TDP-43 maximizes nerve conduction velocity by repressing a cryptic exon for paranodal junction assembly in Schwann cells
Figures
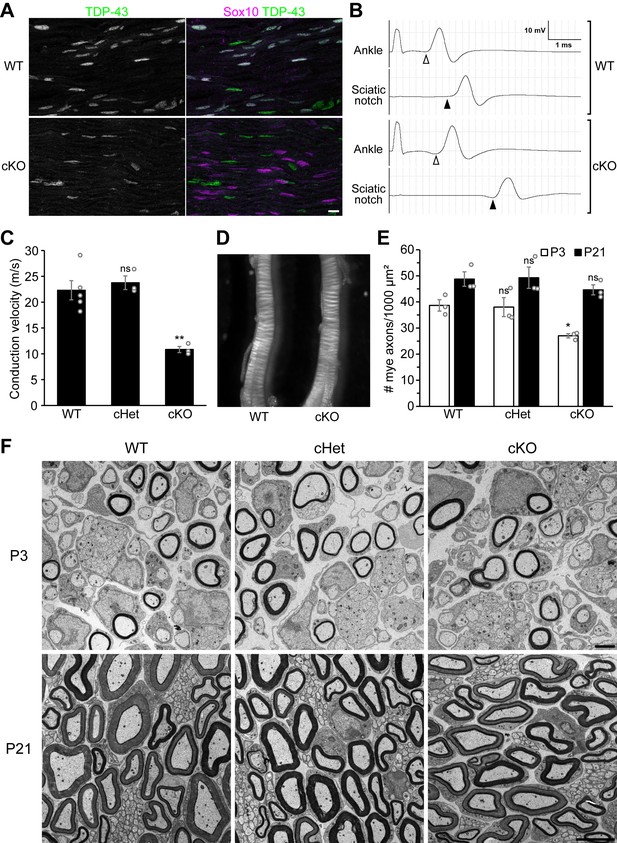
Knockout of TDP-43 in Schwann cells results in a 50% conduction delay without overt alteration of compact myelin.
(A) Longitudinal sections of P28 wild-type (WT) and conditional knockout (cKO) sciatic nerves were immunostained for TDP-43 (green) and Sox10 (magenta). Sox10 labels Schwann cells, which are all TDP-43-negative in the cKO. The cell types other than Schwann cells are Sox10-negative and are still TDP-43-positive in the cKO. Scale bar, 10 μm. (B, C) Motor nerve conduction of P27 mice was measured as compound muscle action potentials at the plantar muscles evoked by stimulation of the nerve at the ankle and sciatic notch. The onset of the compound muscle action potentials is indicated by open arrowheads (ankle stimulation) and solid arrowheads (sciatic notch stimulation) in B. Bars represent mean ± SEM in C. n = 5 mice for WT, 3 for conditional heterozygote (cHet), and 3 for cKO. **p=0.0030 and 0.0028 (WT vs. cKO and cHet vs. cKO, respectively); ns: not significant, p=0.8094 (WT vs. cHet); one-way analysis of variance (ANOVA) with Tukey’s test. (D) Sciatic nerves from P7 WT and cKO mice. (E) The number of myelinated axons per 1000 μm² was quantified with electron micrographs of sciatic nerve cross sections. Bars represent mean ± SEM. n = 3 mice per genotype. *p=0.039 and 0.048 (WT vs. cKO and cHet vs. cKO at P3, respectively); ns: not significant (WT vs. cHet at P3, p=0.9812; P21, p=0.5381); one-way ANOVA with Tukey’s test. (F) Electron micrographs of P3 and P21 sciatic nerve cross sections. Scale bars, 2 μm for P3 and 5 μm for P21. cHet and cKO by Dhh-Cre (A–F).
-
Figure 1—source data 1
Statistical summary for Figure 1C, E and Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/64456/elife-64456-fig1-data1-v1.xlsx
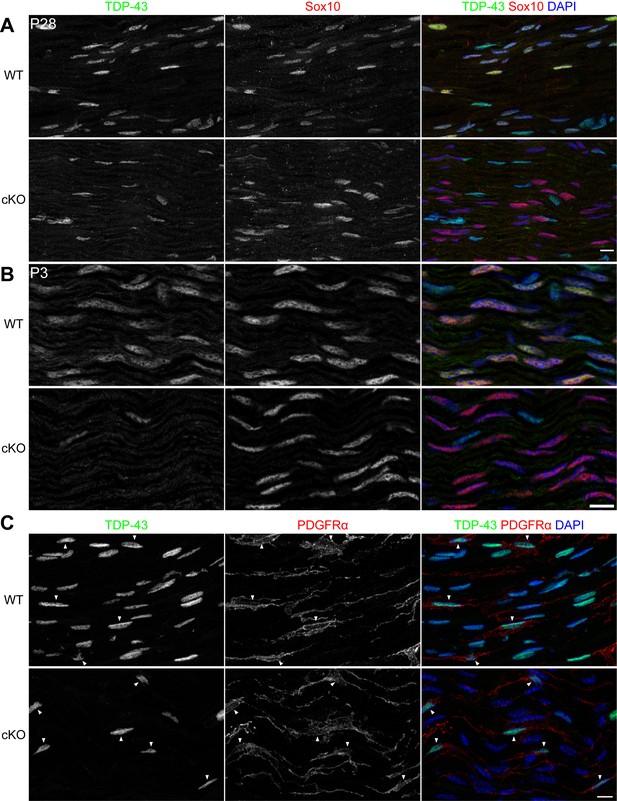
TDP-43 is expressed by wild-type (WT) Schwann cells and specifically ablated from the conditional knockout (cKO) Schwann cells.
(A) Single-channel images and the merged image with DAPI nuclear staining for Figure 1A. Scale bar, 10 μm. (B) Immunostaining of P3 WT and cKO sciatic nerves for TDP-43 (green) and Sox10 (red). Sox10 labels Schwann cells, which are all TDP-43-negative in the cKO. The cell types other than Schwann cells are Sox10-negative and are still TDP-43-positive in the cKO. Scale bar, 10 μm. (C) Immunostaining of P28 WT and cKO sciatic nerves for TDP-43 (green), PDGFRα (red), and DAPI (blue). PDGFRα labels endoneurial fibroblasts, which are TDP-43-positive in the cKO. Scale bar, 10 μm. cKO by Dhh-Cre (A–C).
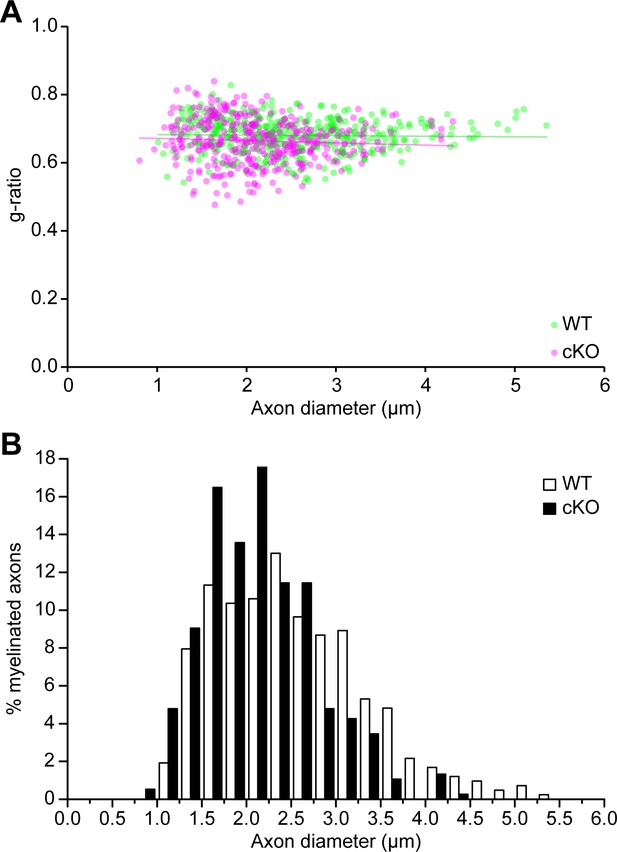
Quantification for the myelin thickness and axon diameters of myelinated axons in the TDP-43-cKO sciatic nerves.
(A) The scatter plot showing the g-ratios (y-axis) in relation to the axon diameters (x-axis) of individual myelinated axons in P21 wild-type (WT) and conditional knockout (cKO) sciatic nerves. The g-ratio was calculated as axon diameter/(axon diameter + myelin thickness ×2). Average g-ratios (mean ± SEM): WT = 0.6809 ± 0.0066; cKO = 0.6640 ± 0.0009. Unpaired two-tailed t-test: p=0.1214. (B) The histogram showing the frequency of different axon populations in each axon diameter range. n = 3 mice (totally 415 axons) for WT; n = 3 mice (totally 376 axons) for cKO. More than 100 axons per mouse were analyzed.
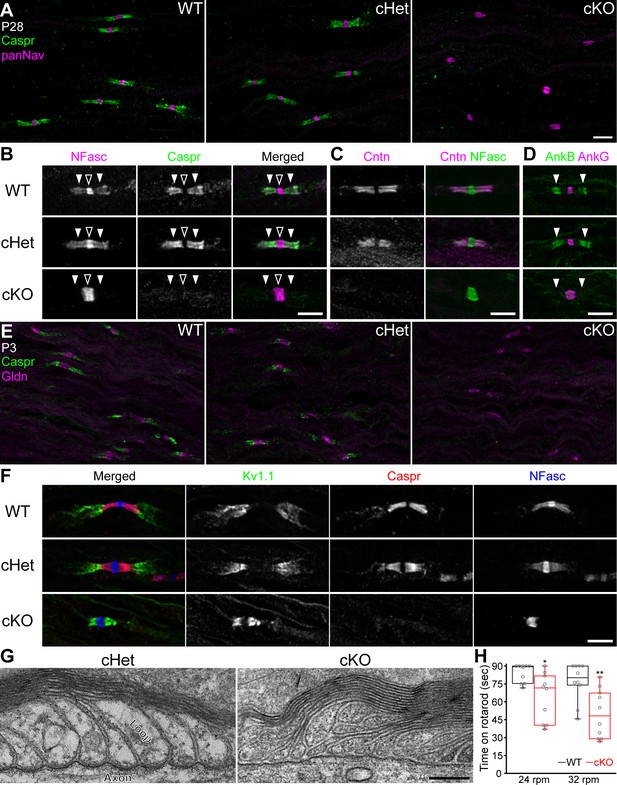
Paranodal axoglial junctions are disrupted in the TDP-43-cKO mice.
(A) Immunostaining of P28 sciatic nerves for contactin-associated protein (Caspr) (green, paranode) and Nav channels (panNav, magenta, node). (B–D) Immunostaining of P28 sciatic nerves for (B) neurofascin (NFasc) (magenta, node and paranode) and Caspr (green), (C) contactin (Cntn, magenta, paranode) and NFasc (green), and (D) ankyrinB (AnkB, green, paranode) and ankyrinG (AnkG, magenta, node). Nodes are indicated by open arrowheads in B. Paranodes are indicated by solid arrowheads in B, D. (E) Immunostaining of P3 sciatic nerves for Caspr (green) and gliomedin (Gldn, magenta, node). (F) Immunostaining of P28 sciatic nerves for Kv1.1 channels (green, juxtaparanode), Caspr (red), and NFasc (blue). (G) Electron micrographs of P21 sciatic nerve longitudinal sections. (H) The time periods for which the mice remained on the rotarod at 24 or 32 rpm. Minima, first quartiles, medians, third quartiles, and maxima are plotted as box-and-whisker plots. n = 10 mice per genotype. *p=0.0115 for 24 rpm; **p=0.0036 for 32 rpm; Mann–Whitney tests. Scale bars, 5 μm (A–F) and 200 nm (G). Conditional heterozygote (cHet) and conditional knockout (cKO) by Dhh-Cre in (A–C) and (E–G), and by Mpz-Cre in (D, H).
-
Figure 2—source data 1
Statistical summary for Figure 2H.
- https://cdn.elifesciences.org/articles/64456/elife-64456-fig2-data1-v1.xlsx
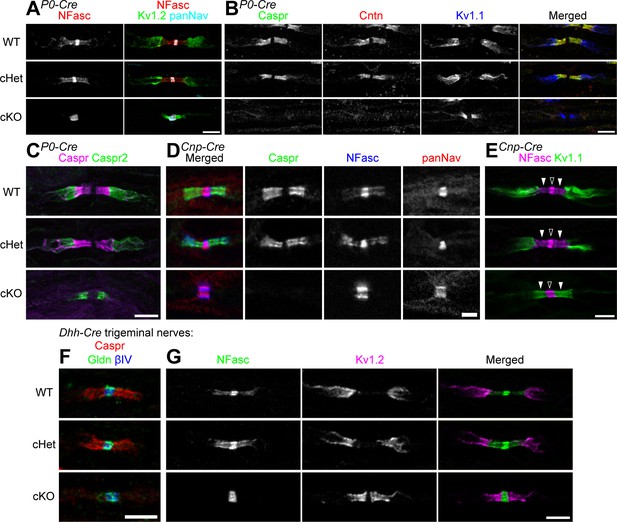
Paranodal axoglial junctions are completely disrupted in the peripheral nervous system of the TDP-43-cKO mice using three different Cre-driver lines.
(A–C) Immunostaining of P28 sciatic nerves for (A) neurofascin (NFasc) (red), Kv1.2 channels (green, juxtaparanode), and Nav channels (panNan) (cyan); (B) contactin-associated protein (Caspr) (green), contactin (Cntn) (red), and Kv1.1 channels (blue); and (C) Caspr (magenta) and Caspr2 (green, juxtaparanode). Conditional heterozygote (cHet) and conditional knockout (cKO) by Mpz-Cre (P0-Cre). Scale bars, 5 μm. (D, E) Immunostaining of P60 sciatic nerves for (D) Caspr (green), NFasc (blue), and Nav channels (panNav) (red); and (E) NFasc (magenta) and Kv1.1 channels (green). cHet and cKO by Cnp-Cre. Scale bars, 2 μm (D) and 5 μm (E). (F, G) Immunostaining of P28 trigeminal nerves for (F) Caspr (red), gliomedin (Gldn) (green), and βIV spectrin (blue, node); and (G) NFasc (green) and Kv1.2 channels (magenta). cHet and cKO by Dhh-Cre. Scale bars, 5 μm.
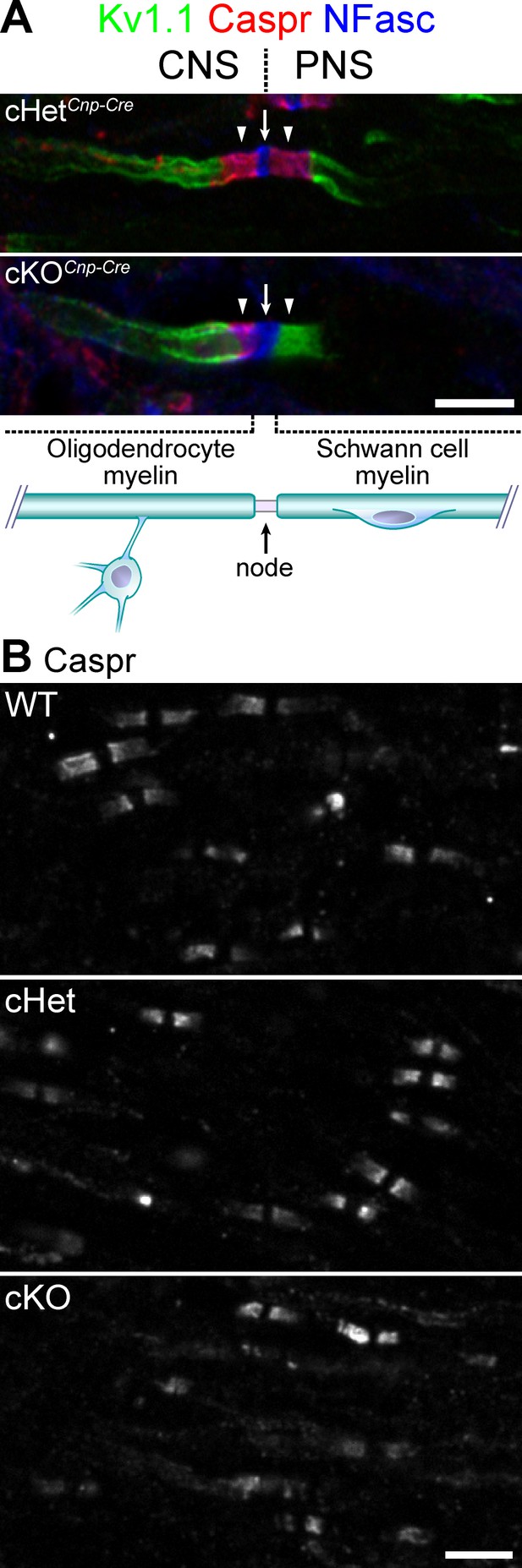
Knockout of TDP-43 in oligodendrocytes displays normal paranodal axoglial junctions in the central nervous system (CNS).
(A) Immunostaining of the dorsal root entry zone from P60 conditional heterozygote (cHet) and conditional knockout (cKO) (by Cnp-Cre) spinal cords for Kv1.1 channels (green), contactin-associated protein (Caspr) (red), and neurofascin (NFasc) (blue). The node at the transition zone is indicated by an arrow and is shared by an oligodendrocyte at the left and a Schwann cell at the right, as illustrated below. The flanking CNS and peripheral nervous system (PNS) paranodes are indicated by arrowheads. Illustration adapted from Figure 1a of Chang et al., 2016, with permission. (B) Immunostaining of P21 spinal cords for Caspr. cHet and cKO by Cnp-Cre. Scale bars, 5 μm (A, B).
© 2016, Springer Nature. Figure 3A is adapted from Figure 1a of Chang et al., 2016, with permission from Springer Nature. Further reproduction of this panel would need permission from the copyright holder.
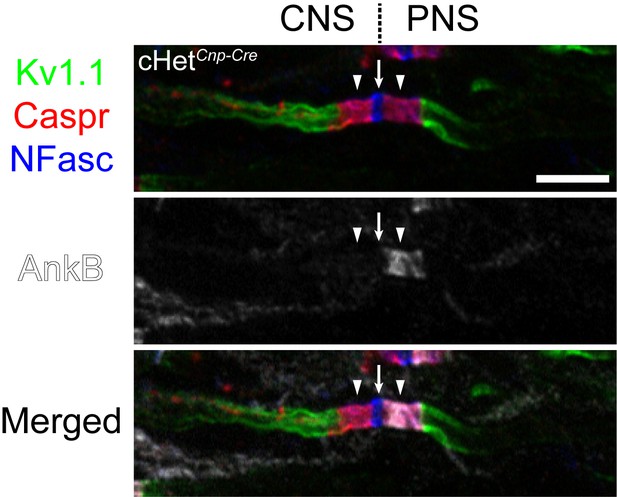
Nodes at the central nervous system–peripheral nervous system (CNS-PNS) transition zone are identified by immunostaining for the PNS paranodal marker ankyrinB (AnkB).
Immunostaining images of the dorsal root entry zone from the P60 conditional heterozygote (cHet) spinal cord shown in Figure 3A with Kv1.1 channels (green), contactin-associated protein (Caspr) (red), neurofascin (NFasc) (blue), and AnkB (white). The node at the transition zone is indicated by an arrow, and the flanking CNS and PNS paranodes are indicated by arrowheads. The PNS paranode, but not the CNS paranode, is labeled by AnkB.
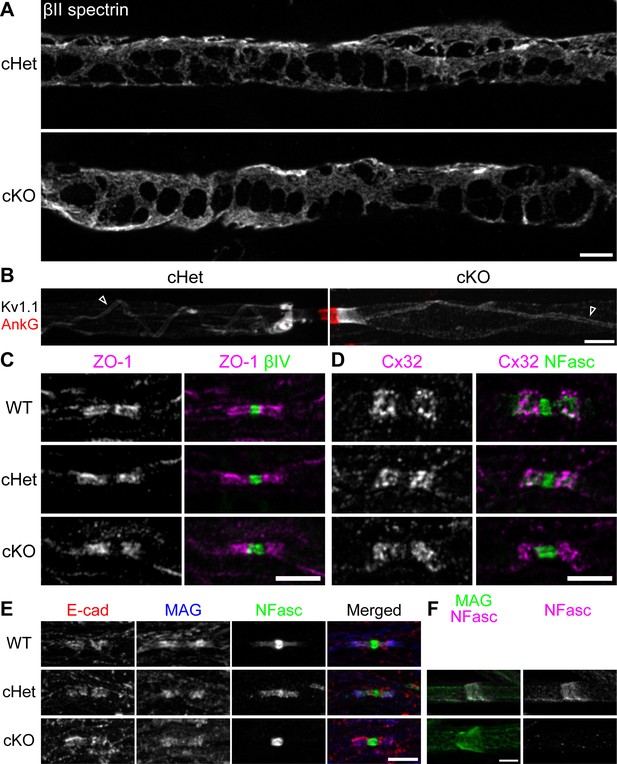
The fine myelin structures are formed normally in the TDP-43-cKO Schwann cells.
(A) Immunostaining of Cajal bands in teased P29 ventral roots with an antibody against βII spectrin. (B) Immunostaining of teased P29 ventral roots for ankyrinG (AnkG) (red, node) and Kv1.1 channels (white). Localization of Kv1.1 channels in the juxtamesaxon is indicated by open arrowheads. (C) Immunostaining of P28 sciatic nerves for zonula occludens-1 (ZO-1) (magenta) and βIV spectrin (green, node). (D) Immunostaining of P28 trigeminal nerves for connexin 32 (Cx32) (magenta) and neurofascin (NFasc) (green). (E) Immunostaining of P28 sciatic nerves for E-cadherin (E-cad) (red), myelin-associated glycoprotein (MAG) (blue), and NFasc (green). (F) Immunostaining of teased P28 sciatic nerves for MAG (green, incisure) and NFasc (magenta). cHet and cKO by Dhh-Cre (A–F). Scale bars, 5 μm (A–F).
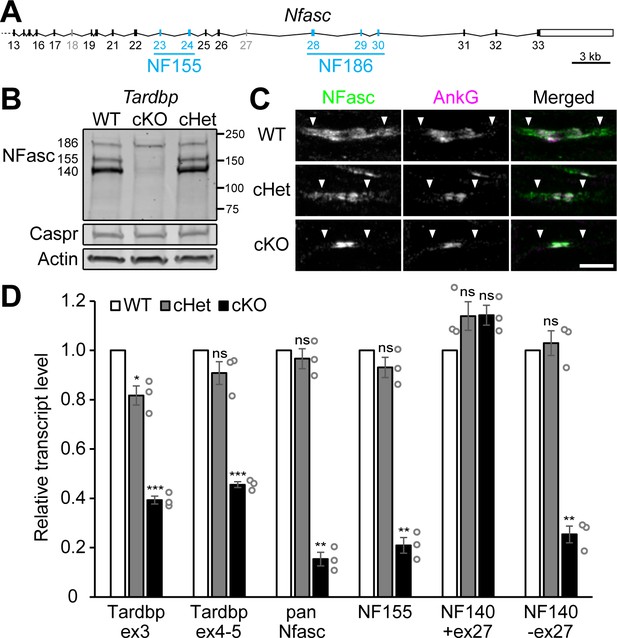
Schwann cell neurofascin (NFasc) expression is abolished in the TDP-43 conditional knockout (cKO).
(A) The schema of mouse Nfasc locus showing the region from exon 13 to exon 33. The coding strand of Nfasc is located on the reverse strand of chromosome 1, as defined in the GRCm38/mm10 mouse genome assembly, and flipped here for reader convenience. Alternative exons specific to NF155 (exons 23 and 24) or NF186 (exons 28–30) are shown in blue while other alternative exons (exons 18 and 27) are shown in gray. NF140 contains neither exons 23 and 24 nor exons 28–30. The open rectangle denotes the untranslated region, and the solid ones the open reading frame. (B) Western blotting of P28 sciatic nerve homogenates probed for NFasc, contactin-associated protein (Caspr), and actin. (C) Immunostaining of P3 sciatic nerves for NFasc (green) and ankyrinG (AnkG) (magenta) showing pairs of approaching heminodes (nodes flanked by myelin sheaths only on one side). The paranodes are indicated by arrowheads. Scale bar, 5 μm. (D) RT-qPCR analysis of P28–P29 sciatic nerves shows the transcript levels relative to those of the wild-type (WT) after normalized to the internal control Polr2a. Exon 3 of Tardbp is the floxed exon, and the primer pair for Tardbp ex3 detects TDP-43 mRNA transcribed from the unrecombined allele. The primer pair for Tardbp exons 4–5 detects the nonfloxed region to confirm mRNA degradation after Cre recombination. Pan-Nfasc is detected by primers spanning exons 12–13, NF155 by primers spanning exons 22–23, NF140+ex27 by primers annealing to the junctions of exons 22/25 and exons 27/31, and NF140-ex27 by primers annealing to the junctions of exons 22/25 and exons 26/31. Bars represent mean ± SEM. n = 3 mice per genotype. *p<0.05; **p<0.01; ***p<0.001; ns, p≥0.05; one-sample unpaired two-tailed t-tests (WT vs. conditional heterozygote [cHet] and WT vs. cKO). cHet and cKO by Dhh-Cre (B–D).
-
Figure 5—source data 1
Statistical summary for Figure 5D.
- https://cdn.elifesciences.org/articles/64456/elife-64456-fig5-data1-v1.xlsx
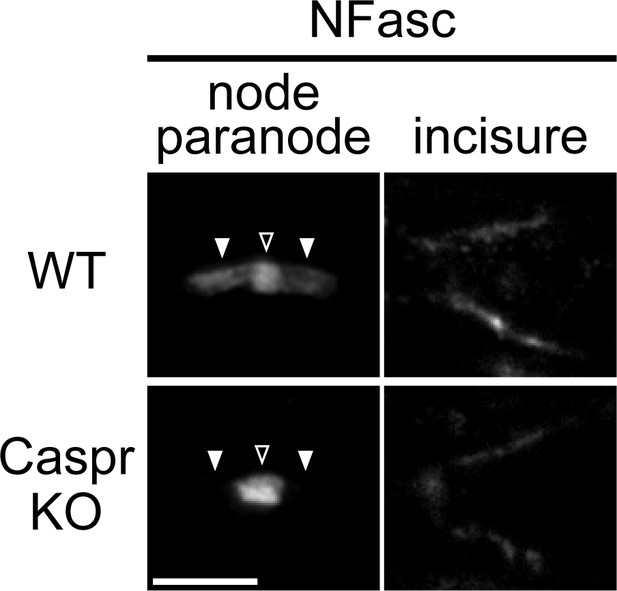
Localization of neurofascin (NFasc) in the Schmidt–Lanterman incisure is not affected in the contactin-associated protein knockout (Caspr KO).
Immunostaining of P80 sciatic nerves for NFasc. Nodes are indicated by open arrowheads, and paranodes by solid arrowheads. Scale bar, 5 μm.
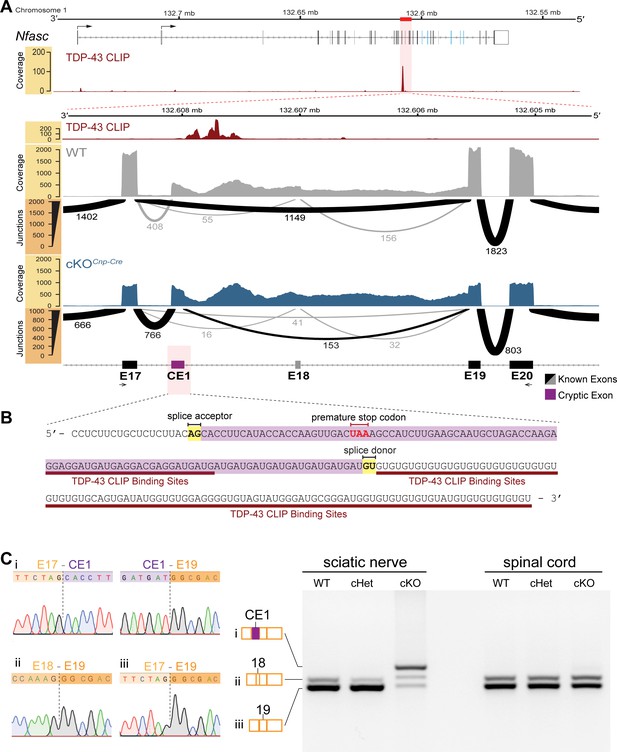
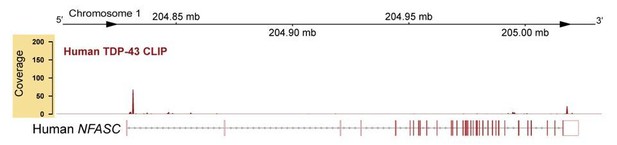
Cryptic exon located upstream of TDP-43-binding sites on Nfasc mRNA is highly expressed in the TDP-43 conditional knockout (cKO).
(A) TDP-43 binds to Nfasc mRNA as seen from the TDP-43 mRNA binding CLIP-seq data from mouse brains, which shows binding peaks between exons 17 and 18 of Nfasc (upper panel). The exon color code in the upper panel follows that in Figure 5A; the bent arrows denote the two alternative transcription start sites from exons 1a and 1b (see Supplementary file 1 for detail). The lower panel is a zoom into this region of interest, with tracks of wild-type (WT) and TDP-43-cKO sciatic nerve RNA-seq read coverage and Sashimi plots to visualize major junction-spanning reads. The arc width is proportionate to the number of reads spanning the junction, which is labeled below each arc. There is a very large proportion of reads spanning from E17 to a new cryptic exonic region, upstream to the TDP-43-binding sites, in the cKO compared to the WT. (B) The RNA sequence transcribed from the cryptic exon (CE1) is given in purple. CE1 contains a stop codon in the open reading frame (ORF), well-defined splice acceptor and donor sites, and a downstream UG-rich TDP-43-binding region, characteristic of TDP-43 cryptic exons. (C) RT-PCR for Nfasc spliceforms using sciatic nerves and spinal cords isolated from P21 mice. Schematic of Nfasc spliceforms shows exon usage: E17–CE1–E19–E20 (i), E17–E18–E19–E20 (ii), and E17–E19–E20 (iii). The bands were excised and sequenced. Chromatograms covering the exon–exon junctions are shown at the left. Conditional heterozygote (cHet) and cKO by Cnp-Cre.
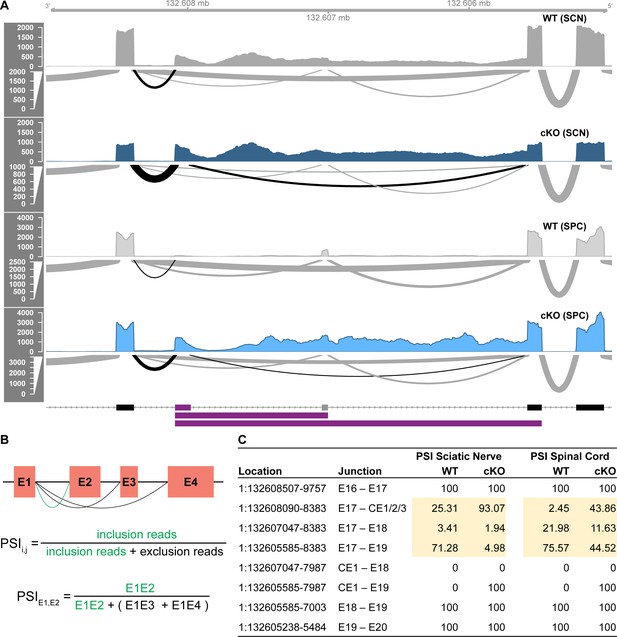
The identified cryptic exon is more highly expressed in sciatic nerves than in spinal cords and is spliced into >90% of Nfasc transcripts in the TDP-43-cKO sciatic nerves.
(A) The Sashimi plot derived from the RNA-seq data of sciatic nerves (SCN) and spinal cords (SPC) is shown for Nfasc E17–E20. The arc width is proportionate to the number of reads spanning the junction. The Sashimi plot of the SCN RNA-seq data is the same as the one shown in Figure 6A and is included here for comparison with the SPC data. (B) The percent spliced in index (PSI) values of exon–exon junctions of interest. (C) Reads from E17 are predominantly spliced to E19 in wild-type (WT) SCN, and this shifts to >90% of reads from E17 spliced to the cryptic exon CE1/2/3 in the TDP-43-cKO SCN. The spliced exon population is tissue-specific. E18 is expressed at a higher proportion in SPC compared to the negligible expression in SCN. The cryptic exon is incorporated into a small proportion of Nfasc transcripts in the WT SCN, but is nominally expressed in the WT SPC. Cryptic exon usage is increased in the conditional knockout (cKO) to different extent in SCN and SPC.
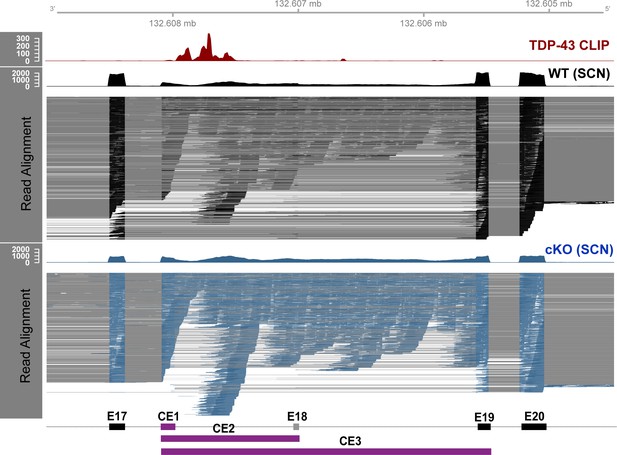
TDP-43-cKO RNA-seq reads align to novel exonic region upstream of TDP-43-binding regions in Nfasc.
Plotted in the region of interest around the TDP-43-binding region of Nfasc, extending from E17 to E20, are the CLIP-seq reads of TDP-43 binding in adult wild-type (WT) mouse brains (red), and RNA-seq data from WT (black) and TDP-43-cKO (blue) sciatic nerves. Read coverage and genomic alignment of the reads are shown, along with annotated exons and novel cryptic exons. The RNA-seq reads spanning exon–exon junctions are connected by gray lines. Reads spanning the junction between E17 and CE1/2/3 or between CE1 and E19 are increased in the TDP-43 conditional knockout (cKO), and the reads spanning the junction of E17 and E19 are dramatically decreased, suggesting that TDP-43 binding to Nfasc pre-mRNA excludes the cryptic exon from the resulting mature transcripts.
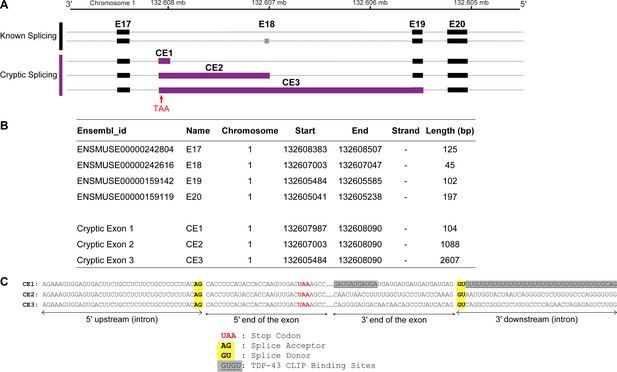
The identified candidate cryptic exons contain a premature stop codon in the open reading frame (ORF).
(A) A diagram showing the known alternative splicing in the Nfasc E17–E20 region and the identified cryptic exon usage. (B) Chromosome location and length of E17–E20 and potential cryptic exons. (C) The sequence of the cryptic exons around the 3′ and 5′ splice sites. All three potential cryptic exons have the same start site, well-defined splice donor and acceptor sites, and a premature stop codon in the ORF.
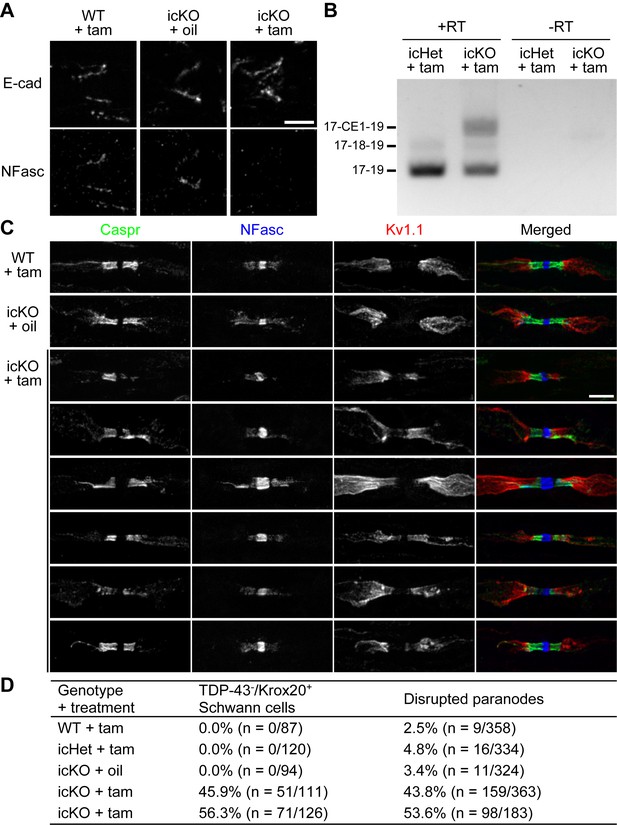
Paranodal junctions are disrupted when TDP-43 is deleted in adult Schwann cells.
(A) Immunostaining of adult sciatic nerves at 3 months after injection of tamoxifen (tam) or corn oil alone (oil) for E-cadherin (E-cad) (incisures) and neurofascin (NFasc). Scale bar, 5 μm. (B) RT-PCR for Nfasc spliceforms using sciatic nerves at 3 months after tamoxifen administration. +RT, reactions with reverse transcriptase; −RT, reactions without reverse transcriptase. (C) Immunostaining of adult sciatic nerves at 5 months after injection of tamoxifen (tam) or corn oil alone (oil) for contactin-associated protein (Caspr) (green), NFasc (blue), and Kv1.1 channel (red). Scale bar, 5 μm. (D) The sciatic nerve sections were immunostained for Krox20 and TDP-43 and quantified for the percentage of Krox20-positive nuclei that are TDP-43-negative. Immunostaining for Caspr and Kv1.1 was quantified for the percentage of Kv1.1-positive paranodes/juxtaparanodes that exhibit paranodal junction disruption defined as paranodal invasion of Kv1.1 and/or fragmented/shortened Caspr clusters. n represents the number of paranodes or nuclei analyzed for each mouse.
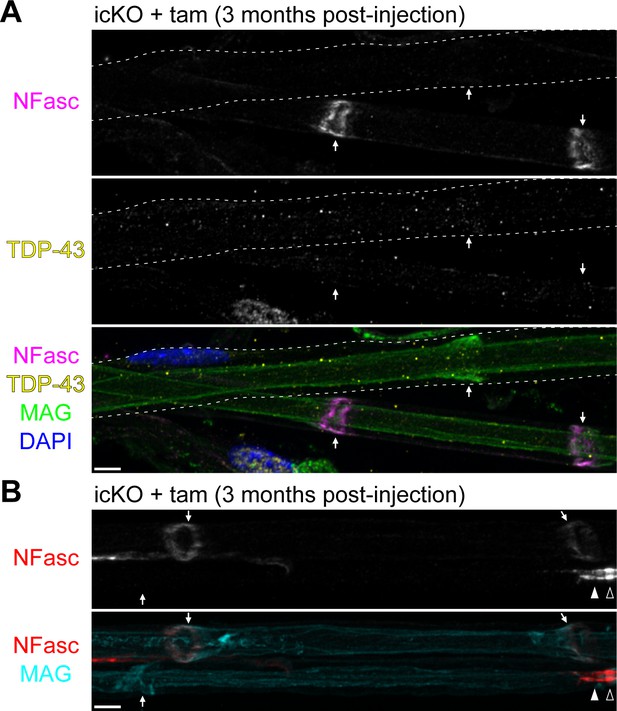
Neurofascin (NFasc) turns over faster at the incisures than at the paranodal junctions.
(A) Immunostaining of teased ventral roots from the TDP-43-icKO mice at 3 months after tamoxifen injection for NFasc (magenta), TDP-43 (yellow), myelin-associated glycoprotein (MAG) (green), and DAPI (blue). The cell contour of a TDP-43-KO Schwann cell is delineated with dashed lines, and NFasc is absent from its incisure. The incisures are indicated by arrows. (B) Immunostaining of teased ventral roots from the TDP-43-icKO mice at 3 months after tamoxifen injection for NFasc (red) and MAG (cyan). NFasc is absent from the incisure of the bottom Schwann cell but is still enriched at its paranode. The incisures are indicated by arrows, the paranode by a solid arrowhead, and the node by an open arrowhead. Scale bars, 5 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | Tardbpfl/fl | Jackson Laboratory | Stock #: 017591; MGI:4834273 | |
| Genetic reagent (M. musculus) | Dhh-Cre | PMID:12782656 | MGI:4359600 | Dr. Dies Meijer |
| Genetic reagent (M. musculus) | Mpz-Cre (P0-Cre) | Jackson Laboratory | Stock #: 017927; MGI:2450448 | |
| Genetic reagent (M. musculus) | Cnp-Cre | PMID:12590258 | MGI:3051635 | Dr. Klaus-Armin Nave |
| Genetic reagent (M. musculus) | Cntnap1-/- (Caspr KO) | PMID:14676309 | MGI:3026869 | |
| Genetic reagent (M. musculus) | Mpz-CreERT2 (P0-CreERT2) | PMID:12727441 | MGI:2663097 | Drs. Ueli Suter and Gabriel Corfas |
| Antibody | Anti-actin (mouse monoclonal C4) | MilliporeSigma | Cat #: MAB1501R; RRID:AB_2223041 | (1:5000) |
| Antibody | Anti-AnkB (mouse monoclonal N105/17) | UC Davis/NIH NeuroMab Facility | Clone: N105/17; RRID:AB_10674432 | (1:10) |
| Antibody | Anti-AnkG (mouse monoclonal N106/36) | UC Davis/NIH NeuroMab Facility | Clone: N106/36; RRID:AB_10697718 | (1:10) |
| Antibody | Anti-AnkG (mouse monoclonal N106/65) | UC Davis/NIH NeuroMab Facility | Clone: N106/65; RRID:AB_10673449 | (1:10) |
| Antibody | Anti-βII spectrin (mouse monoclonal 42) | BD Biosciences | Cat #: 612562; RRID:AB_399853 | (1:200) |
| Antibody | Anti-βIV spectrin (chicken polyclonal) | PMID:20980605 | RRID:AB_2827639 | (1:200) Dr. Matthew N. Rasband |
| Antibody | Anti-βIV spectrin (rabbit polyclonal) | PMID:15317849 | RRID:AB_2315634 | (1:500) Dr. Matthew N. Rasband |
| Antibody | Anti-Caspr (mouse monoclonal Mab275) | PMID:10624965 | RRID:AB_2314218 | (1:200) |
| Antibody | Anti-Caspr (rabbit polyclonal) | PMID:9118959 | RRID:AB_2314220 | (1:800 staining) |
| Antibody | Anti-Caspr (rabbit polyclonal) | Abcam | Cat#: ab34151; RRID:AB_869934 | (1:1000 blotting) |
| Antibody | Anti-Caspr2 (rabbit polyclonal) | PMID:10624965 | (1:500) | |
| Antibody | Anti-Cntn (goat polyclonal) | R&D Systems | Cat #: AF904-SP; RRID:AB_2292070 | (1:500) |
| Antibody | Anti-Cx32 (mouse monoclonal 5F9A9) | Thermo Fisher Scientific | Cat #: 35-8900; RRID:AB_2533228 | (1:200) |
| Antibody | Anti-E-cadherin (mouse monoclonal 36) | BD Biosciences | Cat #: 610181; RRID:AB_397580 | (1:200) |
| Antibody | Anti-Gldn (mouse monoclonal Mab94) | PMID:16039564 | (1:200) | |
| Antibody | Anti-Gldn (rabbit polyclonal) | PMID:17485493 | (1:500) | |
| Antibody | Anti-Krox20 (rabbit polyclonal) | PMID:15282162 | (1:800) Dr. Dies Meijer | |
| Antibody | Anti-Kv1.1 (mouse monoclonal K20/78) | UC Davis/NIH NeuroMab Facility | Clone: K20/78; RRID:AB_10672854 | (1:10) |
| Antibody | Anti-Kv1.2 (rabbit polyclonal) | PMID:7623158 | (1:500) Dr. Matthew N. Rasband | |
| Antibody | Anti-MAG (mouse monoclonal 513) | PMID:2444603 | (1:500) Dr. Marie T. Filbin | |
| Antibody | Anti-NFasc (chicken polyclonal) | R&D Systems | Cat #: AF3235; RRID:AB_10890736 | (1:200 staining) (1:500 blotting) |
| Antibody | Anti-panNav (mouse monoclonal K58/35) | MilliporeSigma | Cat #: S8809; RRID:AB_477552 | (1:200) |
| Antibody | Anti-PDGFRα (rabbit polyclonal) | PMID:8714519 | RRID:AB_2315173 | (1:2000) Dr. William B. Stallcup |
| Antibody | Anti-Sox10 (goat polyclonal) | R&D Systems | Cat #: AF2864; RRID:AB_442208 | (1:200) |
| Antibody | Anti-TDP-43 (mouse monoclonal 3H8) | EnCor Biotechnology | Cat #: MCA-3H8; RRID:AB_2572387 | (1:800) |
| Antibody | Anti-TDP-43 (rabbit polyclonal) | Proteintech | Cat #: 10782-2-AP; RRID:AB_615042 | (1:500) |
| Antibody | Anti-ZO-1 (mouse monoclonal 1A12) | Thermo Fisher Scientific | Cat #: 33-9100; RRID:AB_2533147 | (1:200) |
| Antibody | Anti-chicken AMCA (goat polyclonal) | Jackson ImmunoResearch | Cat#: 103-155-155; RRID:AB_2337385 | (1:200) |
| Antibody | Anti-chicken AlexaFluor 594 (goat polyclonal) | Thermo Fisher Scientific | Cat#: A-11042; RRID:AB_2534099 | (1:1000) |
| Antibody | Anti-chicken DyLight 680 (goat polyclonal) | Rockland | Cat #: 603-144-126; RRID:AB_1057473 | (1:10,000) |
| Antibody | Anti-goat AlexaFluor 594 (donkey polyclonal) | Thermo Fisher Scientific | Cat#: A-11058; RRID:AB_2534105 | (1:1000) |
| Antibody | Anti-mouse IRDye 800CW (goat polyclonal) | LI-COR Biotechnology | Cat #: 925-32210; RRID:AB_2687825 | (1:10,000) |
| Antibody | Anti-mouse IgG1 AlexaFluor 488 (goat polyclonal) | Thermo Fisher Scientific | Cat#: A-21121; RRID:AB_2535764 | (1:1000) |
| Antibody | Anti-mouse IgG1 AlexaFluor 594 (goat polyclonal) | Thermo Fisher Scientific | Cat#: A-21125; RRID:AB_2535767 | (1:1000) |
| Antibody | Anti-mouse IgG1 AlexaFluor 647 (goat polyclonal) | Thermo Fisher Scientific | Cat#: A-21240; RRID:AB_2535809 | (1:1000) |
| Antibody | Anti-mouse IgG2a AlexaFluor 488 (goat polyclonal) | Thermo Fisher Scientific | Cat#: A-21131; RRID:AB_2535771 | (1:1000) |
| Antibody | Anti-mouse IgG2a AlexaFluor 594 (goat polyclonal) | Jackson ImmunoResearch | Cat#: 115-585-206; RRID:AB_2338886 | (1:800) |
| Antibody | anti-mouse IgG2a AlexaFluor 647 (goat polyclonal) | Thermo Fisher Scientific | Cat#: A-21241; RRID:AB_2535810 | (1:1000) |
| Antibody | Anti-mouse IgG2b AlexaFluor 594 (goat polyclonal) | Thermo Fisher Scientific | Cat#: A-21145; RRID:AB_2535781 | (1:1000) |
| Antibody | Anti-mouse IgG2b AlexaFluor 647 (goat polyclonal) | Thermo Fisher Scientific | Cat#: A-21242; RRID:AB_2535811 | (1:1000) |
| Antibody | Anti-rabbit AlexaFluor 488 (donkey polyclonal) | Thermo Fisher Scientific | Cat#: A-21206; RRID:AB_2535792 | (1:1000) |
| Antibody | Anti-rabbit AlexaFluor 488 (goat polyclonal) | Thermo Fisher Scientific | Cat#: A-11034; RRID:AB_2576217 | (1:1000) |
| Antibody | Anti-rabbit AlexaFluor 594 (goat polyclonal) | Thermo Fisher Scientific | Cat#: A-11037; RRID:AB_253409 | (1:1000) |
| Antibody | Anti-rabbit IRDye 800CW (goat polyclonal) | LI-COR Biotechnology | Cat #: 925-32211; RRID:AB_2651127 | (1:10,000) |
Additional files
-
Supplementary file 1
The exon nomenclature of Nfasc used in this study.
The exon information was compiled from the current Ensembl database. The coordinates of each exon in chromosome 1 are shown (GRCm38.p6). Transcription starts from either exon 1a or 1b and terminates at either exon 23T or 33. The alternative exons specific for NF155 (exons 23 or 23L and 24) and the ones for NF186 (exons 28–30) are shaded in blue. Other alternative exons are shaded in gray. The cryptic exon CE1 identified in this study is shaded in pink. The start codon and potential stop codons are framed in red.
- https://cdn.elifesciences.org/articles/64456/elife-64456-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64456/elife-64456-transrepform-v1.pdf