Cardiolipin targets a dynamin-related protein to the nuclear membrane
Figures
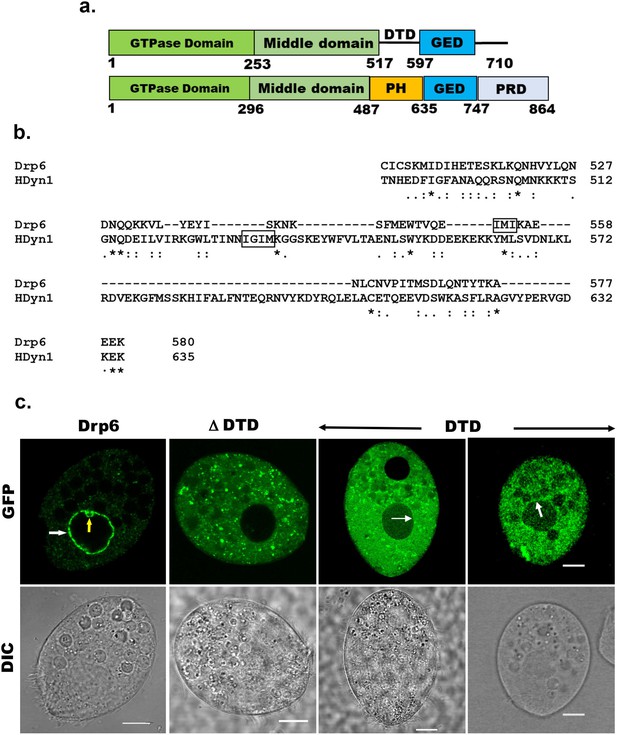
Identification of the region of Drp6 important for nuclear recruitment.
(a) Diagram showing domains of Tetrahymena Drp6 (top) and human dynamin 1 (bottom). Five domains of dynamin indicated as GTPase domain, middle domain, PH domain, GED, and PRD. Drp6 contains three domains but lacks PH domain and PRD. Numbers indicate the position of amino acids in the protein. (b) Sequence alignment of Tetrahymena dynamin-related protein 6 (Drp6) and human dynamin 1 (HDyn1) generated using Clustal Omega. Only the PH domain of HDyn1 and the corresponding aligned region of Drp6 are shown. The hydrophobic patch (IGIM) of PH domain important for membrane insertion is shown within a box. A putative hydrophobic patch (IMI) in Drp6 is also within box. (c) Confocal images of live Tetrahymena cells expressing GFP-drp6 (Drp6), GFP-drp6ΔDTD (ΔDTD), and GFP-drp6-DTD (DTD) are shown. While localization on the nuclear envelope of MAC is indicated by white arrow, yellow arrow indicates MIC. Bar = 10 µm.
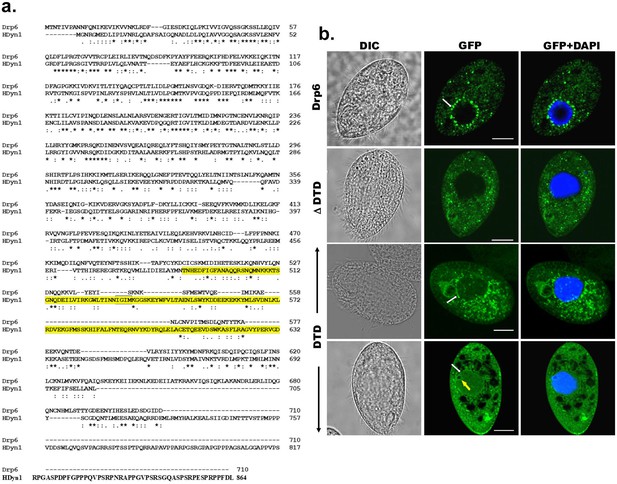
DTD is important for nuclear localization of Drp6.
(a) Alignment of amino acid sequences of Tetrahymena Drp6 (Drp6) and human dynamin 1 (HDyn1). The PH domain of HDyn1 is highlighted in yellow. (b) Confocal images of fixed Tetrahymena cells expressing GFP-drp6 (Drp6), GFP-drp6 ΔDTD (ΔDTD), and GFP-drp6-DTD (DTD). Blue color represents DAPI-stained nucleus. While localization on the nuclear envelope of MAC is indicated by white arrow, yellow arrow indicates MIC. Bar = 10 µm.
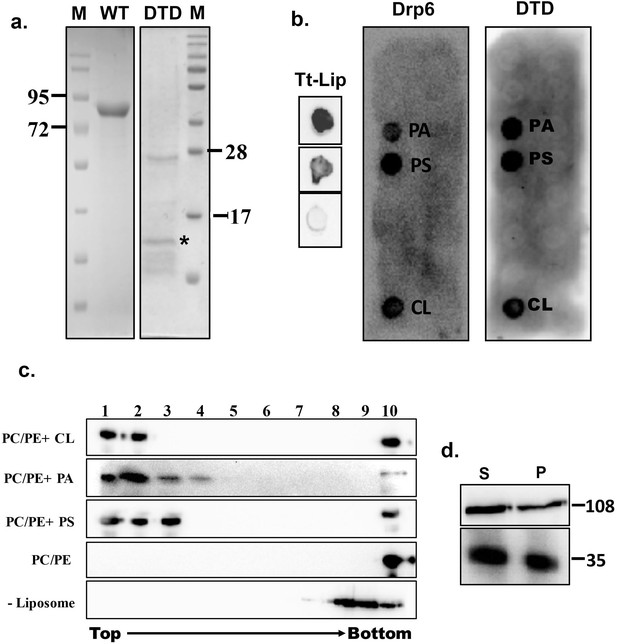
Identification of membrane-binding domain of Drp6.
(a) Coomassie-stained SDS–PAGE gels showing purification of His-drp6 (WT) and His-drp6-DTD (DTD) expressed in Escherichia coli. M is the molecular weight markers. Some of the markers are indicated on the sides. The purification of His-drp6-DTD was partial and contained additional proteins from E. coli, including one prominent band below 28 kDa. The purified His-drp6-DTD appearing below 17 kDa marker is indicated by an asterisk. (b) Lipid overlay assay as detected by western blot analysis using anti-his antibody. (Tt-Lip); total Tetrahymena lipid spotted on nitrocellulose membrane and incubated with His-drp6 in absence (top) or presence (middle) of GTP. The bottom spot is incubated with BSA. Strip spotted with 15 different lipids and incubated either with His-drp6 (Drp6) or with His-drp6-DTD (DTD). Both Drp6 and DTD interacted with PA, PS, and CL are indicated. (c) Floatation assay using liposomes containing 70% phosphatidylcholine and 20% phosphatidylethanolamine additionally supplemented with 10% CL (PC/PE+CL), 10% PA (PC/PE+PA), or 10% PS (PC/PE+PS). While liposomes in (PC/PE) contained 80% phosphatidylcholine and 20% phosphatidylethanolamine, no liposome was added in (–Liposome). His-drp6 was incubated either with different liposomes or without liposomes, overlaid with sucrose gradient, and subjected to ultra-centrifugation. Fractions were collected from top and detected by western blot analysis using anti-his antibody. Drp6 appearing in the top four fractions indicate interaction with liposome. The experiments were repeated at least three times, and representative results are shown here. (d) Lysates of Tetrahymena cells expressing either GFP-drp6 (top) or GFP-drp6-DTD (bottom) were fractionated into soluble (S) and membrane (P) fractions and detected by western blot using anti-GFP antibody. Molecular weights of the proteins are indicated on the right.
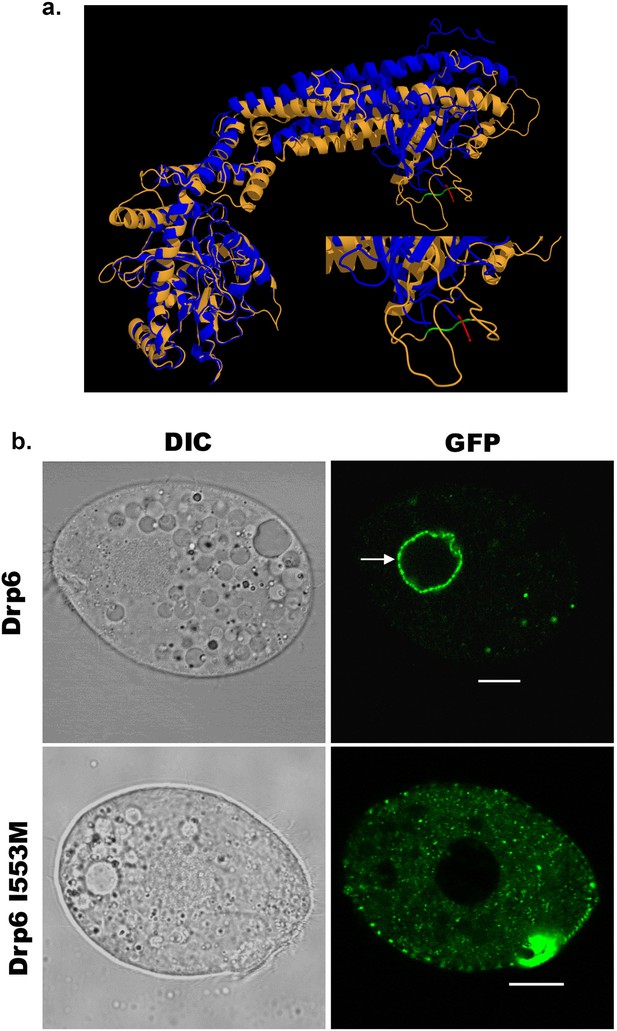
An isoleucine in the membrane-binding domain is important for nuclear localization of Drp6.
(a) Three-dimensional structure of Drp6. Homology model of Drp6 (brown) was generated by I-TASSER using Human Dynamin-1 as template (blue). The part containing the hydrophobic patch (residues 531–534 marked in red) in the PH domain of Human Dynamin-1 important for membrane insertion along with the putative hydrophobic patch (residues 553–555 marked in green) of Drp6 model are shown at the bottom right after enlarging the area. Although far apart in primary sequences, the regions containing hydrophobic patch in both the proteins come to the vicinity in 3-D structure. (b) Confocal images of live Tetrahymena cells expressing GFP-drp6 (top) and GFP-drp6-I553M (bottom). Mutation of isoleucine to methionine at 553rd position leads to loss of nuclear localization. Arrow indicates nuclear envelope. Bar = 10 µm.
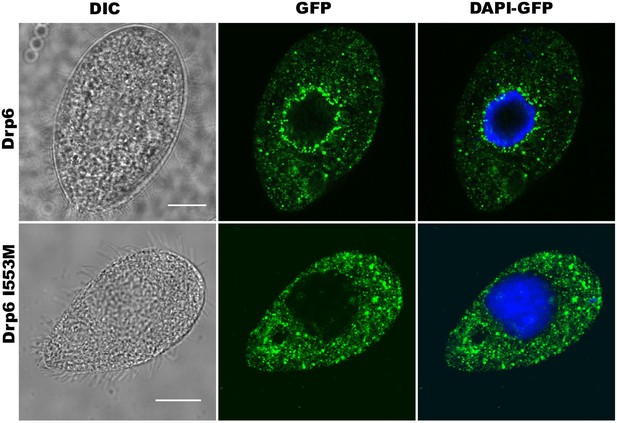
Confocal images of fixed Tetrahymena cells after DAPI-staining either expressing GFP-drp6 (Drp6) or GFP-drp6-I553M (Drp6-I553M).
Bar = 10 µm.
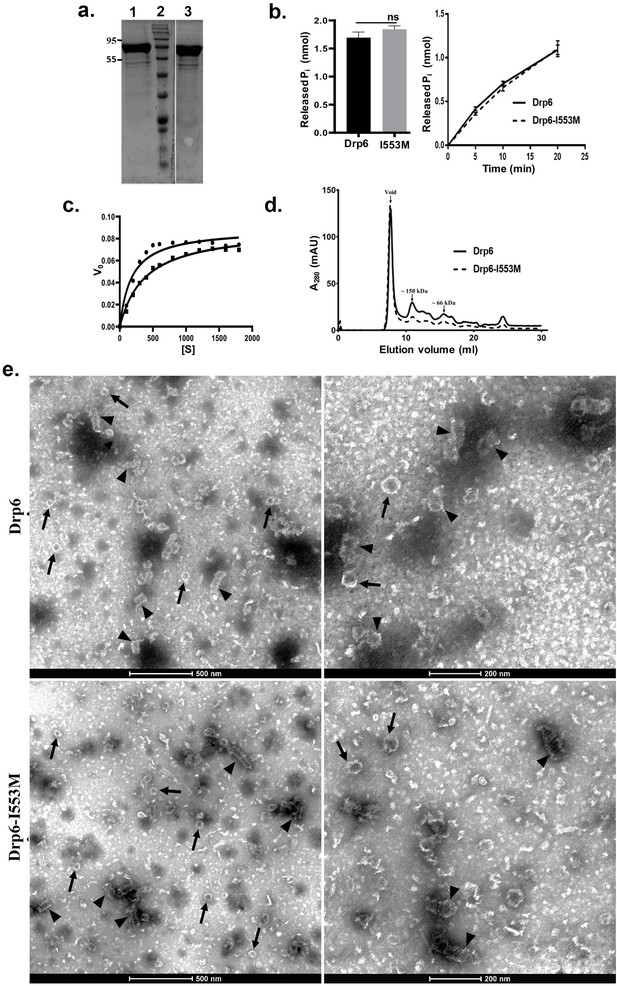
Mutation at I553 does not affect GTP hydrolysis activity and self-assembled structures.
(a) Coomassie-stained SDS–PAGE gel showing purified His-drp6 (lane 1) and His-drp6-I553M (lane 3). Lane 2 is molecular weight marker. The positions of molecular weight are indicated on the left. (b) Graph showing GTP hydrolysis of His-drp6 (Drp6) and His-drp6-I553M (I553M) as measured by phosphate release after 30 min of reaction (left). The graph on right shows reactions carried out for 0–20 min. The statistical analysis was performed using unpaired t-test and the difference was non-significant (p≤0.0001). For both the experiments, n = 3. All the experiments were performed more than three times. (c) Michaelis–Menten plot showing GTP hydrolysis by His-drp6 (circle) and His-drp6-I553M (square). V0 = rate of product formation in nmol Pi/µM protein/min and [S] = GTP concentration in µM. n = 3. (d) Chromatograms depicting elution profiles of His-drp6 and His-drp6-I553M using superdex 200 size exclusion column. The void volume and the positions of molecular weight markers are indicated by arrows. (e) Electron micrographs of negatively stained His-drp6 (Drp6) and His-drp6-I553M (Drp6-I553M) at two different magnifications. Helical spirals and the ring structures are found in both wild-type and mutant proteins and are indicated by arrow head and arrow, respectively.
-
Figure 4—source data 1
Mutation at I553 does not affect GTPase activity and self-assembly of Drp6.
- https://cdn.elifesciences.org/articles/64416/elife-64416-fig4-data1-v2.xlsx
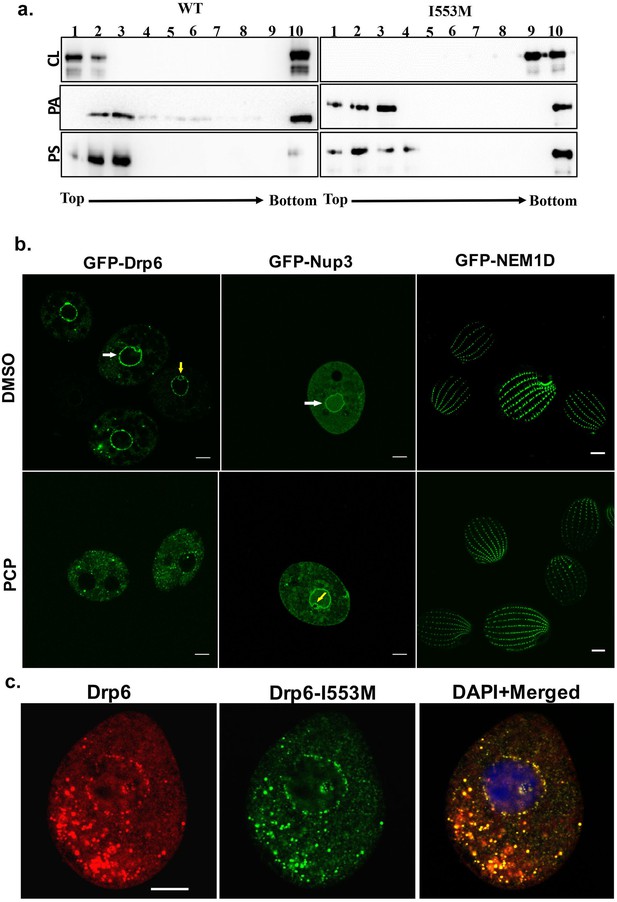
Interaction of CL with membrane-binding domain recruits Drp6 to the nuclear membrane.
(a) Floatation assay was performed using liposomes with same composition and analyzed by western blotting as mentioned in Figure 2c. The assay was performed with His-drp6 (WT) and His-drp6-I553M (I553M). Liposomes supplemented with 10% CL, 10% PA, 10% PS were used for the assay. Fractions collected from top to bottom are indicated. Experiments were repeated at least three times and representative results are shown here. Mutation at I553 lost interaction completely with the liposomes containing CL while retaining interactions with liposomes containing either PS or PA, suggesting isoleucine residue at 553rd position is important for binding with CL in the bilayers. (b) Confocal images of live Tetrahymena cells expressing GFP-drp6 (left panel), GFP-Nup3/MacNup98B (middle panel), and GFP-Nem1D (right panel) either in presence (PCP) or absence (DMSO) of PCP. While localization on the nuclear envelope of MAC is indicated by white arrow, yellow arrow indicates MIC. Bar = 10 µm. (c) Confocal images of fixed Tetrahymena cells co-expressing mCherry-drp6 (left panel) and GFP-drp6-I553M (middle panel). Merged image with DAPI-stained nucleus is shown in right panel. Yellow color in the merged image signifies the presence of both Drp6 and Drp6-I553M in the same complex. Bar = 10 µm.
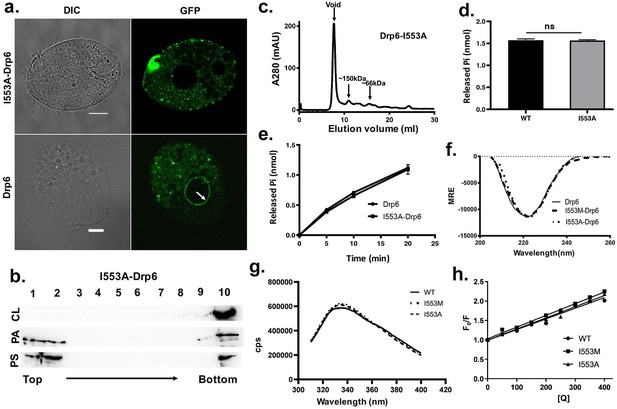
The loss of nuclear localization and CL binding by substitution mutations of I553 are not due to changes in overall structure and conformation.
(a) Confocal images of live Tetrahymena cells expressing either GFP-drp6- I553A (top) or GFP-drp6 (bottom). Localization in the nuclear envelope is indicated by arrow. Bar = 10 µm. (b) Floatation assay showing binding of His-drp6-I553A with liposomes containing 10% CL (top), 10% PA (middle), and 10% PS (bottom) as analyzed by western blotting. Mutation of isoleucine to alanine at 553rd position results in loss of binding specifically with CL. (c) Elution profile of His-drp6-I553A in size exclusion chromatography. Similar to wild-type Drp6, I553A mutant also elutes mostly as higher order oligomeric structures. (d) GTP hydrolysis activities of His-drp6 (WT) and His-drp6-I553A (I553A) as measured after 30 min. The statistical analysis was performed using unpaired t-test and the difference was non-significant (p≤0.0001). n = 3. All the experiments were repeated several times. (e) Same as in (d) except the GTP hydrolysis was carried out 0–20 min. Drp6 = His-drp6 and I553A-Drp6 = His-drp6-I553A. (p≤0.0001). n = 3. All the experiments were repeated several times. (f) The graph showing the CD spectra of His-drp6, His-drp6-I553M, and His-drp6-I553A recorded from 205 nm to 260 nm. (g) Tryptophan fluorescence emission spectra of His-drp6 (WT), His-drp6-I553M (I553M), and His-drp6-I553A (I553A) with excitation at 295 nm. (h) Stern-Volmer plots of tryptophan fluorescence quenching by acrylamide. The emission at 332 nm (emission peak) was plotted.
-
Figure 6—source data 1
Mutations at I553 results in loss of nuclear localization and cardiolipin interactions without affecting GTPase activity, self-assembly, overall folding and 3-D conformation of Drp6.
- https://cdn.elifesciences.org/articles/64416/elife-64416-fig6-data1-v2.xlsx
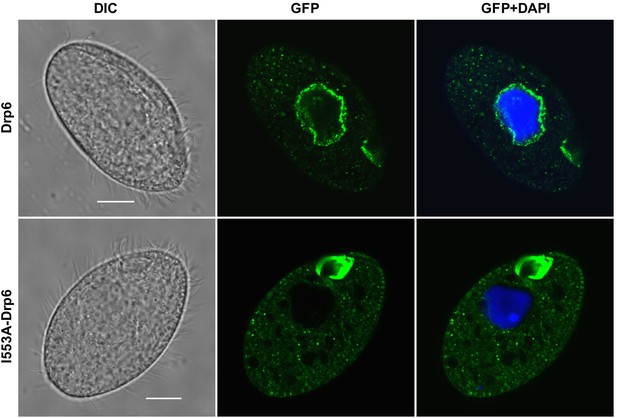
Confocal images of fixed Tetrahymena cells after DAPI-staining either expressing GFP-drp6 (Drp6) or GFP-drp6-I553A (Drp6-I553A).
Bar = 10 µm.
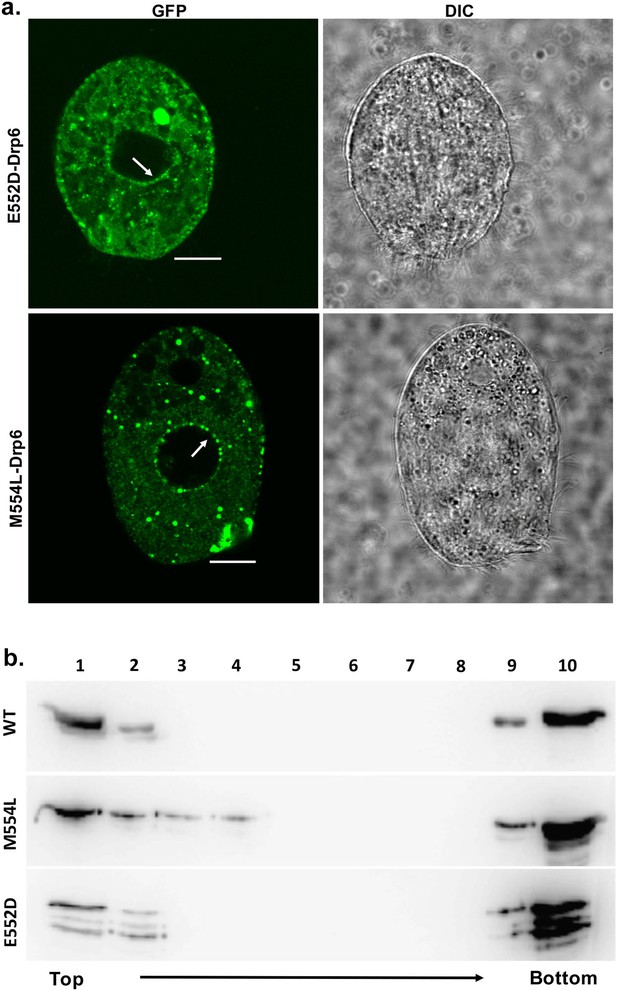
The mutations at E552 and M554 residues do not abrogate CL binding and nuclear localization.
(a) Confocal images of live Tetrahymena cells expressing GFP-drp6-E552D (top) or GFP-drp6-M554L (bottom). Nuclear localization is indicated by arrow. Both E552D and M554L mutants associate with nuclear envelope. Bar = 10 µm. (b) Floatation assay was performed with liposomes containing 10% CL using His-drp6 (WT), His-drp6-M554L (M554L), and His-drp6-E552D (E552D).
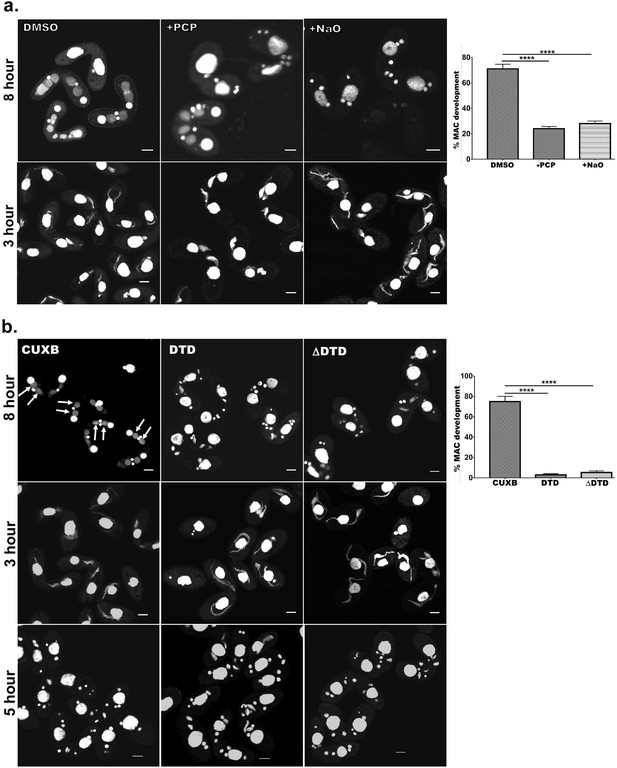
Cardiolipin and membrane-binding domain regulate macronuclear expansion.
(a) Confocal images of fixed and DAPI-stained conjugation pairs of Tetrahymena at 8 hr and 3 hr post-conjugation. Two wild-type strains of Tetrahymena (Cu428 and B2086) were conjugated and treated either with pentachlorophenol (+PCP) or with nonyl acridine orange –D (+NaO) or with DMSO (DMSO). Top panel shows MAC development at 8 hr, and bottom panel shows MIC elongation at 3 hr. Percent MAC development at 8 hr is shown at the right. (b) Confocal images of fixed DAPI-stained Tetrahymena cells conjugated either between CU428 and B2086 (CUXB) or between CU428 and GFP-drp6-DTD-expressing cells (DTD) or between CU428 and GFP-drp6 ΔDTD-expressing cells (ΔDTD). Top panel MAC development stage at 8 hr, middle panel MIC elongation stage at 3 hr, and bottom panel meiotic stage at 5 hr. Percent MAC development at 8 hr is shown at the right. The newly developed MAC is indicated by arrow. For quantitation, three independent experiments were performed and analyzed by unpaired t-test (****p≤0.0001). n > 500.
-
Figure 8—source data 1
Inhibition of Drp6-CL interaction inhibits macronuclear development in Tetrahyemena.
- https://cdn.elifesciences.org/articles/64416/elife-64416-fig8-data1-v2.xlsx
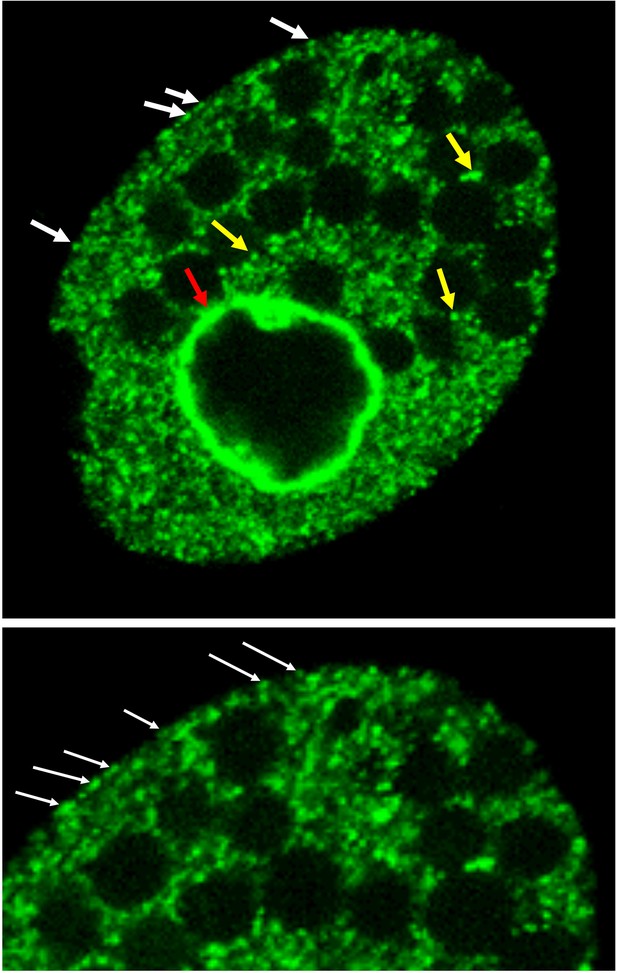
Drp6 localizes to multiple sites.
Top: Confocal image of a live Tetrahymena cell expressing Drp6 as GFP-fusion protein. The localization of Drp6 on nuclear envelope (red arrow), on ER vesicles (yellow arrow), and on plasma membrane (white arrow) is shown. Bottom: Part of the image is enlarged to visualize the localization on the plasma membrane more distinctly.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Tetrahymena thermophila) | CU428.2 | Tetrahymena Stock Center, Cornell University, USA | RRID:TSC_SD00178 | Wild type |
| Strain, strain background (Tetrahymena thermophila) | B2086.2 | Tetrahymena Stock Center, Cornell University, USA | RRID:TSC_SD00709 | Wild type |
| Recombinant DNA reagent | pVGF (plasmid) | Meng-Chao Yao, FHCRC, Seattle, Washington | rDNA-based Tetrahymena expression vector | |
| Recombinant DNA reagent | GFP-drp6 (plasmid) | This study | Drp6 cloned in pVGF expression vector | |
| Recombinant DNA reagent | GFP-drp6-DTD (plasmid) | This study | pVGF vector that expresses aa 517–600 of Drp6 | |
| Recombinant DNA reagent | GFP-drp6-ΔDTD (plasmid) | This study | pIGF vector expressing Drp6 with deletion of aa 517–600 | |
| Recombinant DNA reagent | GFP-drp6-I553M (plasmid) | This study | pIGF vector that expresses Drp6-I553M | |
| Recombinant DNA reagent | GFP-drp6-I553A (plasmid) | This study | pIGF vector that expresses Drp6-I553A | |
| Recombinant DNA reagent | GFP-drp6-E552D (plasmid) | This study | pIGF vector expressing Drp6-E552D | |
| Recombinant DNA reagent | GFP-drp6-M554L (plasmid) | This study | pIGF vector that expresses Drp6-M554L | |
| Recombinant DNA reagent | GFP-Nup3/MacNup98B (plasmid) | Prof. D.L. Chalker, Washington University in St. Louis | NCVB vector expressing GFP-Nup3. Blasticidine resistance | |
| Recombinant DNA reagent | GFP-Nem1D (plasmid) | Previous study in lab (Shukla et al., 2018) | pIGF vector expressing GFP-NEM1D | |
| Recombinant DNA reagent | mCherry-drp6 (plasmid) | This study | NCVB vector that expresses mCherry-DRP6 | |
| Recombinant DNA reagent | His-drp6 (plasmid) | Previous study in the lab (Kar et al., 2018) | pRSETB vector expressing Drp6 as N-terminal histidine tag | |
| Recombinant DNA reagent | His-drp6-DTD (plasmid) | This study | pRSETB vector expressing aa 517 to 600 as N-terminal histidine tag | |
| Recombinant DNA reagent | His-drp6-I553M (plasmid) | This study | pRSETB vector expressing Drp6-I553M as N-terminal histidine tag | |
| Recombinant DNA reagent | His-drp6-I553A (plasmid) | This study | pRSETB vector expressing Drp6-I553A as N-terminal histidine tag | |
| Recombinant DNA reagent | His-drp6-E552D (plasmid) | This study | pRSETB vector expressing Drp6-E552D as N-terminal histidine tag | |
| Recombinant DNA reagent | His-drp6-M554L (plasmid) | This study | pRSETB vector expressing Drp6-M554L as N-terminal histidine tag | |
| Antibody | Anti-His6-peroxidase (mouse monoclonal) | Sigma–Aldrich | Cat#: 11965085001 | WB (1:5000) |
| Antibody | Anti-GFP (rabbit polyclonal) | Sigma–Aldrich | Cat#: AB10145 | WB (1:4000) |
| Antibody | Anti-Rabbit IgG- peroxidase (Goat polyclonal) | Sigma–Aldrich | Cat#: A0545-1ML | WB (1:80,000) |
| Chemical compound, drug | Pentachlorophenol | Sigma–Aldrich | Cat#: 87-86-5 | |
| Chemical compound, drug | Acridine orange 10-nonyl bromide | Invitrogen | Cat#: A1372 | |
| Chemical compound, drug | Phosphatidylcholine | Avanti Polar Lipids | Cat#: 840051 | |
| Chemical compound, drug | Phosphatidylethanolamine | Avanti Polar Lipids | Cat#: 840021 | |
| Chemical compound, drug | Cardiolipin | Avanti Polar Lipids | Cat#: 840012 | |
| Chemical compound, drug | Phosphatidic acid | Avanti Polar Lipids | Cat#: 840101 | |
| Chemical compound, drug | Phosphatidylserine | Avanti Polar Lipids | Cat#: 840032 | |
| Chemical assay or kit | BIOMOL green | Enzo Life Sciences | Cat#: BML-AK111 | |
| Chemical compound | uranyl acetate | MP Biomedicals | Cat#: 181561 | |
| Chemical compound | Ni-NTA agarose | QIAGEN | Cat#: 30210 | |
| Other | Membrane lipid strips | Echelon Biosciences | Cat#: P-6002 | |
| Other | Carbon film 200 mesh copper | Ted Pella Inc | Cat#: CF200-CU | |
| Other | DAPI stain | Invitrogen | Cat#: D1306 | (0.25 μg/ml) |





