Growth-dependent signals drive an increase in early G1 cyclin concentration to link cell cycle entry with cell growth
Figures
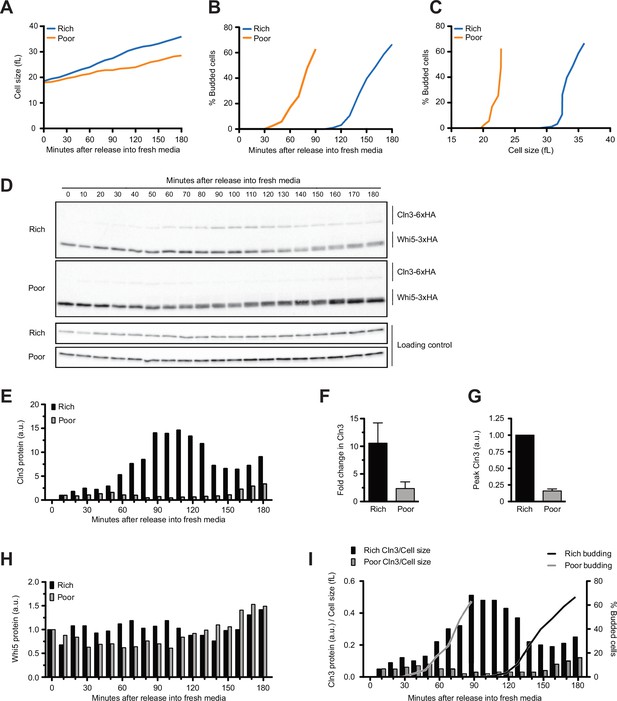
Dynamics of Cln3 and Whi5 proteins during G1 phase in cells growing in rich or poor carbon.
Wild-type cells were grown to mid-log phase in poor carbon (YPG/E), and small unbudded cells were isolated by centrifugal elutriation. Cells were released into either rich carbon (YPD) or poor carbon (YPG/E) at 25°C, and samples were taken at 10 min intervals. All data in the figure are from the same biological replicate. (A) Median cell size was measured using a Coulter counter at 10 min intervals and plotted as a function of time. (B) The percentage of budded cells as a function of time. (C) The percentage of budded cells as a function of median cell size at each time point. (D) The behavior of Cln3-6XHA and Whi5-3XHA was analyzed by western blot on the same blot for each carbon source. An anti-Nap1 antibody was used as a loading control. (E) Quantification of Cln3-6XHA protein levels from (D) as a function of time. At each time point, the relative Cln3 signal was calculated as a ratio to the signal of the 10 min time point. (F) The fold change in peak Cln3 protein signal was calculated as a ratio over the signal from the 10 min time point. The data represent the average of three biological replicates. The error bars represent SEM. (G) The difference in peak Cln3 between rich and poor carbon was calculated by first normalizing the peak Cln3 signal to the loading control in each carbon source, and then comparing the signal between carbon sources. The error bars represent SEM. (H) Quantification of Whi5-3XHA from western blots in (D). The change in relative protein abundance over time was measured by calculating the ratio over the zero time point for each time point. (I) The relative change in Cln3 concentration over time was calculated by taking the ratio of the Cln3 signal over cell size at each time point. The bud emergence data from panel (B) are included for reference. The experiment shown in this figure was repeated for three biological replicates, which gave similar results. In panels (E), (H), and (I), the Cln3 and Whi5 signals were not normalized to a loading control because the loading control signal increases with growth.
-
Figure 1—source data 1
Source data for Figure 1D.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig1-data1-v3.pdf
-
Figure 1—source data 2
Source data for Figure 1D.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig1-data2-v3.pdf
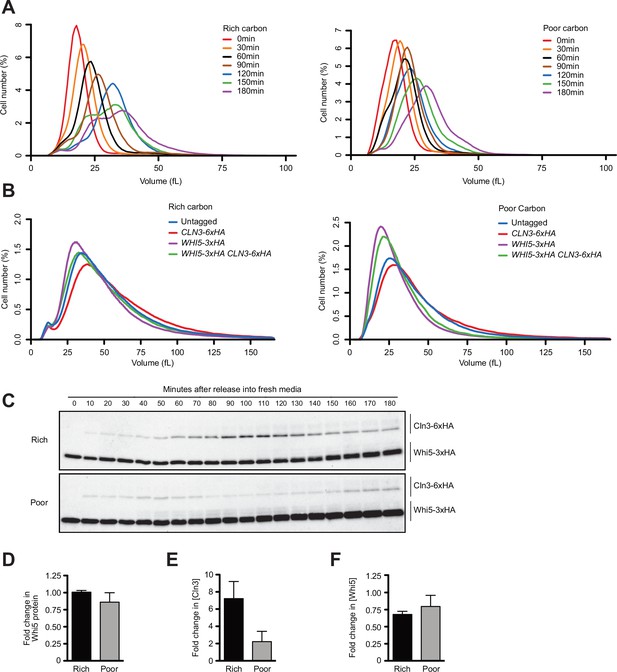
Dynamics of Cln3 and Whi5 proteins during G1 phase in cells growing in rich or poor carbon.
(A) Examples of the cell size plots that were used to create the median cell size plots in Figure 1A. (B) Cells of the indicated genotypes were grown to log phase, and cell size was measured by Coulter counter in rich (YPD) or poor carbon (YPG/E). (C) A longer exposure of the western blots from Figure 1D. (D) The fold change in Whi5 protein levels was calculated by taking the ratio of the Whi5 signal from the time point with peak Cln3 over the Whi5 signal from the 0 min time point. The data show the average of three biological replicates. (E) The fold change in Cln3 concentration before bud emergence was calculated by taking the ratio of the value for peak Cln3 concentration over the value from the 10 min time point. The data show the average of three biological replicates. (F) The fold change in Whi5 concentration before bud emergence was calculated by taking the ratio of the value for Whi5 concentration at peak Cln3 levels over the value from the 0 min time point. The data show the average of three biological replicates. Error bars represent SEM.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1C.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig1-figsupp1-data1-v3.pdf
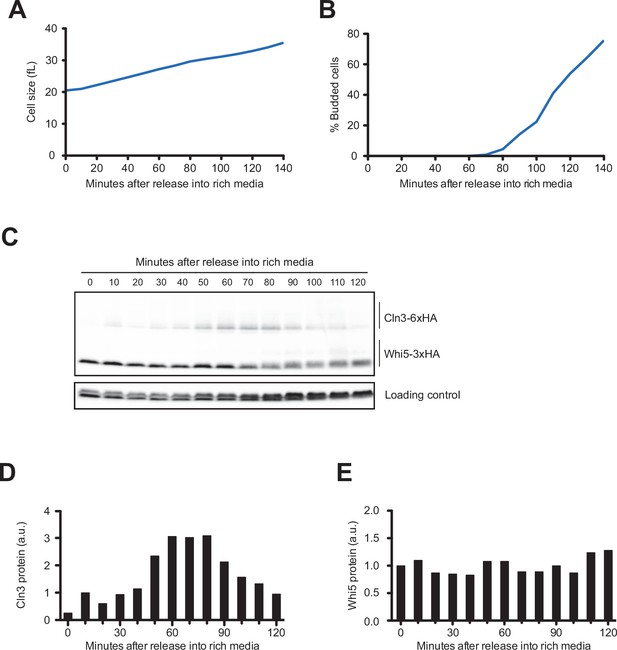
Dynamics of Cln3 and Whi5 during G1 phase in cells grown continuously in complete synthetic media containing dextrose.
Wild-type cells were grown to mid-log phase in complete synthetic media with rich carbon (CSM + 2% dextrose). Small unbudded cells were the isolated by centrifugal elutriation. Cells were released in the same medium at 25°C and samples were collected at 10 min intervals. (A) Median cell size was measured at 10 min intervals and plotted as a function of time. (B) The percentage of budded cells as a function of time. (C) Cln3-6XHA and Whi5-3XHA were analyzed on the same western blot. An anti-Nap1 antibody was used as a loading control. (D) Quantification of Cln3-6XHA protein levels from (C) as a function of time. At each time point, the relative Cln3 signal was calculated as a ratio to the signal of the 10 min time point. (E) Quantification of Whi5-3XHA protein levels from (C) as a function of time. At each time point, the relative Whi5 signal was calculated as a ratio over the signal of the 0 min time point.
-
Figure 1—figure supplement 2—source data 1
Source data for Figure 1—figure supplement 2C.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig1-figsupp2-data1-v3.pdf
-
Figure 1—figure supplement 2—source data 2
Source data for Figure 1—figure supplement 2C.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig1-figsupp2-data2-v3.pdf
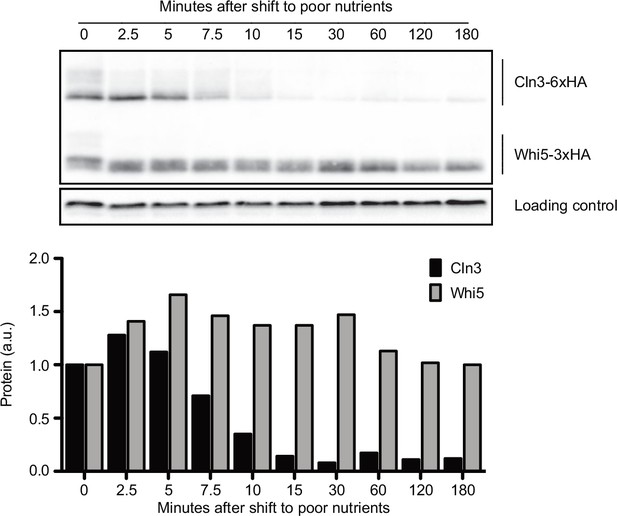
Cln3 protein levels respond rapidly to changes in nutrient availability.
Rapidly growing cells in rich carbon (YPD) were shifted to poor carbon (YPG/E) at 25°C, and the behavior of Cln3-6XHA and Whi5-3XHA was analyzed by western blot on the same blot. The levels of Cln3 and Whi5 protein were quantified relative to the 0 min time point. An anti-Nap1 antibody was used as a loading control. To account for any differences in total protein after shifting to poor carbon, Cln3 and Whi5 signals were calculated as a ratio to that of the loading control, which was quantified relative to the 0 min time point.
-
Figure 2—source data 1
Source data for Figure 2C.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig2-data1-v3.pdf
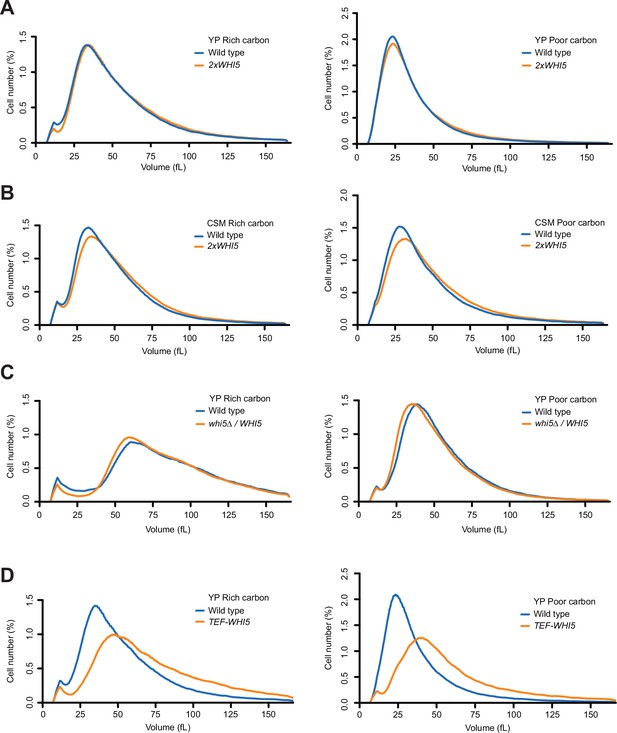
Increased dosage of Whi5 does not cause strong effects on cell size.
(A) Wild-type and 2xWHI5 cells were grown to log phase in rich (YPD) or poor carbon (YPG/E), and cell size was measured using a Coulter counter. (B) Wild-type and 2xWHI5 cells were grown to log phase in complete synthetic medium containing dextrose (CSM-rich carbon) or glycerol/ethanol (CSM-poor carbon). Cell size was measured using a Coulter counter. (C) Wild-type diploid cells and heterozygous WHI5/whi5∆ cells were grown to log phase in rich (YPD) or poor carbon (YPG/E), and cell size was measured using a Coulter counter. (D) Wild-type and TEF1-WHI5 cells were grown to log phase in rich (YPD) or poor carbon (YPG/E), and cell size was measured using a Coulter counter.
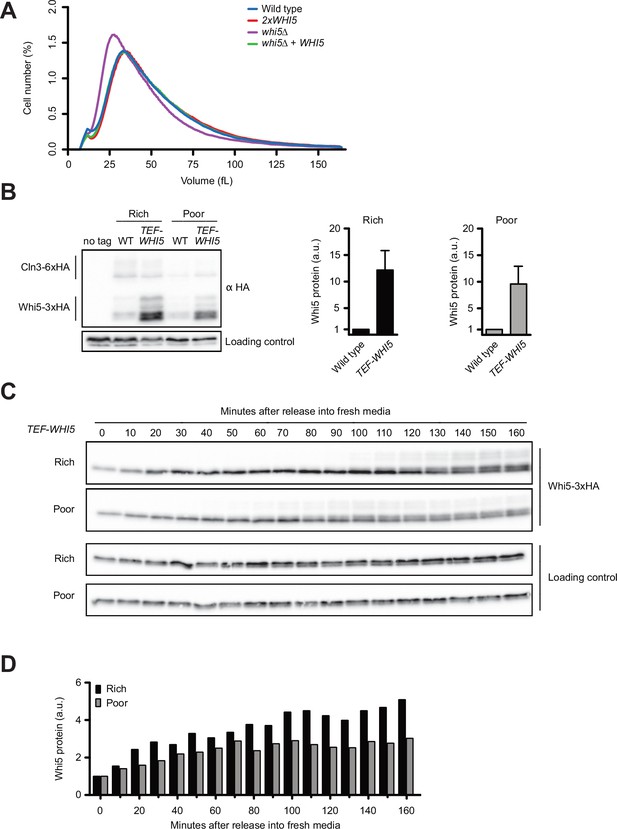
Increased dosage of Whi5 does not cause strong effects on cell size.
(A) Cells of the indicated genotypes were grown to log phase in rich carbon (YPD) or poor carbon (YPG/E), and cell size was measured with a Coulter counter. (B) Cells that express CLN3-6XHA and either WHI5-3XHA or TEF1-WHI5-3XHA were grown to mid-log phase in rich carbon (YPD) or poor carbon (YPG/E). Cln3-6XHA and Whi5-3XHA were imaged on the same blot. An anti-Nap1 antibody was used as a loading control. The TEF1-WHI5-3XHA signal was quantified relative to the Whi5-3XHA signal for the respective carbon source after each signal was normalized to the loading control. Error bars represent the SEM of three biological replicates. (C) TEF1-WHI5-3XHA cells were grown to mid-log phase in poor carbon (YPG/E), and small unbudded cells were isolated via centrifugal elutriation. Cells were then released into rich (YPD) or poor (YPG/E) carbon media, and samples were collected at 10 min intervals. The behavior of Whi5-3XHA was analyzed by western blot. An anti-Nap1 antibody was used as a loading control. (D) Quantification of Whi5-3XHA protein levels from panel (C) as a function of time. At each time point, the relative Whi5 signal was calculated as a ratio over the signal of the zero time point.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1B.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig3-figsupp1-data1-v3.pdf
-
Figure 3—figure supplement 1—source data 2
Source data for Figure 3—figure supplement 1B.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig3-figsupp1-data2-v3.pdf
-
Figure 3—figure supplement 1—source data 3
Source data for Figure 3—figure supplement 1C. This panel also shows data for Cln3-6XHA, which is not discussed.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig3-figsupp1-data3-v3.pdf
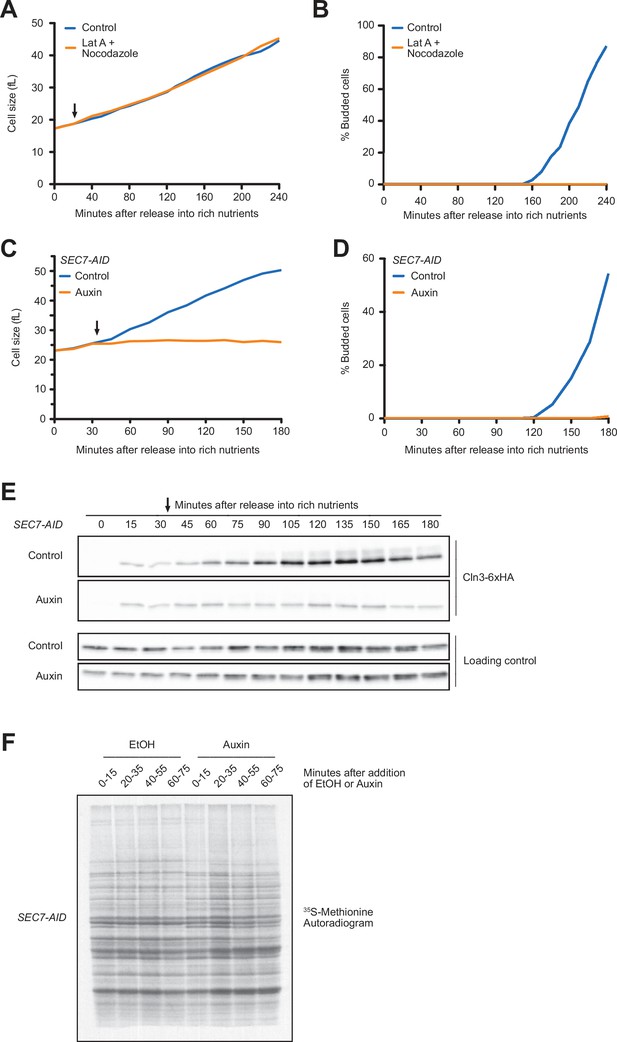
Blocking membrane trafficking events required for cell growth prevents accumulation of Cln3.
(A, B) Wild-type cells were grown to mid-log phase in poor carbon (YPG/E), and small unbudded cells were isolated by centrifugal elutriation. The cells were split into two cultures and were then released into rich carbon (YPD) at 25°C. After 20 min, 100 µM of latrunculin A and 20 µM nocodazole were added to one culture (arrow). (A) Median cell size was measured at 20 min intervals using a Coulter counter and plotted as a function of time. (B) The percentage of budded cells as a function of time. (C–E) SEC7-AID cells were grown to mid-log phase in poor carbon, and small unbudded cells were isolated by centrifugal elutriation. The cells were split and then released into rich carbon at 25°C. After 30 min, 1 mM auxin was added to one culture (arrow). (C) Median cell size was measured at 15 min intervals using a Coulter counter and plotted as a function of time. (D) The percentage of budded cells as a function of time. (E) The behavior of Cln3-6XHA in the SEC7-AID cells was analyzed by western blot. An anti-Nap1 antibody was used as a loading control. (F) Autoradiogram of 35S-methionine labeling to detect de novo protein synthesis. SEC7-AID cells were grown to mid-log phase in -MET synthetic media containing dextrose. After auxin or vehicle addition, samples were labeled with 35S-methionine for 15 min intervals starting every 20 min to measure the rate of protein synthesis within the 15 min intervals.
-
Figure 4—source data 1
Source data for Figure 4E.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig4-data1-v3.pdf
-
Figure 4—source data 2
Source data for Figure 4E.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig4-data2-v3.pdf
-
Figure 4—source data 3
Source data for Figure 4F.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig4-data3-v3.pdf
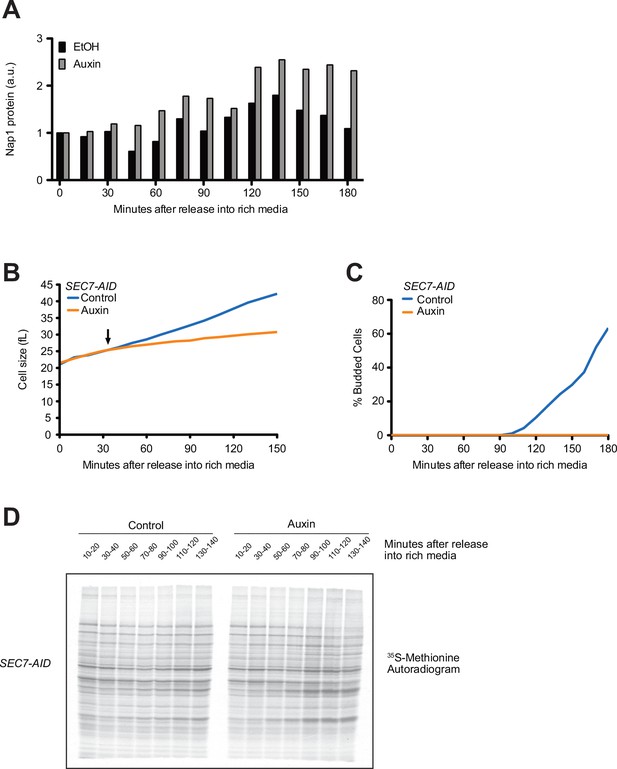
Blocking membrane trafficking events required for cell growth prevents accumulation of Cln3.
(A) Quantification of Nap1 loading control protein levels in the western blots in Figure 4E. Relative Nap1 levels were quantified by taking a ratio of the Nap1 signal at each time point over the Nap1 signal at the 0 min time point. (B–D) SEC7-AID cells were grown in complete synthetic medium with poor carbon (CSM + 2% glycerol/ethanol) to mid-log phase, and small unbudded cells were then isolated by elutriation. Cells were split and released into CSM with rich carbon (CSM + 2% dextrose) without methionine. After 30 min, the vehicle or 1 mM auxin was added to the cultures. (B) Median cell size was measured at 10 min intervals and plotted as a function of time. (C) The percentage of budded cells as a function of time. (D) Autoradiogram of 35S-methionine labeling. To measure de novo protein synthesis, samples were collected every 20 min and labeled for 10 min intervals with 35S-methionine. Each lane represents protein synthesized during the 10 min interval. n = 1.
-
Figure 4—figure supplement 1—source data 1
Source data for Figure 4—figure supplement 1D.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig4-figsupp1-data1-v3.pdf
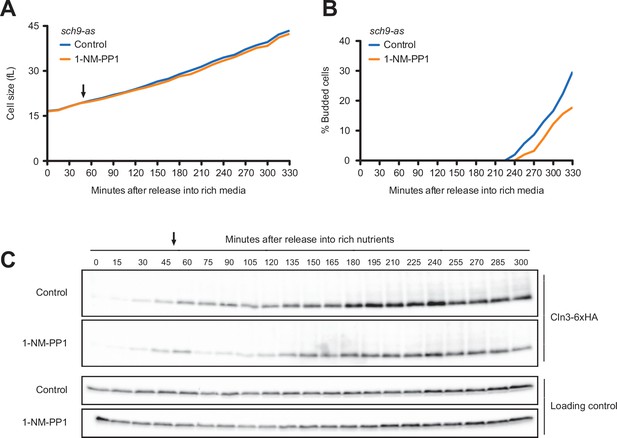
Inhibition of Sch9 does not cause strong effects on accumulation of Cln3 during G1 phase.
sch9-as cells were grown to mid-log phase in poor carbon (YPG/E), and small unbudded cells were isolated via centrifugal elutriation. The cells were split into two cultures and were released into rich carbon (YPD, not supplemented with additional adenine; see Materials and methods). After 45 min, 250 nM 1-NM-PP1 was added to one culture (arrow). (A) Median cell size was measured at 15 min intervals using a Coulter counter and plotted as a function of time. (B) The percentage of budded cells was plotted as a function of time. (C) The behavior of Cln3-6XHA was analyzed by western blot. An anti-Nap1 antibody was used for a loading control.
-
Figure 5—source data 1
Source data for Figure 5C.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig5-data1-v3.pdf
-
Figure 5—source data 2
Source data for Figure 5C.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig5-data2-v3.pdf
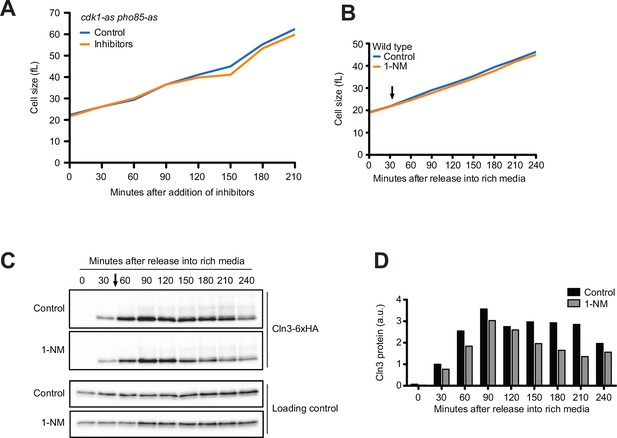
Inhibition of cyclin-dependent kinases does not cause strong effects on growth or accumulation of Cln3 during G1 phase.
(A) cdk1-as pho85-as cells were grown to mid-log phase in poor carbon (YPG/E), and small unbudded cells were isolated via centrifugal elutriation. The cells were divided into two cultures and were then released into rich carbon (YPD, not supplemented with additional adenine;see Materials and methods) at 25°C. 10 µM 1-NA-PP1 and 10 µM 1-NM-PP1 were added to one culture. Median cell size was measured at 30 min intervals using a Coulter counter and plotted as a function of time. n = 1. (B–D) Wild-type cells were grown to mid-log phase in poor carbon (YPG/E), and small unbudded cells were isolated via centrifugal elutriation. Cells were split and released into rich carbon (YPD, not supplemented with additional adenine) and grown at 25°C. After 30 min, 250 nM 1-NM-PP1 was added to one culture (arrow). (B) Median cell size was measured every 30 min and plotted as a function of time. (C) Western blots of Cln3-6XHA. An anti-Nap1 antibody was used as a loading control. (D) Cln3 protein was quantified relative to the 30 min time point in the control cells.
-
Figure 5—figure supplement 1—source data 1
Source data for Figure 5—figure supplement 1C.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig5-figsupp1-data1-v3.pdf
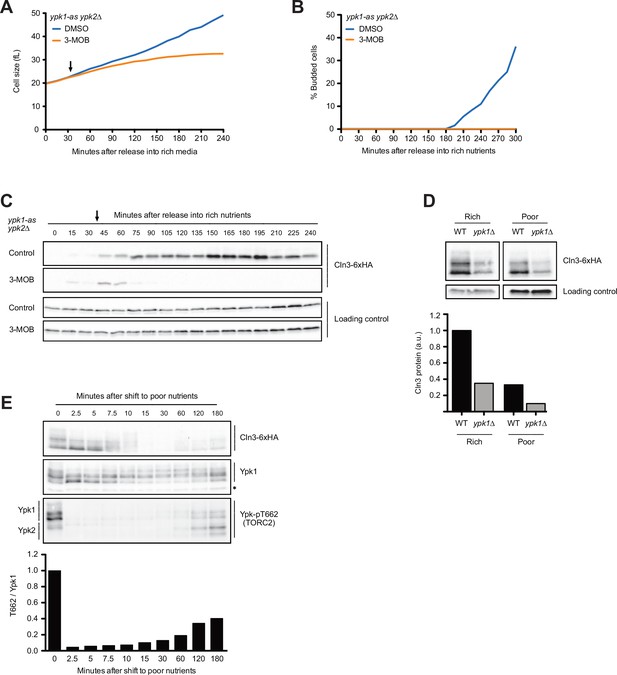
The budding yeast homologs of mammalian SGK kinases are required for accumulation of Cln3 during G1 phase.
(A–C) ypk1-as ypk2∆ cells were grown overnight in poor carbon (YPG/E) to mid-log phase, and small unbudded cells were isolated by centrifugal elutriation. The cells were divided into two cultures and were then released into rich carbon (YPD, not supplemented with additional adenine; see Materials and methods). After 30 min, 25 µM 3-MOB-PP1 was added to one culture (arrow). (A) Median cell size was measured at 15 min intervals using a Coulter counter and plotted as a function of time. (B) The percentage of budded cells as a function of time. (C) The behavior of Cln3-6XHA in the ypk1-as ypk2∆ cells was analyzed by western blot. An anti-Nap1 antibody was used for a loading control. (D) Wild-type and ypk1∆ cells were grown to mid-log phase in rich or poor carbon. The levels of Cln3-6XHA protein were analyzed by western blot. An anti-Nap1 was used for a loading control. Levels of Cln3-6XHA protein were quantified relative to levels of Cln3-6XHA in wild-type cells in rich carbon. Cln3 protein levels were first normalized to the loading control. (E) Wild-type cells were grown to mid-log phase in rich carbon (YPD) and then shifted to poor carbon (YPG/E) at 25°C. The behavior of Cln3-6XHA, TORC2-dependent phosphorylation of Ypk1/2, and total Ypk1 protein was assayed by western blot. A phospho-specific antibody was used to detect a TORC2-dependent phosphorylation site present on both Ypk1 and Ypk2 (referred to as anti-Ypk-pT662). Total Ypk1 protein was detected with an anti-Ypk1 antibody. Asterisk indicates a background band that also serves as a loading control.
-
Figure 6—source data 1
Source data for Figure 6C.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig6-data1-v3.pdf
-
Figure 6—source data 2
Source data for Figure 6C.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig6-data2-v3.pdf
-
Figure 6—source data 3
Source data for Figure 6D.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig6-data3-v3.pdf
-
Figure 6—source data 4
Source data for Figure 6D.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig6-data4-v3.pdf
-
Figure 6—source data 5
Source data for Figure 6D.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig6-data5-v3.pdf
-
Figure 6—source data 6
Source data for Figure 6D.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig6-data6-v3.pdf
-
Figure 6—source data 7
Source data for Figure 6E.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig6-data7-v3.pdf
-
Figure 6—source data 8
Source data for Figure 6E.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig6-data8-v3.pdf
-
Figure 6—source data 9
Source data for Figure 6E.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig6-data9-v3.pdf
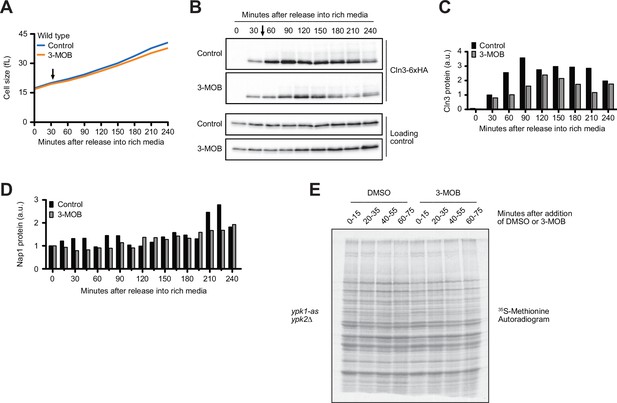
The budding yeast homologs of mammalian SGK kinases are required for accumulation of Cln3 during G1 phase.
(A–B) Wild-type cells were grown to mid-log phase in poor carbon (YPG/E), and small unbudded daughter cells were isolated by centrifugal elutriation. The cells were split and released into rich carbon (YPD, without additional adenine; see Materials and methods) at 25°C. After 30 min, 25 µM 3-MOB-PP1 was added to one culture (arrow). (A) Median cell size was measured at 30 min intervals using a Coulter counter and plotted as a function of time. (B) The behavior of Cln3-6XHA was analyzed by western blot. Nap1 was used as a loading control. (C) Quantification of Cln3 from (B) relative to the 30 min time point. (D) Quantification of anti-Nap1 loading control blots from Figure 6C. Protein levels at each time point represent a ratio over the signal from the 0 min time point. (E) Autoradiogram of 35S-methionine labeling to detect de novo protein synthesis. ypk1-as ypk2∆ cells were grown overnight to mid-log phase in -MET synthetic media containing dextrose. After addition of 3-MOB-PP1 or vehicle, 1.2 mL samples collected from the cultures were labeled with 35S-methionine for 15 min intervals every 20 min after addition of 3-MOB-PP1 to measure the rate of protein synthesis within the 15 min intervals.
-
Figure 6—figure supplement 1—source data 1
Source data for Figure 6—figure supplement 1B.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig6-figsupp1-data1-v3.pdf
-
Figure 6—figure supplement 1—source data 2
Source data for Figure 6—figure supplement 1B.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig6-figsupp1-data2-v3.pdf
-
Figure 6—figure supplement 1—source data 3
Source data for Figure 6—figure supplement 1E.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig6-figsupp1-data3-v3.pdf
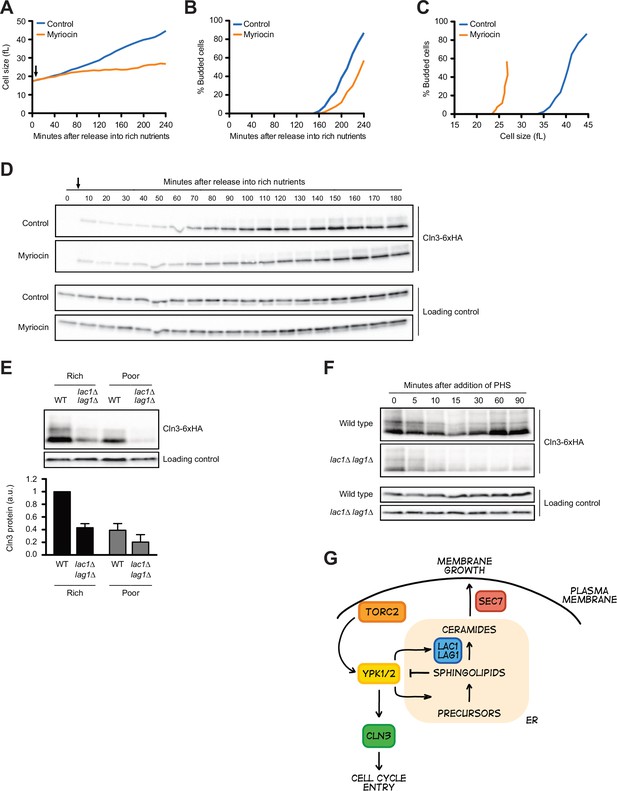
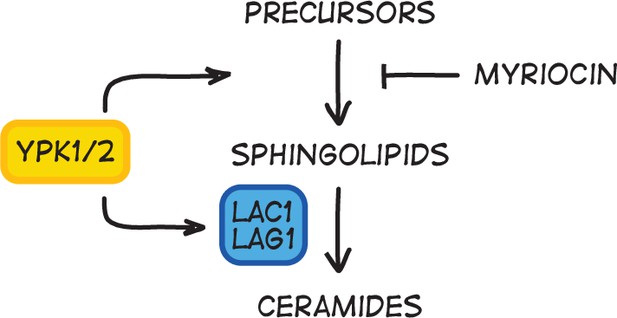
Sphingolipid-dependent signals influence Cln3 levels.
(A–D) Wild-type cells were grown overnight in poor carbon (YPG/E), and small unbudded cells were isolated via centrifugal elutriation. The cells were split into two cultures and released into rich carbon (YPD) at 25°C. After taking the initial time point, 5 µg/mL myriocin was added to one culture (arrow). (A) Median cell size was measured at 20 min intervals using a Coulter counter and plotted as a function of time. (B) The percentage of budded cells as a function of time. (C) The percentage of budded cells plotted as a function of cell size at each time point. (D) The levels of Cln3-6XHA protein were analyzed by western blot. An anti-Nap1 antibody was used for a loading control. (E) Wild-type and lac1∆ lag1∆ cells were grown to mid-log phase in rich (YPD) and poor carbon (YPG/E). The levels of Cln3-6xHA protein were analyzed by western blot. An anti-Nap1 antibody was used as a loading control. Levels of Cln3-6XHA protein were quantified relative to levels of Cln3-6XHA in wild-type cells in rich carbon. Cln3 protein levels were first normalized to the loading control. Error bars represent SEM of three biological replicates. (F) Wild-type and lac1∆ lag1∆ cells were grown to mid-log phase in rich carbon (YPD). 20 µM phytosphingosine (PHS) was added to each culture, and the cultures were incubated at 25°C. The levels of Cln3-6XHA protein were analyzed by western blot. An anti-Nap1 antibody was used as a loading control. (G) A model depicting how Cln3 levels could be modulated by Ypk1/2.
-
Figure 7—source data 1
Source data for Figure 7D.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig7-data1-v3.pdf
-
Figure 7—source data 2
Source data for Figure 7D.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig7-data2-v3.pdf
-
Figure 7—source data 3
Source data for Figure 7E.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig7-data3-v3.pdf
-
Figure 7—source data 4
Source data for Figure 7E.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig7-data4-v3.pdf
-
Figure 7—source data 5
Source data for Figure 7F.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig7-data5-v3.pdf
-
Figure 7—source data 6
Source data for Figure 7F.
- https://cdn.elifesciences.org/articles/64364/elife-64364-fig7-data6-v3.pdf
Tables
Strains with multiple strain numbers indicate multiple independent isolates of the same strain.
| Strain | Genotype | Reference or source | Figures |
|---|---|---|---|
| DK186 | MATa bar1 | Altman and Kellogg, 1997 | Figure 3 |
| DK3092 | MATa bar1 WHI5-3xHA::HPHNT1 CLN3-6xHA::HIS3 | This study | Figure 1 and Figure 1—figure supplement 1, Figure 1—figure supplement 2, Figure 2, Figure 3—figure supplement 1B |
| DK4395 | MATa bar1 WHI5-13MYC::HIS5 | This study | In support of Figure 1, data not shown |
| DK3669DK3670 | MATa bar1 URA3::YIplac211 (URA3) | This study | Figure 3A and B and Figure 3—figure supplement 1A |
| DK3722DK3875DK3876 | MATa bar1 URA3::pRAS1 (WHI5::URA3) | This study | Figure 3A and B and Figure 3—figure supplement 1A |
| DK3726DK3885 | MATa bar1 whi5∆::KANMX URA3::pRAS1 (WHI5::URA3) | This study | Figure 3—figure supplement 1A |
| DK3728 | MATa bar1 whi5∆::KANMX URA3::YIplac211 (URA3) | This study | Figure 3—figure supplement 1A |
| DK3716DK3717DK3718 | MATa/α WHI5/whi5::KANMX | This study | Figure 3C |
| DK3712 | KANMX::TEF1-WHI5 | This study | Figure 3D |
| DK3323 | KANMX::TEF1-WHI5-3xHA::HPHNT1 CLN3-6xHA::HIS3 | This study | Figure 3—figure supplement 1B-D |
| DK2017 | MATa bar1 CLN3-6xHA::HIS3 | Zapata et al., 2014 | Figure 4A and B, Figure 5—figure supplement 1C and D, Figure 6D and E and Figure 6—figure supplement 1A-C, Figure 7A–F |
| DK2907 | MATa bar1 SEC7-V5-AID::KANMX CLN3-6xHA::NATNT2 pTIR2::HIS3 pTIR4::LEU2 | This study | Figure 4C–F and Figure 4—figure supplement 1 |
| DK2936 | MATa bar1 cdc28-as1(F88G) pho85-as1(F82G) CLN3-6xHA::HIS3 | This study | Figure 5—figure supplement 1A and B |
| DK2961 | MATα sch9-as(T492G) CLN3-6xHA::NATNT2 | This study | Figure 5 |
| DK3000 | MATa bar1 ypk1-as (L424A) ypk2∆::HIS3 CLN3-6xHA::NATNT2 | This study | Figure 6A–C and Figure 6—figure supplement 1D and E |
| DK3525 | MATa bar1 ypk1∆::HIS3 CLN3-6xHA::NATNT2 | This study | Figure 6D |
| DK3578 | MATa bar1 lac1∆::HIS3 lag1∆::KANMX CLN3-6xHA::NATNT2 | This study | Figure 7E and F |