetrical NaCl solution (N = 8, black cirlces). Data for both conditions were collected from separate patches/cells. Scatter plots in B–D are mean ± s.e.m.
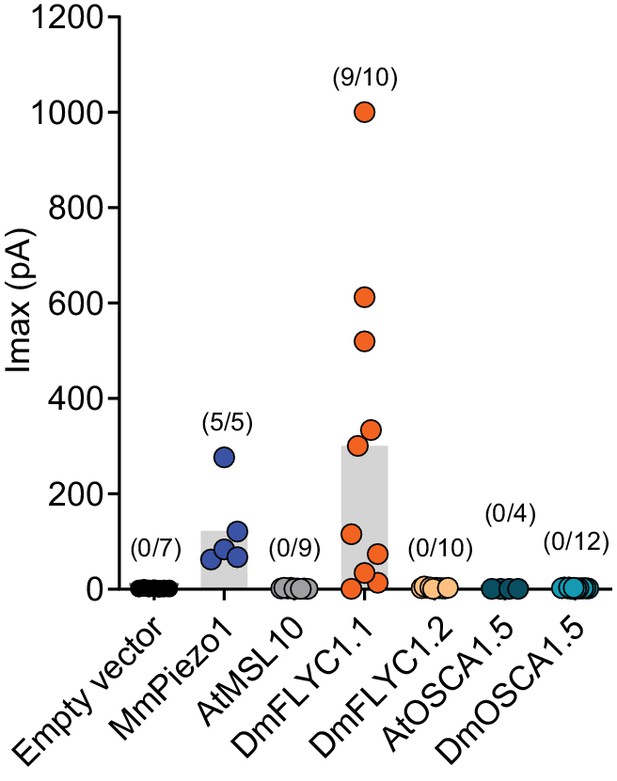
DmFLYC2 and DmOSCA functionality.
Macroscopic stretch-activated currents recorded from HEKP1-KO cells transfected with Mock (N=7), MmPiezo1 (N=5), AtMSL10 (N=9), DmFLYC1 (N=10), DmFLCY2 (N=10), AtOSCA1.5 (N=4), and DmOSCA (N=12) plasmids. Empty vector, MmPiezo1, and DmFLYC1 data is the same as Figure 4A.
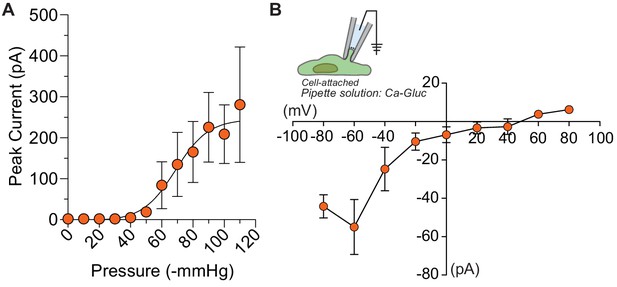
Half-maximal pressure response and chloride permeability in DmFLYC1.
(A) Average pressure response curve across different cells (N=9). Same data as Figure 4B but the peak current is not normalized to maximal response. (B) Average I–V (N=5) of DmFLYC1 stretch-activated currents recorded in cell attached patch clamp configuration. Scatter plots are mean ± s.e.m .
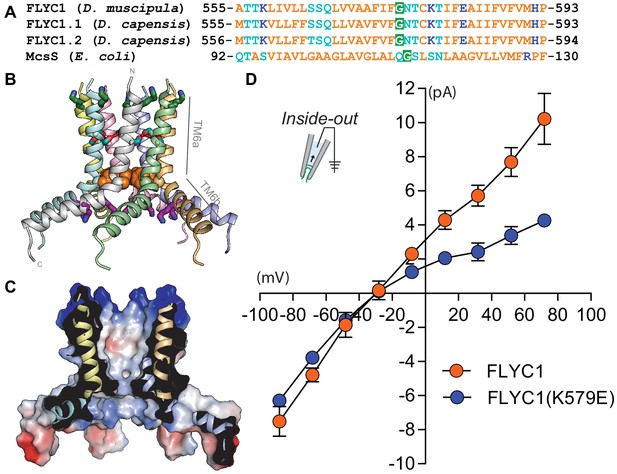
The sequence of a putative pore-forming helix in FLYC1 is compatible with MscS-like channel structure.
(A) Sequence alignment of the putative pore helix of Venus flytrap FLYC1 and Drosera DcFLYC1.1 and DcFLYC1.2 proteins with MSL10 and MscS. Nonpolar, polar, and ionizable residues are orange, light blue, and dark blue, respectively. A glycine predicted to localize at a central bend in the helix is shaded green. (B) Modeled heptameric organization. The sequence of Venus flytrap FLYC1 was threaded on the inner helix of heptameric MscS in a closed conformation (PDB 2OAU), and minimized using the Rosetta energy function while imposing C7 symmetry. Subunits are shown in different colors. Basic residues K558 and K579 are green and purple sticks, respectively. Predicted intersubunit hydrogen bonds between serines S564 and S565 (light blue sticks) of pore segments are shown with red dashes, while a ring of phenylalanines (F572; orange spheres) constrict the pore. Helices TM6a, forming the central pore, and amphipathic helix TM6b are indicated for one pore segment. (C) A cross-section through the protein surface colored by electrostatic potential, showing an uncharged (white) pore, with positive charge (blue) above and below the pore. (D) Average I–V of stretch-activated single-channel currents from FLYC1 (N=7) and FLYC1(K579E) (N=4) in asymmetrical NaCl solution. FLYC1 data is the same as Figure 4D. Scatter plots are mean ± s.e.m.
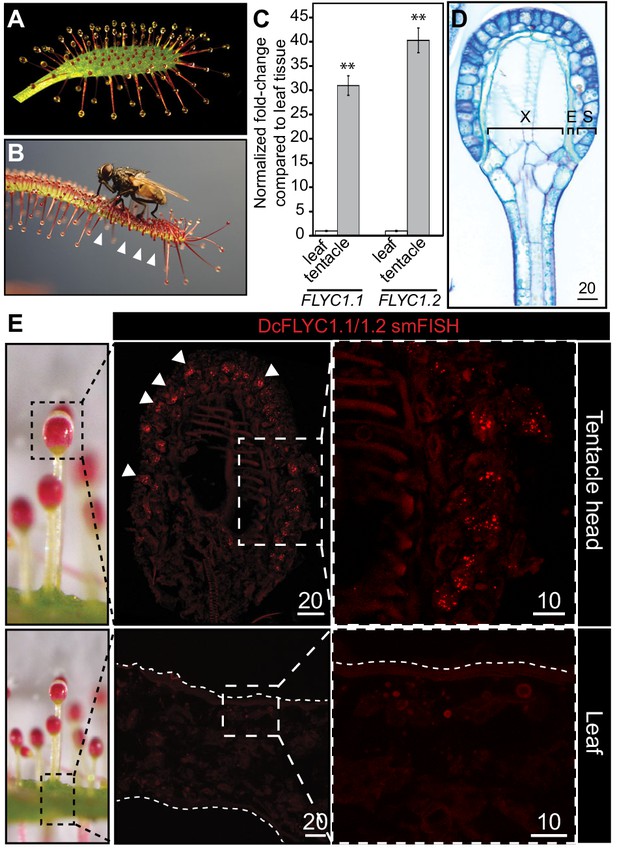
DcFLYC1.1 and DcFLYC1.2 localize to touch-sensitive structures of Drosera.
(A) The Cape sundew leaf, showing tentacle projections with mucilage secretions. (B) Image of tentacle bending in response to insect (house fly) touch. Arrowheads mark examples of tentacles that have bent inward. (C) Relative expression of DcFLYC1.1 and DcFLYC1.2 in sundew tentacles versus tentacle-less leaves by qRT-PCR. **p<0.005, moderated t-test, results from four biological replicates of each tissue type. (D) Toluidine blue–stained longitudinal section through the head and upper neck of a Cape sundew tentacle. The head is composed of xylem (X), an endodermis-like layer (E), and secretory cells (S). (E) Max projection image, at low (left) and high magnification (right), showing collective localization of DcFLYC1.1 and DcFLYC1.2 mRNAs to the outer secretory cells of the tentacle head (top). Arrows indicate example secretory cells with DcFLYC puncta. No signal above background was observed in the leaf (bottom). Scale bar, µm.
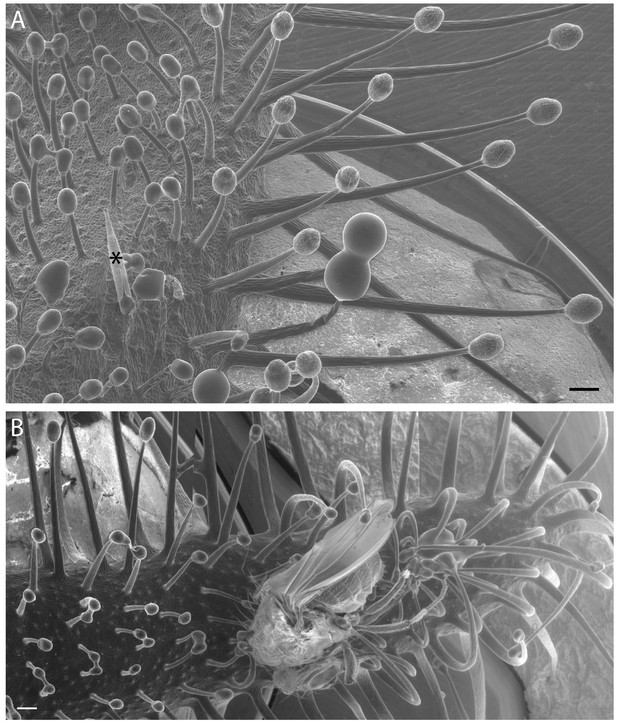
Variation in Cape sundew tentacles.
(A) Scanning electron microscopy (SEM) image showing mucilage-producing tentacles of different lengths on the Cape sundew leaf blade. An asterisk (*) marks the staging needle. Tentacles increase in length towards the leaf edge. (B) A composite image of three SEM micrographs showing tentacle bending around a D. melanogaster fly. Scale bars, 200 μm.
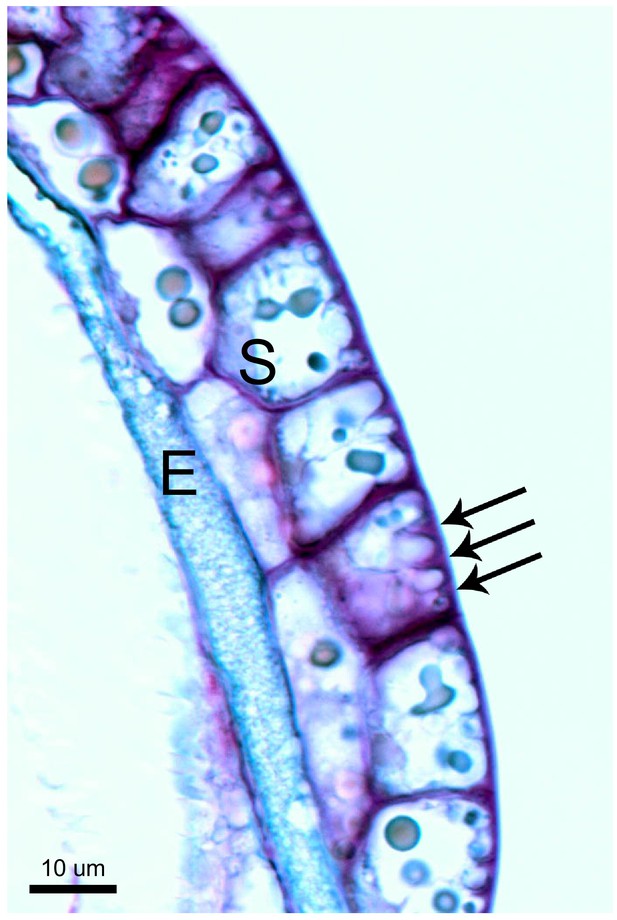
Unique cell morphology of Cape sundew sensory/excretory cells.
Toluidine blue-stained longitudinal section through the head of a Cape sundew tentacle. E, endodermis-like cells; S, outer two layers of secretory cells. Arrows mark the unique cell wall buttresses of a single outer secretory cell, around which membrane crenellations occur. See also Lloyd, 1942.
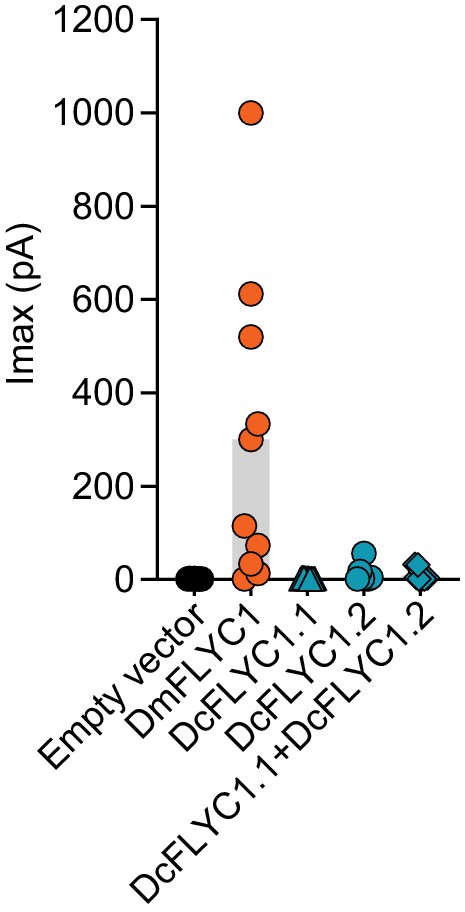
Drosera FLYC functionality.
Macroscopic stretch-activated currents recorded from HEKP1-KO cells transfected with mock (N = 7), DmFLYC1 (N = 10), DcFLCY1.1 (N = 7), DcFLYC1.2 (N = 10), and DcFLYC1.1/1.2 (N = 11) plasmids. Empty vector and DmFLYC1 data are the same as in Figure 4A.
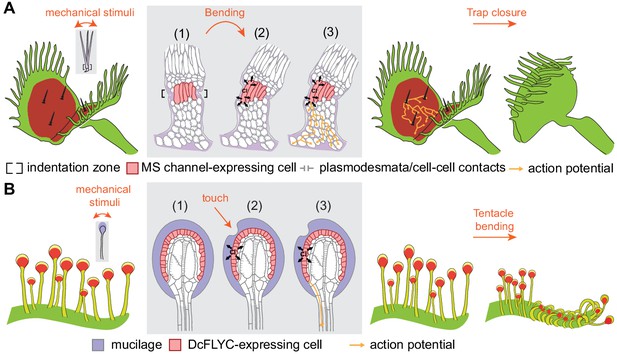
Model of touch-induced movements in carnivorous Droseraceae plants.
(A) In Venus flytrap, mechanical stimulation of the trigger hair by a prey animal causes bending at the indentation zone sensory cells (1), leading to mechanically induced activation of mechanosensitive (MS) channels and chloride efflux (2). This triggers an action potential that propagates from the base of the sensory cells to cells of the podium via plasmodesmata (3) (Williams and Mozingo, 1971). Propagation of action potentials through the lobe of the leaf results in trap closure. (B) In sundew tentacles, the pulling of mucilage could lead to activation of DcFLYC1.1 and DcFLYC1.2 proteins in the outer cell layer of tentacle heads, triggering a propagating action potential down the tentacle stalk.
Videos
Touch response of the Venus flytrap.
Real-time recording of trap closure in response to insect (house fly) touch.
Touch response of the Cape sundew plant.
Time lapse recording of tentacle bending in response to insect (Drosophila melanogaster) touch. Insects were placed directly on the leaf tentacles and the recording started within 10 s thereafter. 1 s represents 5 min.
Additional files
-
Supplementary file 1
Identification of differentially expressed genes in the trigger hair.
(A) Putative protein-coding genes with enriched expression in Venus flytrap trigger hairs. Shown are genes (Trinity components) coding for a transcript with >2-fold enriched expression in the trigger hair relative to trap tissue, and false discovery rate (FDR) < 0.05. CPM, counts per million. TM, transmembrane. (B) Putative protein-coding genes with reduced expression in Venus flytrap trigger hairs. Similar to (A), except showing genes with >2-fold reduction in the trigger hair. (C) Gene ontology (GO) enrichment analysis of genes with higher expression in trigger hairs. Shown are GO term identifiers (GO-ID) and descriptions for all GO terms that are enriched among genes with higher expression in the trigger hair relative to trap tissue (corrected p value<0.05). (D) Gene ontology (GO) enrichment analysis of genes with lower expression in trigger hairs. Similar to (C), except showing enrichment of GO terms associated with genes that have reduced expression in the trigger hair.
- https://cdn.elifesciences.org/articles/64250/elife-64250-supp1-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64250/elife-64250-transrepform-v1.docx





