In vivo reconstitution finds multivalent RNA–RNA interactions as drivers of mesh-like condensates
Figures
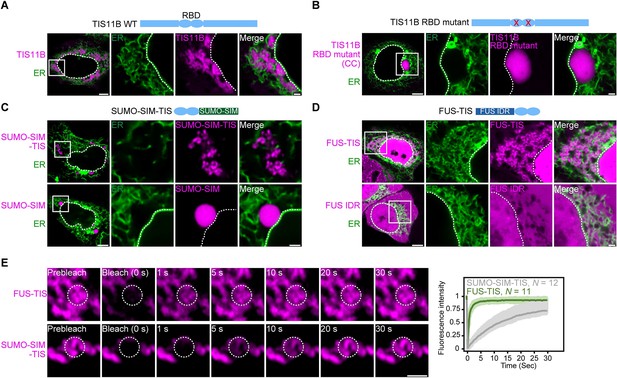
RNA determines the morphology of dynamic mesh-like RNA granules in cells.
(A) Confocal live-cell imaging of HeLa cells after the transfection of mCherry-tagged TIS11B. GFP-SEC61B was co-transfected to visualize the endoplasmic reticulum. The white dotted line demarcates the nucleus. Right: higher magnification of the indicated region. Scale bars, 5 µm (overview) and 1 µm (zoom-in). (B) Same as (A), but after transfection of TIS11B with a mutated RNA-binding domain. See Figure 1—figure supplement 1 for more mutants. CC: C135H/C173H. (C) Same as (A), but after transfection of mCherry-tagged SUMO10-SIM5 or SUMO-SIM-TIS chimera. 73% (N = 52) of SUMO-SIM-TIS granules are mesh-like. (D) Same as (A), but after transfection of mGFP-tagged FUS IDR (amino acids 1–214) or FUS-TIS chimera. All granules are mesh-like. (E) Fluorescence recovery after photobleaching of FUS-TIS and SUMO-SIM-TIS 16 hr after transfection of mCherry-SUMO-SIM-TIS or mGFP-FUS-TIS into HeLa cells. Scale bar, 1 µm.
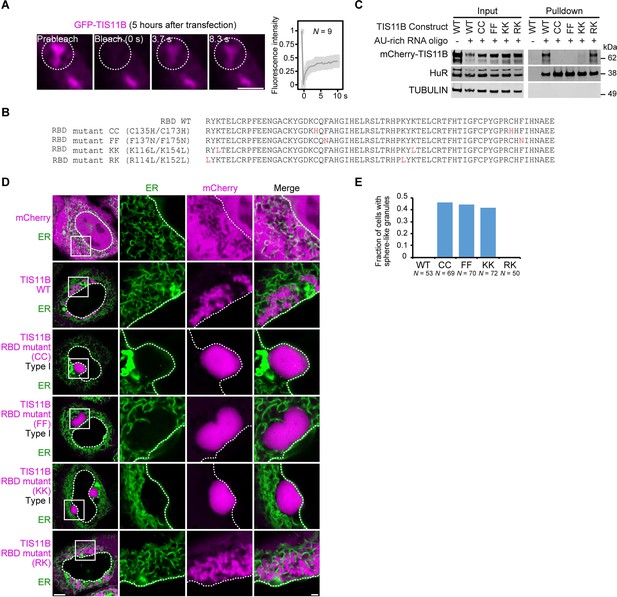
Mutation of the TIS11B RNA-binding domain generates sphere-like granules in cells.
(A) Fluorescence recovery after photobleaching of TIS11B 5 hr after transfection of mGFP-TIS11B into HeLa cells. Scale bar, 1 µm. (B) Shown is the amino acid sequence of the wild-type (WT) TIS11B RNA-binding domain (RBD), together with the introduced point mutations (red). CC: C135H/C173H; FF: F137N/F175N; KK: K116L/K154L; RK: R114L/K152L. (C) Western blot showing RNA oligonucleotide pulldown after co-transfection of the indicated mCherry-tagged TIS11B constructs and an AU-rich RNA oligo (TNFα AU-rich element) into HeLa cells. RNA-binding is disrupted in CC, FF, and KK mutants. HuR also binds to the AU-rich element. Tubulin was used as loading control. 2.5% of the input was loaded. (D) Confocal live-cell imaging of HeLa cells after transfection of the indicated constructs described in (B). GFP-SEC61B was co-transfected to visualize the endoplasmic reticulum. Scale bar, 1 µm. (E) Quantification of the data shown in (D). Shown is the fraction of cells with sphere-like granules formed by the indicated TIS11B proteins.
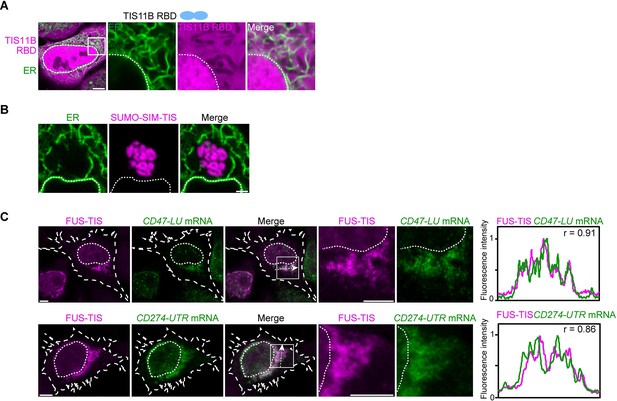
In the context of various multivalent domains, the TIS11B RNA-binding domain generates mesh-like condensates in vivo.
(A) Confocal live-cell imaging of HeLa cells after the transfection of mCherry-tagged TIS11B RNA-binding domain (RBD). GFP-SEC61B was co-transfected to visualize the endoplasmic reticulum (ER). The white dotted line demarcates the nucleus. Right: higher magnification of the indicated region. Scale bars, 5 µm (overview) and 1 µm (zoom-in). (B) Confocal live-cell imaging of HeLa cells after transfection of mCherry-tagged-SUMO-SIM-TIS. GFP-SEC61B was co-transfected to visualize the ER. Scale bar, 1 µm. (C) Representative images obtained from RNA-FISH (green) against GFP after transfection of GFP-CD47-LU or GFP-CD274-3′UTR in HeLa cells. BFP-FUS-TIS (magenta) was co-transfected. The white dotted lines demarcate the cell border and the nucleus, respectively. Right: line profiles of fluorescence intensities including the Pearson’s correlation coefficient (r). The arrow indicates the plane used for line profile generation. Scale bar, 5 µm.
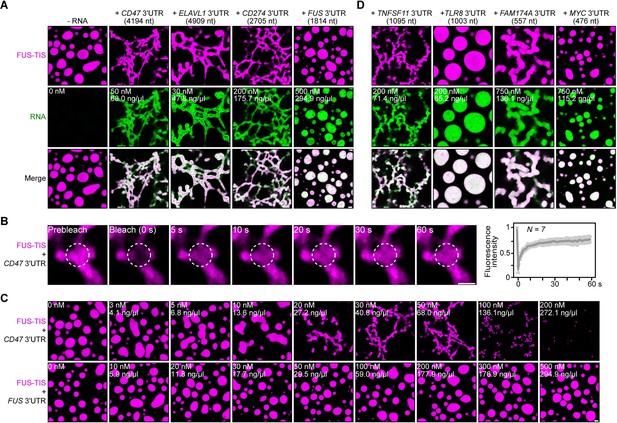
Specific RNAs induce formation of dynamic mesh-like condensates in vitro.
(A) Representative confocal images of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the absence or presence of the indicated in vitro transcribed RNAs after 16 hr of incubation. Scale bar, 2 µm. Five percent dextran was added into the phase separation buffer as crowding agent in all experiments. (B) Fluorescence recovery after photobleaching of mGFP-FUS-TIS (10 µM) mixed with CD47 3′UTR (50 nM) after 2 hr of incubation. Scale bar, 1 µm. (C) Same as (A), but in the presence of different RNA concentrations. (D) Same as (A), but additional RNAs are shown.
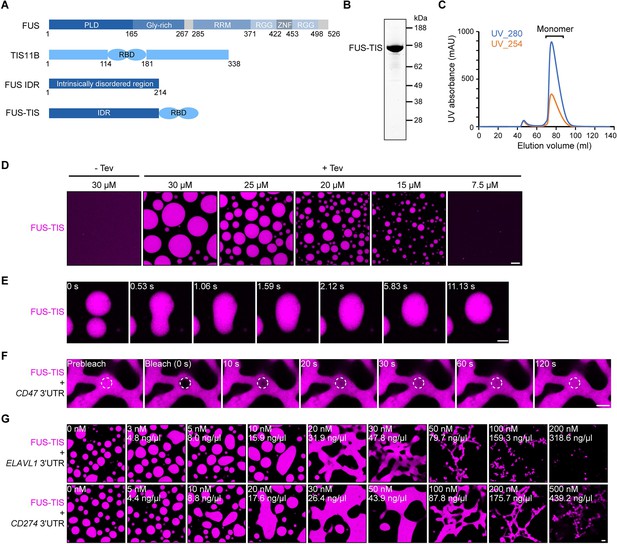
Specific RNAs induce mesh-like condensates in vitro.
(A) Schematic of the FUS-TIS chimeric protein. (B) SDS-PAGE of the purified FUS-TIS protein. FUS-TIS was tagged with His-MBP-mGFP at the N-terminus and with Strep-tag II at the C-terminus and purified from E. coli. mGFP: monomeric GFP. (C) Size exclusion chromatography of FUS-TIS protein. (D) Phase separation of purified mGFP-FUS-TIS protein at the indicated concentrations was induced by reducing the salt concentration from 600 mM to 150 mM through dilution; shown is a 2 hr time point. The His-MBP tag was cleaved off before the phase separation experiment using TEV protease. Scale bar, 10 µm. (E) Snapshots showing a fusion event of mGFP-FUS-TIS condensates at 2 hr. (F) Fluorescence recovery after photobleaching of mGFP-FUS-TIS (10 µM) mixed with CD47 3′UTR (50 nM) after 16 hr of incubation. Scale bar, 2 µm. (G) Representative confocal images of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the absence or presence of the indicated in vitro transcribed RNAs after 16 hr of incubation. Scale bar, 2 µm.
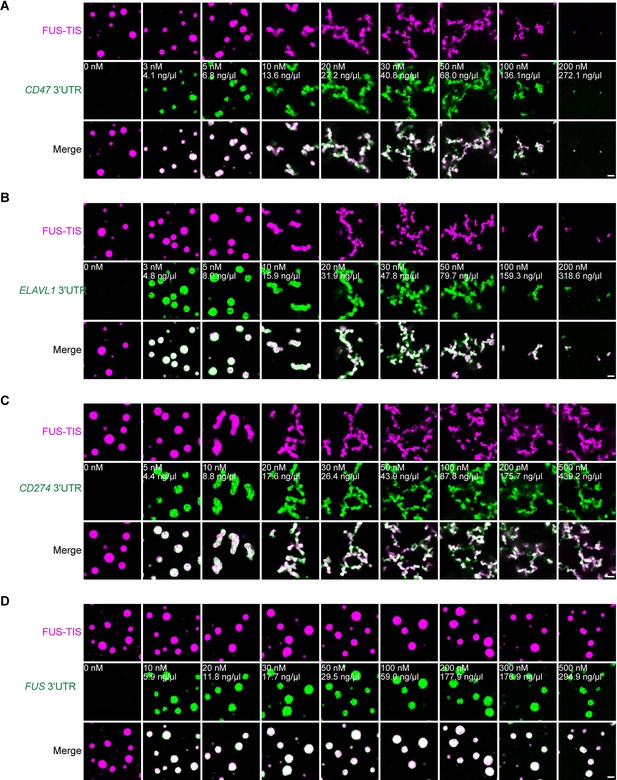
Specific RNAs induce mesh-like condensates in vitro.
(A–D) Representative confocal images of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the absence or presence of the indicated in vitro transcribed RNAs after 2 hr of incubation. Scale bar, 2 µm.
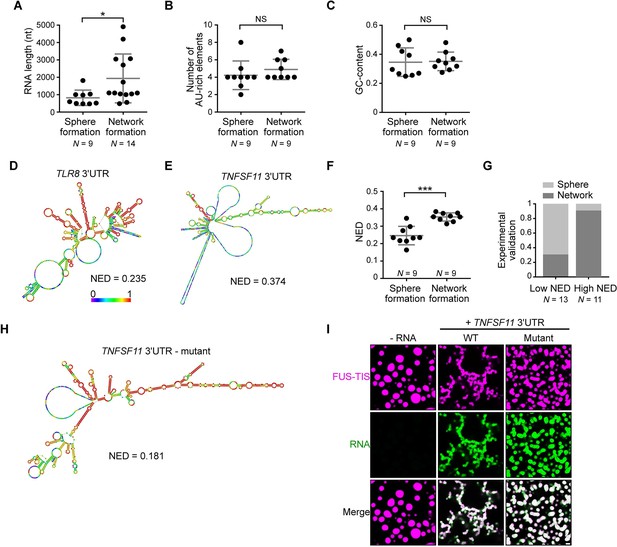
RNAs predicted to have large disordered regions have a high propensity to induce network formation in vitro.
(A) Distribution of length of sphere- and network-forming RNAs. Mann–Whitney test, Z = −2.76, p=0.004. See also Figure 3—source data 1. (B) Number of AU-rich elements in sphere- and network-forming RNAs with a length shorter than 2000 nt. See also Figure 3—source data 1. Mann–Whitney test, Z = 0.190, p=0.258, NS, not significant. (C) Distribution of GC-content of sphere- and network-forming RNAs with a length shorter than 2000 nt. See also Figure 3—source data 1. Mann–Whitney test, Z = 0.566, p=0.605. (D) Centroid RNA secondary structure of TLR8 3′UTR predicted by RNAfold. The color code represents base-pairing probability. (E) Same as (D), but the TNFSF11 3′UTR is shown. (F) Normalized ensemble diversity (NED) values of sphere- and network-forming RNAs. See Figure 3—source data 1. Mann–Whitney test, Z = −3.3, ***p<0.0003. (G) Experimental validation of N = 24 in vitro transcribed RNAs whose ability for network formation was predicted by NED. Sphere formation is indicated in dark gray, whereas network formation is indicated in light gray. See Figure 3—source data 1. Mann–Whitney test was performed on the experimental validation, Z = −2.8, ***p=0.004. (H) Same as (D), but the mutant TNFSF11 3′UTR is shown. (I) Representative confocal images of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the presence of 150 nM of the indicated in vitro transcribed RNAs after 16 hr of incubation. Scale bar, 2 µm.
-
Figure 3—source data 1
Length, number of AU-rich elements, GC-content, and normalized ensemble diversity values of the 47 experimentally tested 3′UTRs.
- https://cdn.elifesciences.org/articles/64252/elife-64252-fig3-data1-v3.xlsx
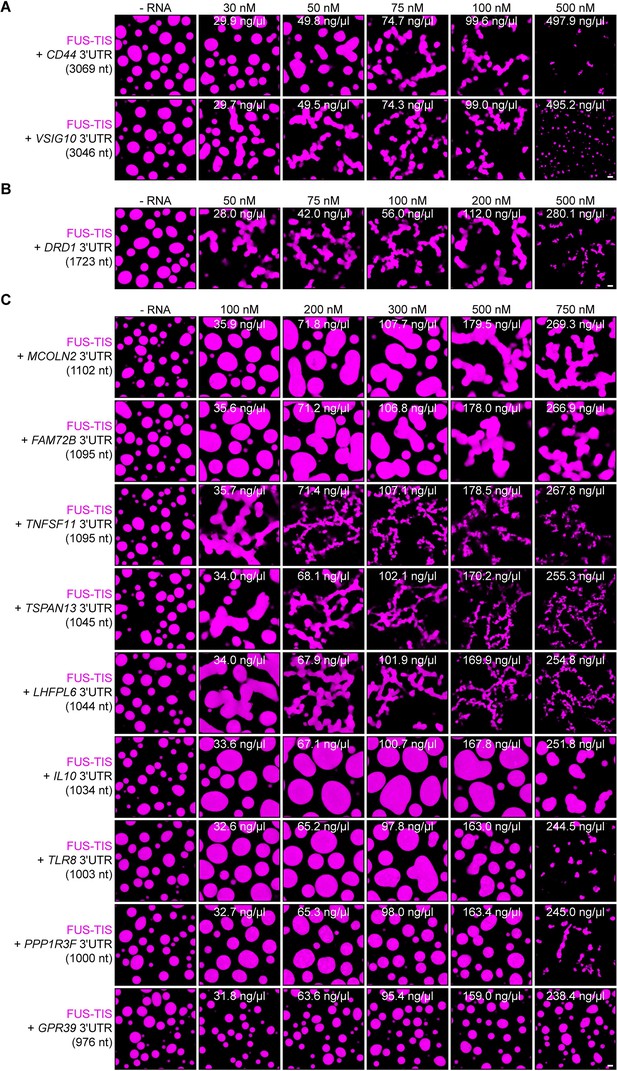
Specific RNAs with various lengths induce mesh-like condensates in vitro.
Representative confocal images of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the absence or presence of the indicated in vitro transcribed RNAs after 16 hr of incubation. Scale bar, 2 µm. (A) RNAs with a length of approximately 3000 nt are shown. (B) RNAs with a length between 1500 and 2000 nt are shown. (C) RNAs with a length of approximately 1000 nt are shown.
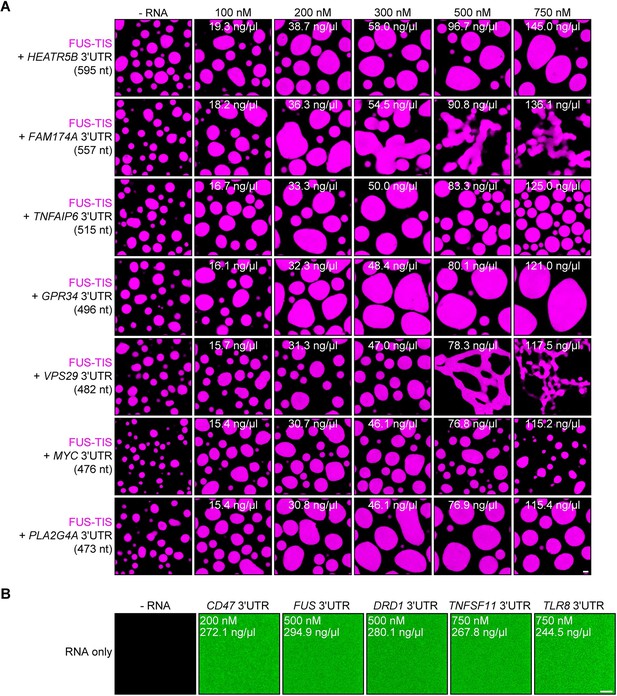
RNA alone does not induce phase separation in vitro.
Representative confocal images of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the absence or presence of the indicated in vitro transcribed RNAs after 16 hr of incubation. Scale bar, 2 µm. (A) RNAs with a length of approximately 500 nt are shown. (B) Representative confocal images of Cy5-labeled RNAs at the indicated concentrations. Scale bar, 5 µm.
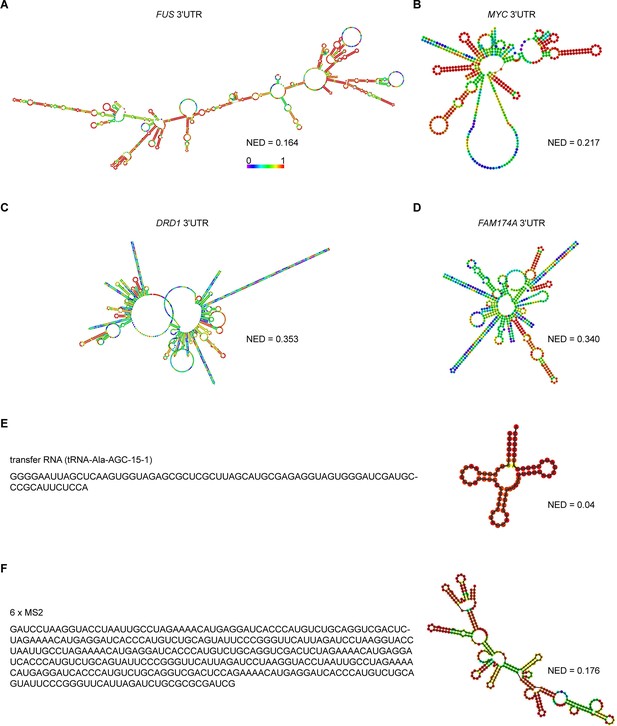
Predicted RNA secondary structures and their corresponding normalized ensemble diversity values for examples of sphere-forming, network-forming, and highly structured RNAs.
(A, B) Centroid RNA secondary structure of two sphere-forming 3′UTRs predicted by RNAfold. The color code represents base-pairing probability. See also the URLs in Figure 3—source data 1. (C, D) As in (A), but centroid RNA secondary structure of two network-forming 3′UTRs predicted by RNAfold. (E) As in (A), but sequence and centroid RNA secondary structure of a tRNA is shown. (F) As in (A), but sequence and centroid RNA secondary structure of six MS2-binding sites is shown.
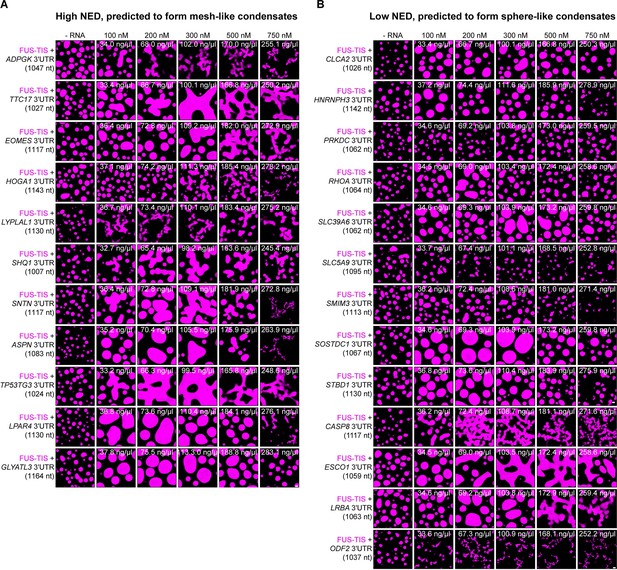
The normalized ensemble diversity (NED) value of RNAs is highly predictive for their ability to form sphere- or mesh-like condensates.
(A) Totally 10 out of 11 RNAs with a high NED value were predicted correctly to induce network formation. Representative confocal images of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the absence or presence of the indicated in vitro transcribed RNAs after 16 hr of incubation. Scale bar, 2 µm. The length of all RNAs is approximately 1000 nt. The minimum concentration for induction of network formation varies. The GLYATL3 3′UTR is unable to induce network formation even at high concentrations. (B) Totally 9 out of 13 RNAs with a low NED value were predicted correctly to induce sphere-like condensate formation. As in (A).

In a size-restricted dataset, the number of AU-rich elements does not predict mesh-like condensate formation.
(A) Number of AU-rich elements in RNAs with high normalized ensemble diversity (NED) or low NED values. See also Figure 3—source data 1. Mann–Whitney test, NS, not significant. (B) Nucleotide sequence of the TNFSF11 3′UTR mutant. Two 15-nt oligonucleotides (red and magenta bars) that are complementary to upstream sequences (red and magenta fonts) were added into the TNFSF11 3′UTR. This reduces the unstructured regions and increases the local secondary structure.
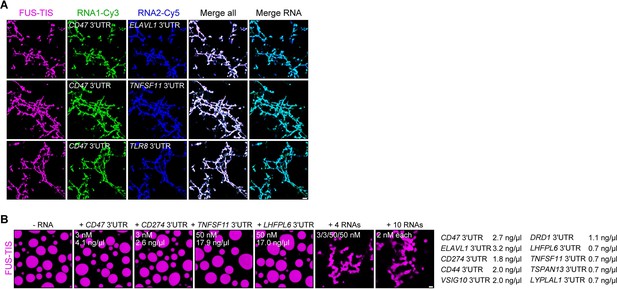
RNAs work additively to induce formation of granule networks in vitro.
(A) RNAs co-localize in mesh-like condensates. Representative confocal images of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the presence of two different in vitro transcribed RNAs that were labeled with Cy3 or Cy5 fluorescent dye, respectively. Images were taken after 16 hr of incubation. Scale bar, 5 µm. (B) Representative confocal images of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the presence of a single network-forming RNA at suboptimal concentration or in the presence of 4 or 10 network-forming RNAs, each at suboptimal concentration. Images were taken after 16 hr of incubation. Scale bar, 2 µm.
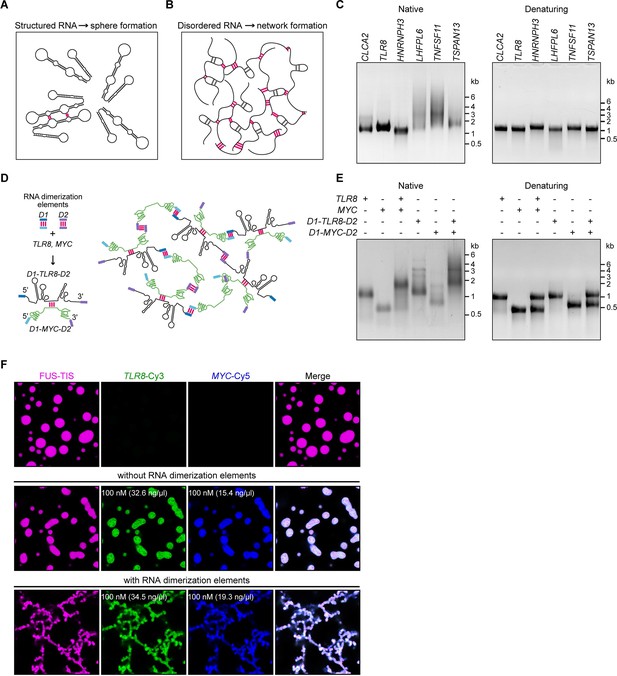
A multivalent RNA matrix is responsible for mesh-like condensate formation in vitro.
(A) Schematic of RNAs with strong local secondary structures that are predicted to induce spherical condensates. (B) Schematic of RNAs with large disordered regions that form extensive intermolecular RNA–RNA interactions that are predicted to form network-like condensates. (C) Native and denaturing agarose gel electrophoresis of sphere-forming (lanes 1–3) and network-forming (lanes 4–6) RNAs (5 µM, each [1.7, 1.6, 1.9, 1.7, 1.8, 1.7 µg/µl]). (D) Schematic of a complex RNA network characterized by extensive intermolecular RNA–RNA interactions mediated by two dimerization elements (D1 and D2) that were added to structured RNAs. (E) Native and denaturing agarose gel electrophoresis of the indicated RNAs (1 µM, each [326, 154, 345, 193 ng/µl]). (F) Representative images of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the presence of the indicated Cy3- or Cy5-labeled RNAs generated by in vitro transcription after 16 hr of incubation. Scale bar, 2 µm.
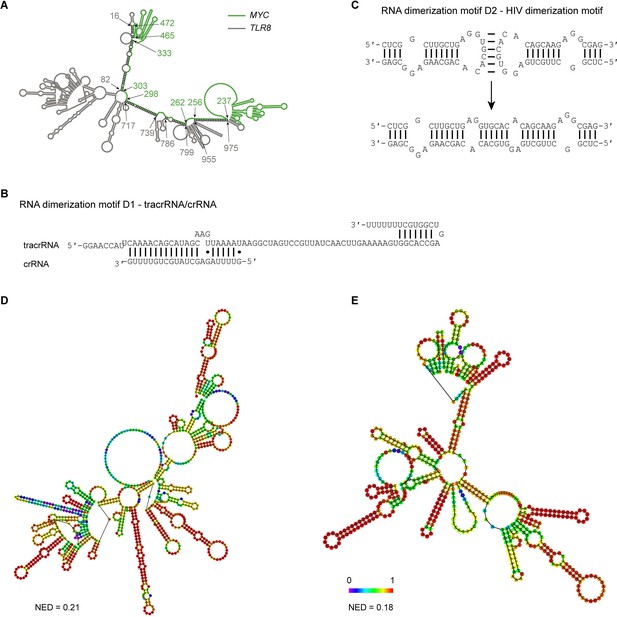
Extensive intermolecular RNA–RNA interactions are responsible for formation of mesh-like condensates in vitro.
(A) The TLR8 3′UTR (gray) and the MYC 3′UTR (green) are predicted by RNAfold to dimerize. The secondary structure prediction shows extensive base-pairing between the two RNAs, indicated by the black marks. The nucleotide positions that demarcate the interaction sites are indicated by arrows, and the numbers show their positions in their respective 3′UTRs. The MYC 3′UTR was added directly downstream of the TLR8 3′UTR for structure prediction. Although both RNAs are able to base-pair, they are unable to form a complex RNA network. (B) Schematic of the D1 RNA dimerization motif. Shown is the sequence and base-pairing of the directional RNA dimerization motif D1 that was obtained from the CRISPR/Cas9 system and represents tracrRNA/crRNA. The two components form a heterologous RNA–RNA interaction. (C) Schematic of the D2 RNA dimerization motif. Shown is the sequence and base-pairing of the RNA dimerization motif D2, which is the HIV dimerization motif. It forms a homologous RNA–RNA interaction. (D) Centroid RNA secondary structure of the TLR8 3′UTR with the RNA dimerization elements D1 and D2 as predicted by RNAfold is shown. The color code represents base-pairing probability. NED: normalized ensemble diversity. (E) As in (D), but centroid RNA secondary structure of the MYC 3′UTR with the RNA dimerization elements D1 and D2 as predicted by RNAfold is shown.
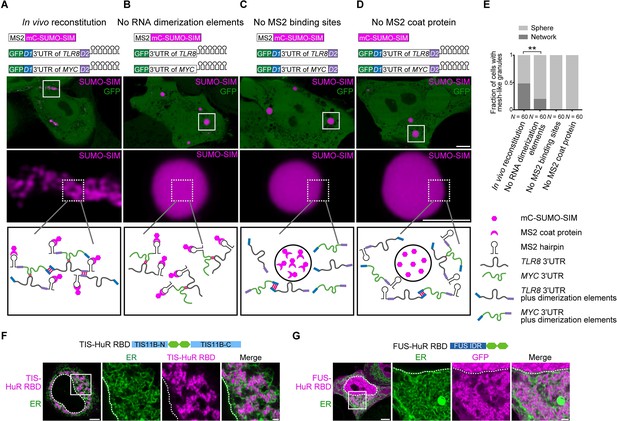
Formation of an extensive mRNA network is sufficient for the reconstitution of mesh-like condensates in vivo.
(A) Representative confocal image of in vivo reconstitution of mesh-like condensates using the MS2 system. mCherry-SUMO-SIM fused to the MS2 coat protein was transfected into HeLa cells. Constructs containing eGFP-fused 3ʹUTRs of TLR8 and MYC with MS2-binding sites and with the RNA dimerization elements D1 and D2 were co-transfected. Bottom: higher magnification of the indicated regions. Scale bars, 5 µm (overview) and 2 µm (zoom-in). Images were taken 16 hr after transfection. mC: mCherry. (B-D) As in (A), but one of the indicated components was omitted. (E) Quantification of the fraction of cells with mesh-like granules in the conditions shown in (A–D). Mann–Whitney test: Z = −3.5, **p=0.001. (F) Confocal live-cell imaging of HeLa cells after the transfection of mCherry-tagged TIS-HuR chimera, containing the RRM1/2 of HuR as well as the N- and C-terminal regions of TIS11B. All granules are mesh-like. Scale bars, 5 µm (overview) and 1 µm (zoom-in). (G) Same as (F), but after transfection of mGFP-tagged FUS-HuR chimera, containing the RRM1/2 of HuR as well as the FUS IDR. All granules are mesh-like.
-
Figure 6—source data 1
Transcriptome-wide analysis on normalized ensemble diversity values of 3′UTRs.
- https://cdn.elifesciences.org/articles/64252/elife-64252-fig6-data1-v3.xlsx
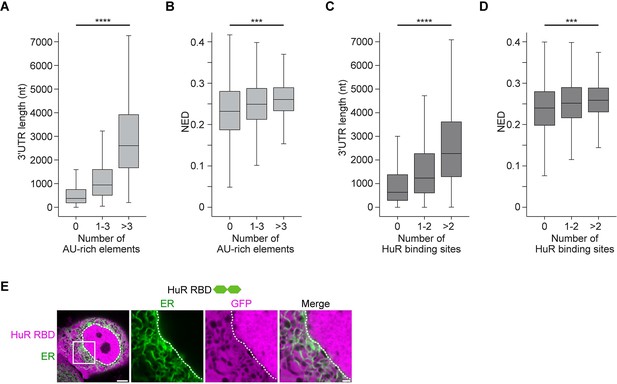
mRNAs with large disordered regions are enriched in AU-rich elements.
(A) The number of AU-rich elements in 3′UTRs correlates with 3′UTR length. The dataset contains all mRNAs expressed in HeLa cells. 3′UTRs without AU-rich elements, N = 2504, 3′UTRs with 1–3 AU-rich elements, N = 3081, 3′UTRs with more than three AU-rich elements, N = 2889 are shown. Kruskal–Wallis test: Χ2 = 4414, p=0. (B) As in (A). The number of AU-rich elements in 3′UTRs correlates with normalized ensemble diversity (NED). Kruskal–Wallis test: Χ2 = 315, p=4×10−69. (C) The number of HuR-binding sites in 3′UTRs as assessed by PAR-CLIP correlates with 3′UTR length. 3′UTRs without HuR-binding sites, N = 3860, 3′UTRs with one or two HuR-binding sites, N = 2030, 3′UTRs with more than two HuR-binding sites, N = 2584 are shown. Kruskal–Wallis test: Χ2 = 1538, p=0. (D) As in (C). The number of HuR-binding sites correlates with NED. Kruskal–Wallis test: Χ2 = 169, p=4×10−37. (E) Confocal live-cell imaging of HeLa cells after the transfection of mGFP-tagged HuR RNA-binding domain. mCherry-SEC61B was co-transfected to visualize the endoplasmic reticulum. The white dotted line demarcates the nucleus. Right: higher magnification of the indicated region. Scale bars, 5 µm (overview) and 1 µm (zoom-in).
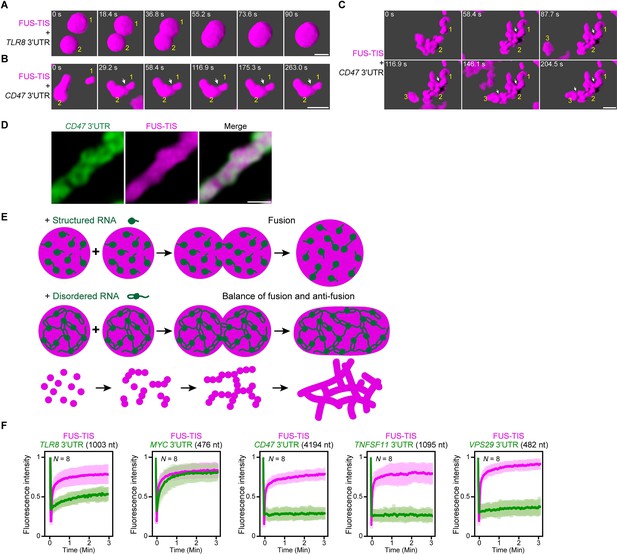
An RNA matrix prevents full fusion of spherical condensates, thus promoting arrangement into filamentous structures in vitro.
(A) Confocal 3D time-lapse imaging of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the presence of the TLR8 3′UTR (200 nM) after 30 min of incubation. Scale bar, 2 µm. Snapshots show a fusion event of two FUS-TIS condensates. (B) Same as (A), but in the presence of the CD47 3′UTR (30 nM). Snapshots show the contact of two FUS-TIS condensates. As they do not fully mix, they grow into condensates with irregular shapes. The contact site is indicated by the white arrow. (C) Same as (B). Snapshots show two fusion events between FUS-TIS condensates 1 and 2 and between 2 and 3 and demonstrate how irregularly shaped condensates grow into large filamentous networks. (D) Representative high-resolution confocal images of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the presence of Cy5-labeled CD47 3ʹUTR RNA after 16 hr of incubation. Scale bar, 1 µm. (E) Model showing how structured RNAs induce spherical condensates and how RNAs with large unstructured regions induce formation of filamentous and mesh-like condensates. Purple indicates the condensate. For details, see text. (F) Fluorescence recovery after photobleaching of mGFP-FUS-TIS and the indicated Cy5-labeled RNAs performed at 2 hr after setting up the phase separation experiments.
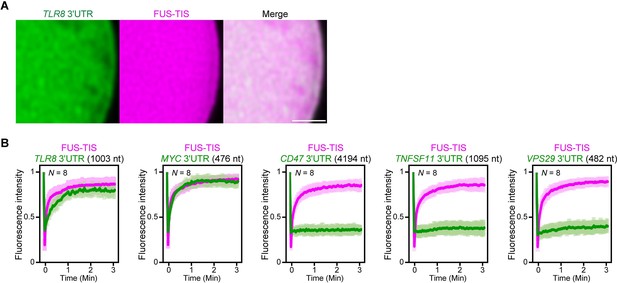
Extensive intermolecular RNA–RNA interactions are responsible for formation of mesh-like condensates in vitro.
(A) Representative high-resolution confocal images of phase separation experiments using purified mGFP-FUS-TIS (10 µM) in the presence of Cy5-labeled TLR8 3′UTR RNA after 16 hr of incubation. Scale bar, 1 µm. (B) Fluorescence recovery after photobleaching of mGFP-FUS-TIS and the indicated Cy5-labeled RNAs at 16 hr after setting up the phase separation experiments.
Tables
Chimeric proteins investigated for mesh-like condensate formation.
RBDmut, RNA-binding domain mutant.
| Multivalent domain | RNA-binding domain | RNA | Diffusive pattern | Sphere-like condensate | Mesh-like condensate |
|---|---|---|---|---|---|
| TIS11B | √ | ||||
| HuR | √ | ||||
| FUS IDR | √ | ||||
| SUMO-SIM | √ | ||||
| TIS11B-N/C | TIS11B RBDmut | √ | |||
| TIS11B-N/C | TIS11B | √ | |||
| TIS11B-N/C | HuR | √ | |||
| FUS IDR | TIS11B | √ | |||
| FUS IDR | HuR | √ | |||
| SUMO-SIM | TIS11B | √ | |||
| SUMO-SIM | MS2 | √ | |||
| SUMO-SIM | MS2 | Singlevalent RNA | √ | ||
| SUMO-SIM | MS2 | Multivalent RNA | √ |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HeLa | Jonathan S. Weissman | N/A | A human cervical cancer cell line (female origin). |
| Strain, strain background (Escherichia coli) | BL21(DE3) | NEB | C2527H | Chemically competent E. coli cells. |
| Antibody | Anti-α-tubulin (mouse monoclonal) | Sigma-Aldrich | Cat# T9026, RRID:AB_477593 | WB (1:5000). |
| Antibody | Anti-mCherry (mouse monoclonal) | Abcam | Cat# ab125096, RRID:AB_11133266 | WB (1:5000). |
| Antibody | Anti-HuR (rabbit polyclonal) | Millipore | Cat# 07-1735, RRID:AB_1977173 | WB (1:2000). |
| Antibody | IRDye 680RD anti-rabbit IgG secondary antibody (donkey polyclonal) | LI-COR Biosciences | Cat# 926-68073, RRID:AB_10954442 | WB (1:10,000). |
| Antibody | IRDye 800CW anti-mouse IgG secondary antibody (donkey polyclonal) | LI-COR Biosciences | Cat# 926–32212, RRID:AB_621847 | WB (1:10,000). |
| Transfected construct (human) | pcDNA-SP-GFP-CD47-LU | Berkovits and Mayr, 2015 | N/A | See Materials and methods. |
| Transfected construct (human) | pcDNA-GFP-ELAVL1-LU | Ma and Mayr, 2018 | N/A | See Materials and methods. |
| Transfected construct (human) | pcDNA-SP-GFP-CD274-UTR | Ma and Mayr, 2018 | N/A | See Materials and methods. |
| Transfected construct (human) | pcDNA-SP-GFP-FUS-UTR | Ma and Mayr, 2018 | N/A | See Materials and methods. |
| Transfected construct (human) | pcDNA-GFP-SEC61B | Ma and Mayr, 2018 | N/A | See Materials and methods. |
| Transfected construct (human) | pcDNA-mCherry-SEC61B | Ma and Mayr, 2018 | N/A | See Materials and methods. |
| Transfected construct (human) | pcDNA-mCherry-TIS11B | Ma and Mayr, 2018 | N/A | See Materials and methods. |
| Transfected construct (human) | pcDNA-mCherry-TIS11B CC | This paper | N/A | See Materials and methods. |
| Transfected construct (human) | pcDNA-mCherry-TIS11B FF | This paper | N/A | See Materials and methods. |
| Transfected construct (human) | pcDNA-mCherry-TIS11B KK | This paper | N/A | See Materials and methods. |
| Transfected construct (human) | pcDNA-mCherry-TIS11B RK | This paper | N/A | See Materials and methods. |
| Transfected construct (human) | pcDNA-mCherry-TIS-HuR RBD | This paper | N/A | See Materials and methods. |
| Transfected construct (human) | pmCherry-SUMO10-SIM5 | Liam J. Holt (NYU) | N/A | See Materials and methods. |
| Transfected construct (human) | pmCherry-SUMO10-SIM5-TIS | This paper | N/A | See Materials and methods |
| Transfected construct (human) | pcDNA-mGFP-FUS-TIS | This paper | N/A | See Materials and methods. |
| Recombinant DNA reagent | pET28a | Dirk Remus (MSKCC) | N/A | Bacterial expression vector. |
| Recombinant DNA reagent | pDZ2087 | Addgene | Cat# 92414 | Bacterial expression of TEV protease. |
| Recombinant DNA reagent | pET28a-6xHis-MBP-mGFP-FUS-TIS-Strep-Tag II | This paper | N/A | Bacterial expression of 6xHis-MBP-mGFP-FUS-TIS-Strep Tag II. See Materials and methods. |
| Recombinant DNA reagent | T7-CD47 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-ELAVL1 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-CD274 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-FUS 3′UTR | This paper | N/A | T7 RNA polymerase- based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-CD44 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-VSIG10 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-IL10 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-TNFSF11 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-GPR39 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-TLR8 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-GPR34 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-TNFAIP6 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-MYC 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-PLA2G4A 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-HEATR5B 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-PPP1R3F 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-DRD1 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-FAM72B 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-MCOLN2 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-TSPAN13 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-LHFPL6 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-FAM174A 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-VPS29 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-ADPGK 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-ASPN 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-CASP8 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-CLCA2 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-EOMES 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-ESCO1 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-GLYATL3 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-HNRNPH3 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-HOGA1 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-LPAR4 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-LRBA 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-LYPLAL1 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-ODF2 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-PRKDC 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-RHOA 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-SHQ1 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-SLC39A6 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-SLC5A9 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-SMIM3 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-SNTN 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-SOSTDC1 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-STBD1 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-TP53TG3 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Recombinant DNA reagent | T7-TTC17 3′UTR | This paper | N/A | T7 RNA polymerase-based in vitro transcription. See Materials and methods. |
| Sequence-based reagent | Biotinylated RNA oligo, TNFα ARE-1 | Ma and Mayr, 2018 | RNA oligonucleotides | 5′-CACUUGUGAUUAUUUAUUAUUUAUUUAUUAUUUAUUUAUUUA −3′ |
| Peptide, recombinant protein | FUS-TIS | This paper | N/A | Recombinant 6xHis-MBP-mGFP-FUS-TIS-Strep Tag II protein purified from bacteria. See Materials and methods. |
| Peptide, recombinant protein | Bovine serum albumin (BSA) | New England Biolab | Cat# B9000S | |
| Commercial assay or kit | Streptavidin C1 beads | Invitrogen | Cat# 65002 | Streptavidin pulldown assay. |
| Commercial assay or kit | QuikChange Lightning Multi Site-Directed Mutagenesis Kit | Agilent Technologies | Cat# 210513 | Site-directed mutagenesis. |
| Commercial assay or kit | MEGAscript T7 Transcription Kit | Invitrogen | Cat# AMB13345 | In vitro T7 transcription. |
| Commercial assay or kit | Quick Star Bradford Protein Assay Kit | Bio-Rad | Cat# 5000202 | Bradford assay –protein quantitation. |
| Chemical compound, drug | Lipofectamine 2000 | Invitrogen | Cat# 11668019 | |
| Chemical compound, drug | Dextran T500 | PHARMACOSMOS | Cat# 40030 | |
| Chemical compound, drug | Desthiobiotin | Sigma-Aldrich | Cat# D1411-1G | |
| Chemical compound, drug | Zinc chloride | Sigma-Aldrich | Cat# 793523-100G | |
| Chemical compound, drug | Imidazole | Sigma-Aldrich | Cat# I2399-100G | |
| Chemical compound, drug | IPTG | Gold Biotechnology | Cat# I2481-EZ10 | |
| Chemical compound, drug | PMSF | Sigma-Aldrich | Cat# 11359061001 | |
| Chemical compound, drug | DTT | Sigma-Aldrich | Cat# 10708984001 | |
| Software, algorithm | FIJI | NIH | https://fiji.sc/ | |
| Software, algorithm | ZEN | ZEISS | https://www.zeiss.com/microscopy/int/downloads/zen.html | |
| Software, algorithm | GraphPad Prism 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism | |
| Software, algorithm | Odyssey | LI-COR Biosciences | https://www.licor.com/bio/products/imaging_systems/odyssey/ | |
| Other | Ni-NTA Agarose | Qiagen | Cat# 30230 | His tag purification. |
| Other | StrepTrap column | GE Healthcare | Cat# 28907547 | Strep tag II purification. |
| Other | Amicon Ultra-centrifugal filters-50K | EMD Millipore | Cat# UFC905024 | Concentrating protein samples. |
| Other | 384-well glass-bottom microplate | Greiner Bio-One | Cat# M4437-16EA | Glass-bottom microplate for confocal imaging. |





