Robo recruitment of the Wave regulatory complex plays an essential and conserved role in midline repulsion
Figures
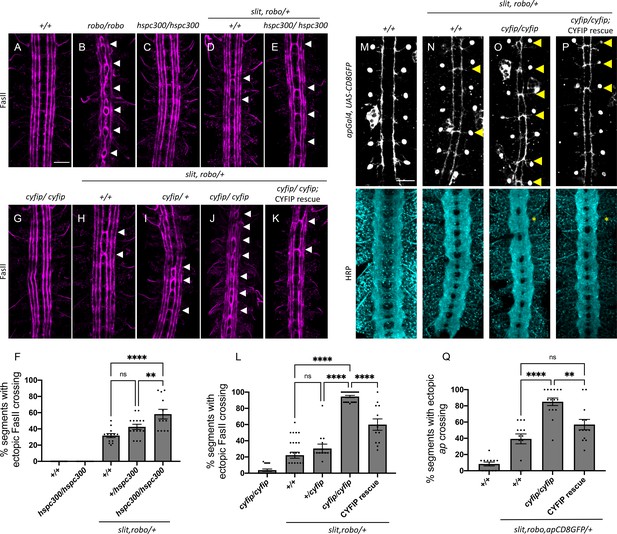
The wave regulatory complex genetically interacts with slit and robo.
(A–E, G–K) Stage 17 Drosophila embryos stained with anti-FasII to label ipsilateral axons. (A, C) Wild-type and hspc300 homozygous mutant embryos show three FasII-positive tracts that do not cross the midline. (B) Robo homozygous mutants show severe ectopic FasII crossing defects in 100% of segments (white arrowheads). (D) Slit, robo transheterozygous embryos show a mild loss-of-repulsion phenotype with ectopic FasII crossing in 31% of nerve cord segments. (E) Hscp300 homozygous mutants that are simultaneously heterozygous for slit and robo show ectopic FasII crossing in significantly more segments of the nerve cord (58%). (G) Cyfip embryos have almost no ectopic crossing defects and appear like wild-type embryos. (H) Double heterozygous slit, robo embryos show a mild loss-of-repulsion phenotype with ectopic FasII crossing in 22% of nerve cord segments. Removing (I) one and (J) two copies of cyfip in a slit, robo background results in a dose-dependent enhancement of the ectopic FasII crossing defects (30 and 95%, respectively). (K) Driving UAS-CYFIP expression in neurons using the pan-neuronal elav-Gal4 driver results in a partial rescue of the ectopic FasII crossing defects (60%). (F, L) Quantitation shows the percentage of segments in which FasII axons ectopically cross the midline. Data are presented as mean ± SEM, number of embryos, n = 15, 10, 15, 15, 12 (for F) and 17, 27, 13, 21, 12 (for L). Significance was assessed using one-way ANOVA with Tukey’s multiple comparisons test. (M–P) Stage 17 embryos carrying apGal4 and UAS-CD8GFP transgenes stained with anti-GFP, which labels the apterous (ap) cell bodies and axons, and anti-HRP, which labels all central nervous system (CNS) axons. (M) Wild-type embryos show ap axons that normally project ipsilaterally without crossing the midline. (N) Double heterozygous slit, robo embryos show a mild ectopic ap crossing phenotype of 39% (yellow arrowheads) while HRP (Horse radish peroxidase) depicts a wild type arrangement of longitudinal and commissural axon pathways. (O) Cyfip homozygous mutants in a slit, robo background show a strong enhancement of the ectopic ap crossing defects to 85% and HRP shows abnormal thickening and fusion of the commissures (asterisk). (P) Ap-specific expression of UAS-CYFIP significantly rescues the ectopic ap crossing defects (57%) but not the pan-neuronal HRP defects. (Q) Quantitation shows percentage of segments with ectopic apterous crossing defects. Data are presented as mean ± SEM, number of embryos, n = 12, 13, 15, 13. Significance was assessed using one-way ANOVA with Tukey’s multiple comparisons test. Scale bars in (A) and (M) represent 20 µm.
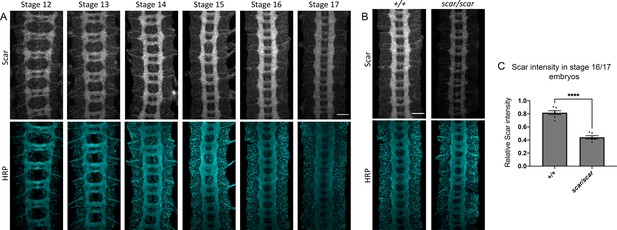
Scar expression in wild-type and scar mutant embryos.
(A) Wild-type embryos across developmental stages 12–17 stained with anti-Scar and anti-HRP show Scar enrichment in developing central nervous system (CNS) axons. (B) Stage 16 wild-type and scar mutant embryos stained with anti-Scar and anti-HRP. Scar protein levels are reduced in scar mutant embryos. (C) Quantification of relative fluorescence intensity of Scar in CNS axons of stage 16/17 embryos calculated as Scar fluorescence intensity normalized to HRP fluorescence intensity. Data are presented as mean ± SEM, number of embryos, n = 7, 6. Significance was assessed using Student’s t-test. Scale bars in (A) and (B) represent 20 µm.
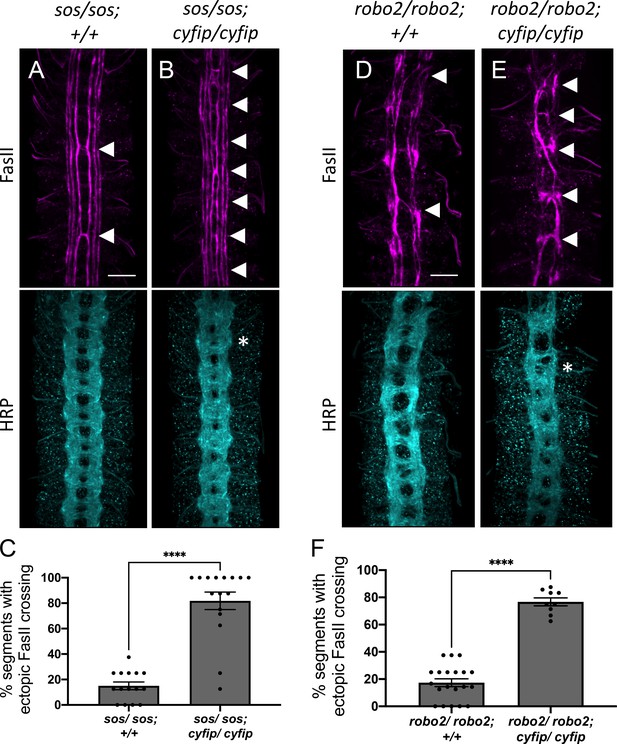
The wave regulatory complex genetically interacts with sos and robo2.
(A, B, D, E) Stage 17 embryos stained with anti-FasII and anti-HRP. (A) Sos embryos show mild ectopic crossing defects of 15% in FasII axons (arrowheads) and no phenotype in HRP. (B) Simultaneous removal of sos and cyfip results in a very strong enhancement of the ectopic FasII crossing defects to 82% and a strong HRP phenotype with thickening and fusion of commissures (asterisk). Similarly, (D) robo2 mutants show mild ectopic crossing defects of 17% in FasII axons and a mildly disorganized axon scaffold in HRP while (E) double mutants for robo2 and cyfip show strong ectopic FasII crossing defects of 77% and thickening and fusion of commissures in HRP. (C, F) Quantitation shows the percentage of segments in which FasII axons ectopically cross the midline. Data are presented as mean ± SEM, number of embryos, n = 15 and 16 (for E) and 20 and 9 (for F). Significance was assessed using Student’s t-test. Scale bars in (A) and (D) represent 20 µm.
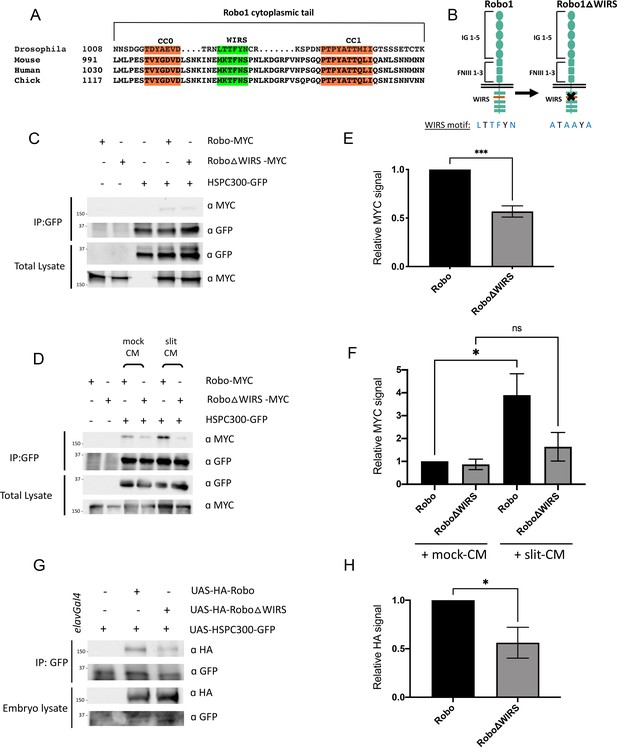
Slit-dependent recruitment of the WAVE regulatory complex (WRC) to Robo1 requires the WRC-interacting receptor sequence (WIRS) motif.
(A) Sequence alignments of the cytoplasmic tail of Robo1 showing that the WIRS motif is conserved across species. (B) Schematic depicting the residues of the WIRS motif that are mutated in the Robo1ΔWIRS variant. (C) Drosophila S2R+ cell lysates co-expressing HSPC300-GFP with either wild-type Robo1-MYC or Robo1ΔWIRS-MYC were immunoprecipitated with an anti-GFP antibody. The first three lanes show the individual proteins expressed alone. The fourth lane shows wild-type Robo1 co-immunoprecipitating with HSPC300 while the fifth lane shows that mutating the WIRS motif decreases this binding. (D) Cell lysates were immunoprecipitated with anti-GFP following a 12 min bath application of mock conditioned media or conditioned media obtained from Slit-expressing cells. The interaction between wild-type Robo1 and HSPC300 is increased in the presence of Slit; however, no significant increase is noted with Robo1ΔWIRS. (E, F) Quantitation of band intensities of the MYC-tagged Robo1 variants in the immunoprecipitates normalized to wild-type Robo1-MYC. Data were normalized to lysate levels of the Robo1 variants and HSPC300 levels in the immunoprecipitates. Error bars represent SEM. Number of trials, n = 4. Significance was assessed using Student’s t-test (for E) and one-way ANOVA with Tukey’s multiple comparisons test (for F). (G) Lysates from Drosophila embryos with elavGal4 pan-neuronally driving the expression of HSPC300-GFP alone (lane 1), with wild-type HA-Robo1 (lane 2) or with HA-Robo1ΔWIRS (lane 3), were immunoprecipitated with anti-GFP. Wild-type Robo1 co-immunoprecipitates with HSPC300 and mutating the WIRS motif decreases this binding. (H) Quantitation of band intensities of the HA-tagged Robo1 variants in the imunnoprecipitates normalized to wild-type HA-Robo1. Data were normalized to the lysate levels of the Robo1 variants and HSPC300 levels in the immunoprecipitates. Error bars represent SEM. Number of trials, n = 5. Significance was assessed using Student’s t-test. Normalized values for the co-immunoprecipitation data are provided in Figure 3—source data 1.
-
Figure 3—source data 1
Normalized values of co-immunoprecipitation data.
Related to Figure 3E, F, H. Also, Figure 3—figure supplement 1C.
- https://cdn.elifesciences.org/articles/64474/elife-64474-fig3-data1-v2.xlsx
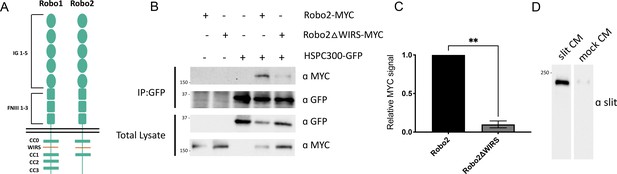
Robo2 interaction with the WAVE regulatory complex (WRC) is entirely dependent on its WRC-interacting receptor sequence (WIRS) motif.
(A) Schematic of Drosophila Robo1 and Robo2 receptors. Both Robo1 and Robo2 have a WIRS motif between CC0 and CC1 but Robo2 lacks the CC2 and CC3 motifs that are present in Robo1. (B) Drosophila S2R+ cell lysates co-expressing HSPC300-GFP with either wild-type Robo2-MYC or Robo2ΔWIRS -MYC were immunoprecipitated with an anti-GFP antibody. The first three lanes show the individual proteins expressed alone. The fourth lane shows wild-type Robo2 co-immunoprecipitating with HSPC300 while the fifth lane shows that mutating the WIRS motif completely abolishes this binding. (C) Quantitative representations of band intensities of the MYC-tagged Robo2 variants in the immunoprecipitates normalized to wild-type Robo2-MYC. Data were normalized to lysate levels of the Robo2 variants and HSPC300 levels in the immunoprecipitates. Error bars represent SEM. Number of trials, n = 2. Significance was assessed using Student’s t-test. (D) Western blotting of proteins harvested from the media of Drosophila S2R+ cells transfected with or without a Slit construct with an anti-Slit antibody reveals low levels of Slit in mock conditioned media.
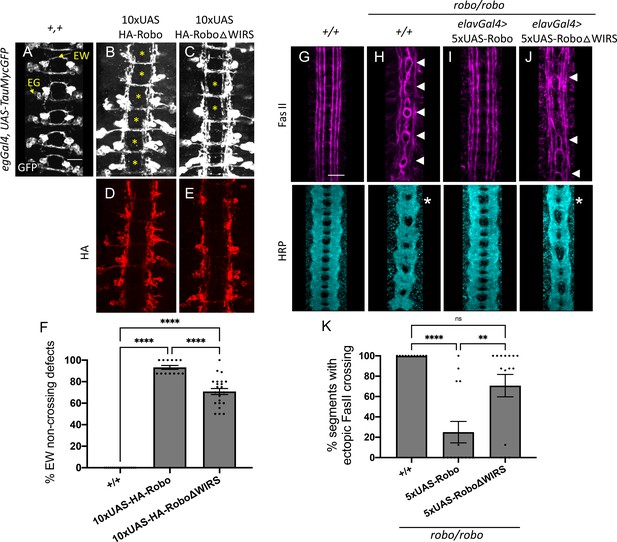
The WRC-interacting receptor sequence (WIRS) motif is essential for Robo1 function in vivo.
(A–C) Stage 16 Drosophila embryos carrying egGal4 and UAS-TauMycGFP transgenes stained with anti-GFP, which labels cell bodies and axons of the eagle neurons (EG and EW). EG neurons project through the anterior commissure of each segment while EW neurons project through the posterior commissure. (A) EW neurons cross in 100% of segments in wild-type embryos. (B) Misexpression of wild-type HA-tagged Robo1 in eagle neurons results in a strong disruption of midline crossing where EW axons fail to cross in almost all segments of the nerve cord (93%; asterisk). (C) Misexpressing HA-tagged Robo1ΔWIRS results in a significantly milder disruption with fewer segments showing EW non-crossing defects (71%). (D, E) Embryos stained with anti-HA show comparable expression of the HA-tagged Robo1 variants that were inserted into the same genomic locus. (F) Quantitation shows the percentage of segments in which EW axons fail to cross the midline. Data are presented as mean ± SEM, number of embryos, n = 17, 13, 23. Significance was assessed using one-way ANOVA with Tukey’s multiple comparisons test. (G–J) Stage 17 embryos stained with anti-FasII and anti-HRP. (G) Wild-type embryos show no ectopic FasII crossing defects and no phenotype in HRP. (H) Robo mutants show severe ectopic FasII crossing defects in 100% of segments (arrowheads) and a strong HRP phenotype with thickening and fusion of commissures (asterisk). (I) Pan-neuronal expression of wild-type 5XUAS-Robo1 significantly rescues the robo mutant phenotype in FasII (to 25%) as well as HRP; however, (J) 5XUAS-Robo1ΔWIRS fails to rescue the robo mutant phenotype as efficiently as wild-type Robo1 with frequent ectopic crossing in FasII (71%) and thickened commissures in HRP still evident in these embryos. (K) Quantitation shows the percentage of segments in which FasII axons ectopically cross the midline. Data are presented as mean ± SEM, number of embryos, n = 11, 14, 15. Significance was assessed using one-way ANOVA with Tukey’s multiple comparisons test. Scale bars in (A) and (G) represent 20 µm.
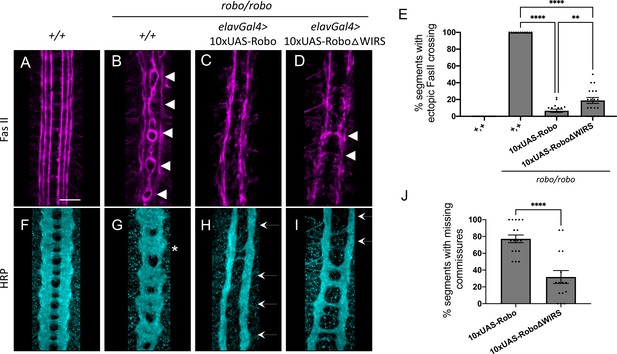
10XUAS-Robo1 rescue of the robo mutant phenotype.
(A–D) Stage 17 embryos stained with anti-FasII. (A) Wild-type embryos show no phenotype in FasII. (B) Robo mutants show severe ectopic FasII crossing defects in 100% of segments (arrowheads). (C) Pan-neuronal expression of wild-type 10xUAS-Robo1 completely rescues the robo mutant phenotype in FasII. (D) Pan-neuronal expression of 10xUAS-Robo1ΔWIRS results in a mildly weaker rescue with a small number of FasII bundles still crossing the midline. (E) Quantitation shows the percentage of segments in which FasII axons ectopically cross the midline. Data are presented as mean ± SEM, number of embryos, n = 14, 11, 16, 18. Significance was assessed using one-way ANOVA with Tukey’s multiple comparisons test. (F–I) Stage 17 embryos stained with anti-HRP. (F) Wild-type embryos show no phenotype in HRP. (G) Robo mutants show a strong HRP phenotype with thickening and fusion of commissures (asterisk). (H) Pan-neuronal expression of wild-type 10xUAS-Robo1 gives the opposite phenotype with strong ectopic repulsion of commissural axons resulting in segments with a complete absence of commissures (arrows). (I) In contrast, pan-neuronal expression of 10xUAS-Robo1ΔWIRS shows a significantly reduced ability to induce ectopic repulsion in commissural axons, resulting in much fewer segments with missing commissures. (J) Quantitation shows the percentage of segments with missing commissures. Data are presented as mean ± SEM, number of embryos, n = 16, 18. Significance was assessed using Student’s t-test. Scale bar in (A) represents 20 µm.
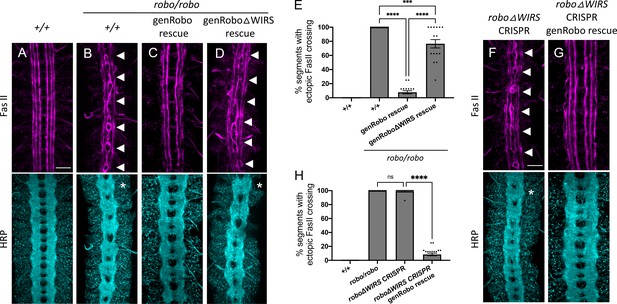
Mutating the endogenous WRC-interacting receptor sequence (WIRS) motif disrupts Robo1 function in vivo.
(A–D) Stage 17 embryos stained with anti-FasII and anti-HRP. (A) Wild-type embryos showing no phenotype in FasII or HRP. (B) Robo mutants show severe ectopic FasII crossing defects in 100% of segments (arrowheads) and a strong HRP phenotype with thickening and fusion of commissures (asterisk). (C) The strong FasII and HRP phenotypes seen in robo mutant embryos can be completely rescued with one copy of a wild-type genomic Robo1 rescue construct (genRobo) that contains additional upstream and downstream regulatory regions of robo1, more closely mimicking the endogenous Robo1 expression pattern (8%). (D) In contrast, the genomic Robo1 rescue construct containing mutations in the WIRS motif of Robo1 (genRoboΔWIRS) fails to rescue the robo mutant phenotype in both FasII (77%) and HRP. (E) Quantitation shows the percentage of segments in which FasII axons ectopically cross the midline. Data are presented as mean ± SEM, number of embryos, n = 14, 11, 16, 16. Significance was assessed using one-way ANOVA with Tukey’s multiple comparisons test. (F, G) Stage 17 embryos stained with anti-FasII and anti-HRP. (F) CRISPR embryos with mutations in the endogenous WIRS motif of robo1 show severe phenotypes in FasII and HRP bearing strong resemblance to robo mutants. (G) The phenotypes seen in these CRISPR roboΔWIRS embryos can be completely rescued with one copy of the wild-type genomic Robo1 rescue construct (8%). (H) Quantitation shows the percentage of segments in which FasII axons ectopically cross the midline. Data are presented as mean ± SEM, number of embryos, n = 14, 11, 14, 20. Significance was assessed using one-way ANOVA with Tukey’s multiple comparisons test. Scale bars in (A) and (F) represent 20 µm.
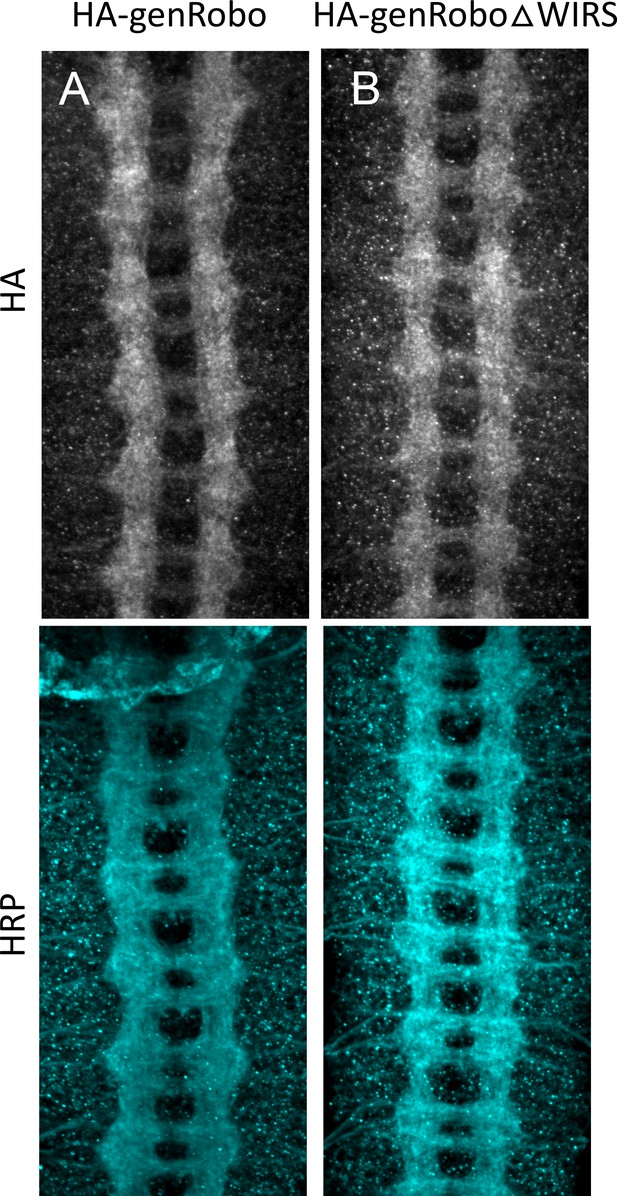
Comparable expression of the genomic rescue transgenes.
(A) Stage 17 embryo expressing the genomic HA-tagged Roundabout (Robo) rescue transgene stained with anti-HA and anti-HRP. The HA expression pattern closely resembles that of endogenous Robo. (B) Stage 17 embryo expressing the genomic HA-tagged RoboΔWIRS rescue transgene shows comparable HA staining to wild-type genomic HA-tagged Robo.
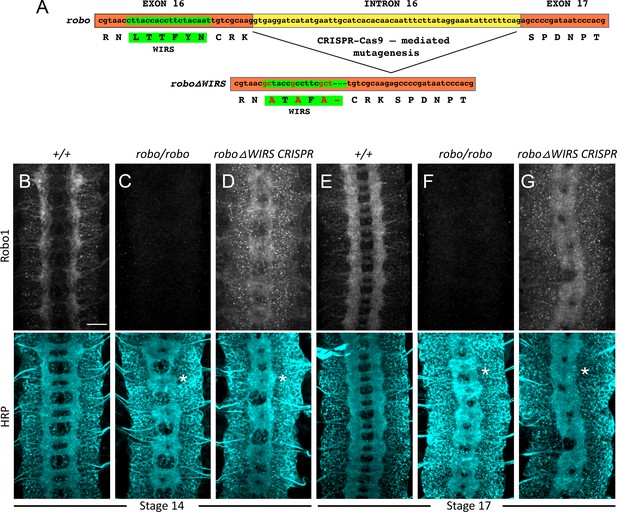
Schematic for CRISPR-Cas9 mutagenesis and Robo1 staining in CRIPR roboΔWIRS embryos.
(A) Schematic for CRSIPR-Cas9 mutagenesis of the WRC-interacting receptor sequence (WIRS) motif in the endogenous robo1 locus. Four amino acids in the WIRS motif were mutated, and deletion of the small intron 16 occurred during mutagenesis. (B–D) Stage 14 embryos stained with anti-Robo1 and anti-HRP. (E–G) Stage 17 embryos stained with anti-Robo1 and anti-HRP. (B, E) Wild-type embryos show Robo1 staining enriched in the longitudinal tracts and no HRP phenotype. (C, F) robo mutant embryos show a complete loss of Robo1 staining along with a strong loss-of-repulsion phenotype in HRP with thickening and fusion of commissures (asterisk). (D, G) In contrast, CRISPR roboΔWIRS embryos show a strong loss-of-repulsion phenotype in HRP but strong Robo1 staining on longitudinal tracts as well as on commissures, demonstrating that the phenotype is not due to loss of protein production. Scale bar in (A) represents 20 µm.
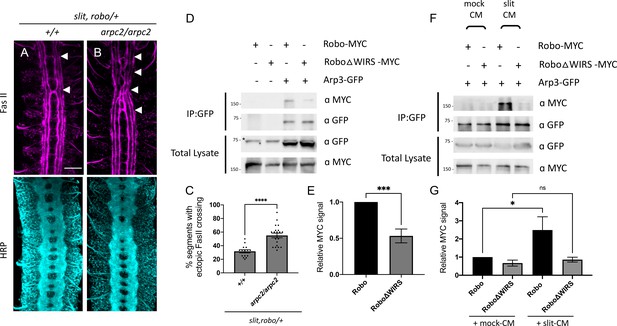
The Arp2/3 complex interacts genetically and physically with the Slit-Robo pathway.
(A, B) Stage 17 Drosophila embryos stained with anti-FasII and anti-HRP. (A) Slit, robo transheterozygous embryos show a mild loss-of-repulsion phenotype with ectopic FasII crossing in 31% of nerve cord segments (arrowheads). (B) Arpc2 homozygous mutants that are simultaneously heterozygous for slit and robo show ectopic FasII crossing in significantly more segments of the nerve cord (55%). (C) Quantitation shows the percentage of segments in which FasII axons ectopically cross the midline. Data are presented as mean ± SEM, number of embryos, n = 15 and 20. Significance was assessed using Student’s t-test. Scale bar in (A) represents 20 µm. (D) Drosophila S2R+ cell lysates co-expressing Arp3-GFP with either wild-type Robo1-MYC or Robo1ΔWIRS-MYC were immunoprecipitated with an anti-GFP antibody. The first two lanes show the individual Robo1 variants expressed alone. The third lane shows wild-type Robo1 co-immunoprecipitating with Arp3 while the fourth lane shows that mutating the WIRS motif decreases this binding. Asterisk indicates non-specific bands. (F) Cell lysates were immunoprecipitated with anti-GFP following a 12 min bath application of mock conditioned media or conditioned media obtained from Slit-expressing cells. The interaction between wild-type Robo1 and Arp3 is increased in the presence of Slit; however, no significant increase is noted with Robo1ΔWIRS. (E, G) Quantitation of band intensities of the MYC-tagged Robo1 variants in the immunoprecipitates normalized to wild-type Robo1-MYC. Data were normalized to lysate levels of the Robo1 variants and Arp3 levels in the immunoprecipitates. Error bars represent SEM. Number of trials, n = 7. Significance was assessed using Student’s t-test (for E) and one-way ANOVA with Tukey’s multiple comparisons test (for G). Normalized values for the co-immunoprecipitation data are provided in Figure 6—source data 1.
-
Figure 6—source data 1
Normalized values of co-immunoprecipitation data.
Related to Figure 6E, G.
- https://cdn.elifesciences.org/articles/64474/elife-64474-fig6-data1-v2.xlsx
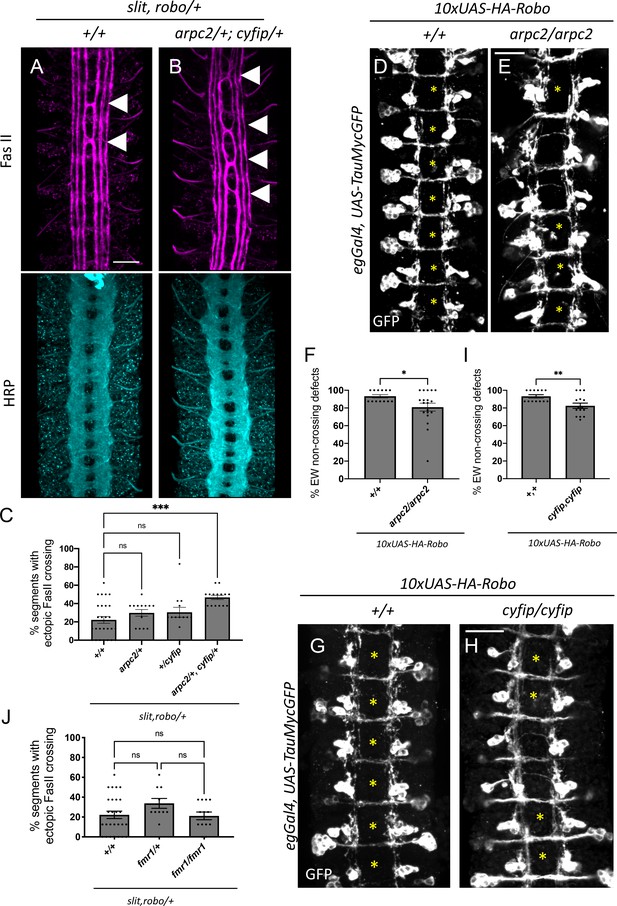
arpc2 mutants genetically interact with the Slit-Robo pathway.
(A, B) Stage 17 embryos stained with anti-FasII and anti-HRP. (A) Double heterozygous slit, robo embryos show a mild loss-of-repulsion phenotype with ectopic FasII crossing in 22% of nerve cord segments (arrowheads). (B) Removing one copy of arpc2 and one copy of cyfip in embryos that are simultaneously heterozygous for slit and robo show ectopic FasII crossing in significantly more segments of the nerve cord (47%). (C) Quantitation shows the percentage of segments in which FasII axons ectopically cross the midline. Data are presented as mean ± SEM, number of embryos, n = 27, 13, 13, 15. Significance was assessed using one-way ANOVA with Tukey’s multiple comparisons test. (D, E, G, H) Stage 16 Drosophila embryos carrying egGal4 and UAS-TauMycGFP transgenes stained with anti-GFP, which labels cell bodies and axons of the eagle neurons. (D, G) Misexpression of wild-type HA-tagged Robo1 in eagle neurons results in a strong disruption of midline crossing where EW axons fail to cross in almost all segments of the nerve cord (93%; asterisk). (E) In contrast, overexpressing Robo1 in arpc2 mutants results in a small but significant suppression of this phenotype with fewer segments showing EW non-crossing defects (81%). (H) Similarly, overexpressing Robo1 in cyfip mutants results in a similar suppression of this phenotype (83%). (F, I) Quantitation shows the percentage of segments in which EW axons fail to cross the midline. Data are presented as mean ± SEM, number of embryos, n = 13, 19 (for F) and 13, 15 (for I). Significance was assessed using Student’s t-test. (J) Quantitation shows the percentage of segments in which FasII axons ectopically cross the midline for fmr1mutants in a slit, robo sensitized background. Removing one or both copies of fmr1 has no effect on the slit, robo ectopic FasII crossing phenotype. Data are presented as mean ± SEM, number of embryos, n = 27, 10, 13. Significance was assessed using one-way ANOVA with Tukey’s multiple comparisons test. Scale bars in (A), (E), and (H) represent 20 µm.
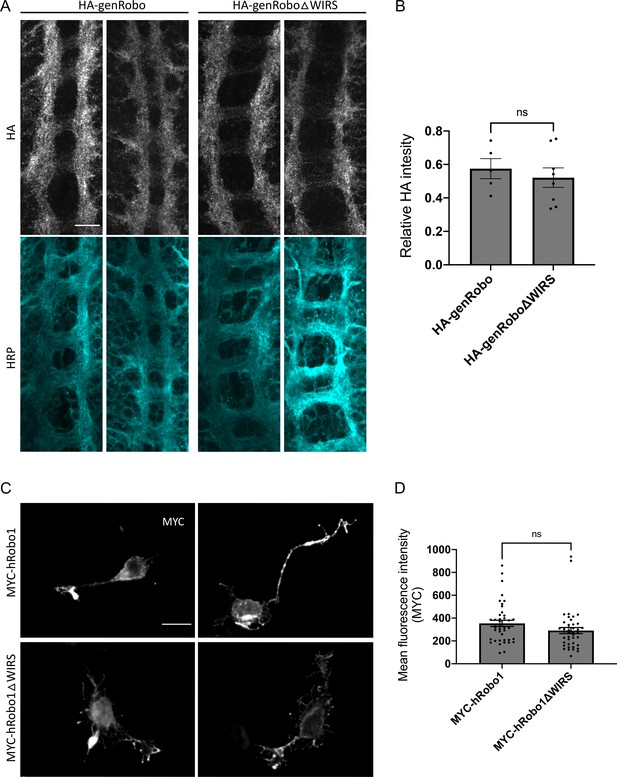
Comparable surface expression of the wild-type and WRC-interacting receptor sequence (WIRS) mutant forms of Robo1.
(A) Drosophila embryos expressing genomic HA-tagged Roundabout (Robo) rescue transgenes stained for surface HA and HRP. Embryos were dissected live, and surface expression of Robo was visualized by staining the N-terminal HA tag before fixation and permeabilization. (B) Quantitation of surface Robo represented as mean fluorescence intensity of HA normalized to HRP shows no difference in surface expression of wild-type Robo and RoboΔWIRS. Data are presented as mean ± SEM, number of embryos, n = 5 and 8. Significance was assessed using Student’s t-test. Scale bars represent 5 mm in (A) and 20 mm in (C). (C) E12 dorsal commissural neurons electroporated either with MYC-tagged hRobo1 or with MYC-tagged hRobo1ΔWIRS. Following a 30 min treatment with Slit, surface expression of hRobo1 was visualized by staining the N-terminal MYC tag before fixation and permeabilization. (D) Quantitation of surface hRobo1 represented as mean fluorescence intensity of MYC shows no difference in surface expression of wild-type hRobo1 and hRobo1ΔWIRS. Only Robo3-positive commissural neurons were quantified for MYC intensity. Data are presented as mean ± SEM, number of neurons, n = 42 (from two independent trials). Significance was assessed using Student’s t-test.
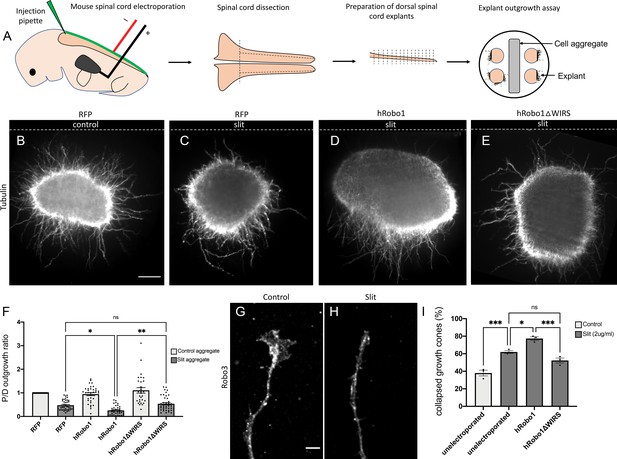
The WRC-interacting receptor sequence (WIRS) motif is required for Slit-dependent Robo1 repulsion in mouse spinal commissural axons.
(A) Schematic of electroporation and culture of spinal cord explants. Dotted lines show cut sites to obtain dorsal spinal cord explants. The image on the right depicts the arrangement of explants cultured around a 293 T cell aggregate (control or Slit-expressing) embedded in collagen. (B–E) E12 dorsal spinal cord explants labeled with anti-tubulin to visualize axon outgrowth. Dotted lines indicate the position of the cell aggregate. (B) RFP electroporated explant cultured next to a mock cell aggregate shows uniform outgrowth on all sides of the explant. (C) RFP electroporated explant cultured next to a Slit-expressing cell aggregate shows decreased outgrowth on the quadrant proximal to the aggregate as compared to the quadrant distal to it (0.47). (D) Explant electroporated with wild-type hRobo1 cultured next to a Slit-expressing cell aggregate shows even less outgrowth on the proximal quadrant demonstrating increased responsiveness to Slit (0.14). (E) Explant electroporated with hRobo1ΔWIRS cultured next to a Slit-expressing cell aggregate shows no such increase in Slit responsiveness as the proximal: distal outgrowth ratio is similar to that seen for RFP electroporated explants (0.54). (F) Quantification shows the proximal:distal outgrowth ratio for explants cultured next to control cell aggregates (white) and Slit-expressing cell aggregates (gray). Data are presented as mean ± SEM, number of explants, n = 29, 39, 33, 39, 29, 41 (from three independent experiments). Significance was assessed using one-way ANOVA with Tukey’s multiple comparisons test. (G, H) Growth cone collapse in response to Slit in E12-dissociated commissural axons. Growth cone morphology was examined by staining for the commissural marker Robo3. (I) Quantification shows percentage of axons with collapsed growth cones. Unelectroporated neurons show an increased level of collapse when treated with Slit (from 38% without Slit to 62% with bath application of Slit). Neurons electroporated with wild-type hRobo1 show a gain-of-function response to Slit with an even higher collapse level (77%). In contrast, neurons electroporated with hRobo1ΔWIRS show no gain-of-function and a collapse level similar to unelectroporated neurons (52%). For neurons electroporated with the MYC-tagged hRobo1 variants, only Robo3- and MYC-positive axons were analyzed. Data are presented as mean ± SEM, number of trials, n = 3 (over 30 neurons for each condition/trial). Significance was assessed using one-way ANOVA with Tukey’s multiple comparisons test. Scale bars represent 100 µm in (B) and 5 µm in (G).
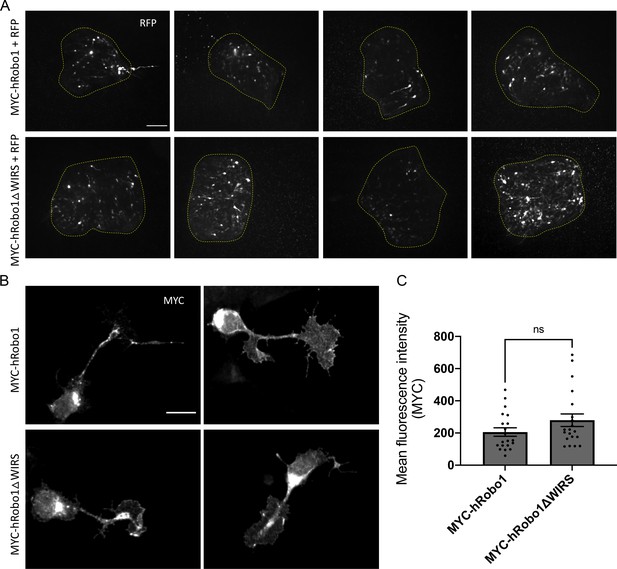
Expression of hRobo1 variants electroporated into dorsal spinal commissural neurons.
(A) E12 dorsal spinal cord explants electroporated with RFP and either MYC-tagged hRobo1 or with MYC-tagged hRobo1ΔWIRS show comparable levels of RFP staining across explants. (B) E12 dorsal commissural neurons electroporated either with MYC-tagged hRobo1 or with MYC-tagged hRobo1ΔWIRS stained with anti-MYC. (C) Quantitation of total levels of hRobo1 represented as mean fluorescence intensity of MYC shows comparable expression levels of wild-type hRobo1 and hRobo1ΔWIRS in dissociated neurons. Only Robo3-positive commissural neurons were quantified for MYC intensity. Data are presented as mean ± SEM, number of neurons, n = 20 (from two independent trials). Significance was assessed using Student’s t-test. Scale bars represent 100 µm in (A) and 5 µm in (B).
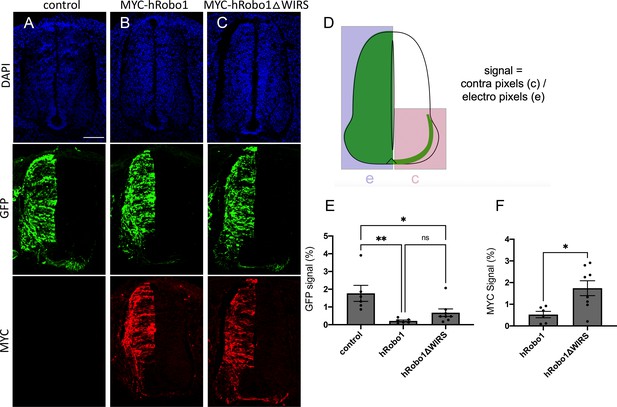
The WRC-interacting receptor sequence (WIRS) motif is required for vertebrate Robo1-mediated ectopic repulsion in vivo.
(A–C) Transverse sections of Hamburger–Hamilton (HH) stage 22–23 chicken spinal cords electroporated with GFP alone or together with MYC-tagged hRobo1 or hRobo1ΔWIRS and stained with DAPI, anti-GFP, and anti-MYC. (A) Electroporation of GFP alone shows numerous GFP-positive axons crossing the midline with a GFP crossing index of 1.8%. (B) Electroporation of GFP together with MYC-hRobo1 shows far fewer GFP-positive axons crossing the midline and very few MYC-positive axons on the contralateral side, with GFP and MYC crossing indices of 0.21 and 0.53%, respectively. (C) In contrast, electroporation of GFP along with MYC-hRobo1ΔWIRS shows substantially more GFP- and MYC-positive axons crossing the midline with higher GFP and MYC crossing indices of 0.68 and 1.7%, respectively. (D) Crossing index (signal) is the GFP or MYC fluorescence signal in the contralateral side of the spinal cord expressed as a fraction of total GFP or MYC fluorescence signal in the electroporated side. (E, F) Quantitation of crossing index for GFP and MYC signal. Data are presented as mean ± SEM, number of embryos, n = 6, 6, 8 (for E) and 6, 8 (for F). Significance was assessed using one-way ANOVA with Tukey’s multiple comparisons test (for E) and Student’s t-test (for F). Scale bar represents 100 µm in (A).
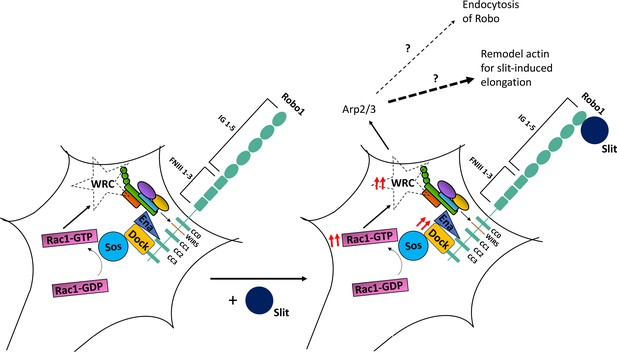
A model of WAVE regulatory complex (WRC) function in Robo1 signaling.
In our proposed model, the WRC binds to Robo1 partly via its WRC-interacting receptor sequence (WIRS) motif. Rac1 is activated downstream of Robo1 (Fan et al., 2003; Wong et al., 2001), which likely activates the complex. Slit binding induces increased WIRS-dependent recruitment of the WRC to Robo1, which is vital to Robo1-mediated repulsive signaling. WRC functions downstream of Robo1 by activating Arp2/3 to remodel the actin cytoskeleton. We hypothesize that these WIRS-WRC-mediated actin rearrangements are more likely to facilitate an initial extension of Slit-induced filopodia than endocytosis or recycling of the Robo1 receptor.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | 293T | ATCC | ATCC CRL-3216 | RRID:CVCL_0063 Authenticated via STR profiling using ATCC services |
| Cell line (Drosophila melanogaster) | S2R+ | Drosophila Genomics Resource Center | Cat#150 | RRID:CVCL_Z831 Authenticated by morphology and doubling time |
| Genetic reagent (Mus musculus) | CD-1 line | Charles River | Stock#022 | RRID:IMSR_CRL:022 |
| Genetic reagent (D. melanogaster) | D. melanogaster: w1118 | Chance and Bashaw, 2015 | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster: roboGA285 | Chance and Bashaw, 2015 | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster: slit2 | Chance and Bashaw, 2015 | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster: sos4G | Yang and Bashaw, 2006 | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster: robo2x123 | Evans and Bashaw, 2010 | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster: scarΔ37 | Bloomington Drosophila Stock Center | BDSC: 8754 | RRID:BDSC_8754 |
| Genetic reagent (D. melanogaster) | D. melanogaster: arpc2KG04658 | Bloomington Drosophila Stock Center | BDSC: 13978 | RRID:BDSC_13978 |
| Genetic reagent (D. melanogaster) | D. melanogaster:hspc300Δ54.3 | Kind gift from A. Giangrande | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster:cyfipΔ85.1 | Kind gift from A. Giangrande | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster:fmr13 | Kind gift from T. Jongens | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster:apGal4 | Evans and Bashaw, 2010 | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster:egGal4 | Evans and Bashaw, 2010 | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster: UAS-CD8GFP | Evans and Bashaw, 2010 | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster: UAS-TauMycGFP | Evans and Bashaw, 2010 | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster: 10XUAS-HA-Robo1 86 F8 | Evans and Bashaw, 2010 | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster: 10XUAS-HA-Robo1ΔWIRS 86 F8 | This paper | N/A | Available from Bashaw lab; methods: genetic stocks |
| Genetic reagent (D. melanogaster) | D. melanogaster: 5XUAS-HA-Robo1 86 F8 | Chance and Bashaw, 2015 | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster: 5XUAS-HA-Robo1ΔWIRS 86 F8 | This paper | N/A | Available from Bashaw Lab; methods: genetic stocks |
| Genetic reagent (D. melanogaster) | D. melanogaster: UAS-CYFIP | Kind gift from A. Giangrande | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster: robo1::HArobo1 28E7 | Kind gift from T. Evans | N/A | |
| Genetic reagent (D. melanogaster) | D. melanogaster: robo1::HArobo1ΔWIRS 28E7 | This paper | N/A | Available from Bashaw lab; methods: genetic stocks |
| Genetic reagent (D. melanogaster) | D. melanogaster: robo1ΔWIRS CRISPR | This paper | N/A | Available from Bashaw Lab; methods: genetic stocks |
| Recombinant DNA reagent | Plasmid: pCAG-MYC-hRobo1 | This paper | N/A | Available from Bashaw lab; methods: molecular biology |
| Recombinant DNA reagent | Plasmid: pCAG-MYC-hRobo1ΔWIRS | This paper | N/A | Available from Bashaw lab; methods: molecular biology |
| Recombinant DNA reagent | Plasmid: pCAG-RFP | Kind gift from A. Jaworski | N/A | |
| Recombinant DNA reagent | Plasmid: pSecTagB-hSlit2-MYC | Kind gift from A. Chedotal | N/A | |
| Recombinant DNA reagent | Plasmid: p10UASTattB-HA-Robo1 | Evans and Bashaw, 2010 | N/A | |
| Recombinant DNA reagent | Plasmid: p10UASTattB-HA-Robo1ΔWIRS | This paper | N/A | Available from Bashaw Lab; methods: molecular biology |
| Recombinant DNA reagent | Plasmid: p10UAST-HSPC300-GFP | This paper | N/A | Available from Bashaw Lab; methods: molecular biology |
| Recombinant DNA reagent | Plasmid: p5UASTattB-HA-Robo1 | Chance and Bashaw, 2015 | N/A | |
| Recombinant DNA reagent | Plasmid: p10UASTattB-HA-Robo1ΔWIRS | This paper | N/A | Available from Bashaw Lab; methods: molecular biology |
| Recombinant DNA reagent | Plasmid: pUAST-Slit | Chance and Bashaw, 2015 | N/A | |
| Recombinant DNA reagent | Plasmid: pMT-Gal4 | Chance and Bashaw, 2015 | N/A | |
| Recombinant DNA reagent | Plasmid: robo1 genomic rescue construct | Kind gift from T. Evans | N/A | |
| Recombinant DNA reagent | Plasmid: robo1ΔWIRS genomic rescue construct | This paper | N/A | Available from Bashaw Lab; methods: molecular biology |
| Recombinant DNA reagent | Plasmid: pCFD3-dU6:3gRNA | Addgene | Plasmid#49410 | RRID:Addgene_49410 |
| Recombinant DNA reagent | Plasmid: p10UASTattB-Robo1-MYC | This paper | N/A | Available from Bashaw Lab; methods: molecular biology |
| Recombinant DNA reagent | Plasmid: p10UASTattB-Robo1ΔWIRS-MYC | This paper | N/A | Available from Bashaw Lab; methods: molecular biology |
| Recombinant DNA reagent | Plasmid: p10UASTattB-Robo2-MYC | This paper | N/A | Available from Bashaw Lab; methods: molecular biology |
| Recombinant DNA reagent | Plasmid: p10UASTattB-Robo2ΔWIRS-MYC | This paper | N/A | Available from Bashaw Lab; methods: molecular biology |
| Antibody | Mouse monoclonal anti-MYC | DSHB | Cat#9E10-C | IF (1:500), WB (1:1000), RRID:AB_2266850 |
| Antibody | Mouse monoclonal anti-HA | BioLegend | Cat#901502 | IF (1:500), WB (1:1000), RRID:AB_2565007 |
| Antibody | Mouse monoclonal anti-beta tubulin | DSHB | Cat#E7-S | IF (1:300), WB (1:1000), RRID:AB_528499 |
| Antibody | Chick polyclonal anti-beta gal | Abcam | Cat#ab9361 | IF (1:500), RRID:AB_307210 |
| Antibody | Mouse monoclonal anti-Fasciclin II | DSHB | 1D4 | IF (1:50), RRID:AB_528235 |
| Antibody | Rabbit polyclonal anti-GFP | Invitrogen | Cat#a11122 | IF (1:250), WB (1:500), IP (1:500), RRID:AB_221569 |
| Antibody | Rabbit polyclonal anti-dsRed | Takara | Cat#632496 | IF (1:200), RRID:AB_10013483 |
| Antibody | Mouse monoclonal anti-Scar (supernatant) | DSHB | Cat#P1C1 | IF (1:50), RRID:AB_2618386 |
| Antibody | Mouse monoclonal anti-Robo (supernatant) | DSHB | Cat#13C9 | IF (1:50), RRID:AB_2181861 |
| Antibody | Mouse monoclonal anti-Slit (supernatant) | DSHB | Cat#C555.6D | WB (1:100), RRID:AB_528470 |
| Antibody | Goat polyclonal anti-Robo3 | R&D Systems | Cat#AF3076 | IF (1:200), RRID:AB_2181865 |
| Antibody | Alexa647 Goat polyclonal anti-HRP | Jackson Immunoresearch | Cat#123-605-021 | IF (1:500), RRID:AB_2338967 |
| Antibody | Goat polyclonal anti-Mouse HRP | Jackson Immunoresearch | Cat#115-035-146 | WB (1:10,000), RRID:AB_2307392 |
| Antibody | Goat polyclonal anti-Rabbit HRP | Jackson Immunoresearch | Cat#111-035-003 | WB (1:10,000), RRID:AB_2313567 |
| Antibody | 488 Donkey polyclonal anti-Mouse | Jackson Immunoresearch | Cat#715-545-150 | IF (1:500), RRID:AB_2340846 |
| Antibody | 488 Donkey polyclonal anti-Goat | Jackson Immunoresearch | Cat#705-165-147 | IF (1:500), RRID:AB_2307351 |
| Antibody | Alexa488 Goat polyclonal anti-Rabbit | Invitrogen | Cat#A11034 | IF (1:500), RRID:AB_2576217 |
| Antibody | Alexa488 Goat polyclonal anti-Mouse | Invitrogen | Cat#A11029 | IF (1:500), RRID:AB_138404 |
| Antibody | Alexa488 Goat polyclonal anti-Chick | Invitrogen | Cat#A11039 | IF (1:500), RRID:AB_142924 |
| Antibody | Cy3 Goat polyclonal anti-Mouse | Jackson Immunoresearch | Cat#115-165-003 | IF (1:500), RRID:AB_2338680 |
| Antibody | Cy3 Goat polyclonal anti-Rabbit | Jackson Immunoresearch | Cat#111-165-003 | IF (1:500), RRID:AB_2338000 |
| Antibody | Cy3 Goat polyclonal anti-Chick | Abcam | Cat#ab97145 | IF (1:500), RRID:AB_10679516 |
| Peptide, recombinant protein | Poly-D-lysine | Sigma Aldrich | Cat#P6407 | |
| Peptide, recombinant protein | N-Cadherin | R&D Systems | Cat#1388-NC | |
| Peptide, recombinant protein | Netrin-1 | R&D Systems | Cat#1109-N1/CF | |
| Peptide, recombinant protein | Slit2N | R&D Systems | Cat#5444-SL-050 | |
| Chemical compound, drug | KCl | Thermo Fisher | Cat#BP366-1 | |
| Chemical compound, drug | MgCl2 | Thermo Fisher | Cat#BP214-500 | |
| Chemical compound, drug | HEPES | Thermo Fisher | Cat#BP299-1 | |
| Chemical compound, drug | L15 media | Gibco | Cat#11415-064 | |
| Chemical compound, drug | Horse serum | Gibco | Cat#16050122 | |
| Chemical compound, drug | Fastgreen dye | Thermo Fisher | Cat#F99-10 | |
| Chemical compound, drug | Opti-MEM | Gibco | Cat#31985-070 | |
| Chemical compound, drug | F12 | Gibco | Cat#11765-054 | |
| Chemical compound, drug | Glucose | Thermo Fisher | Cat#D16-500 | |
| Chemical compound, drug | 100x Pen/Strep/Glutamine | Gibco | Cat#10378-016 | |
| Chemical compound, drug | HBSS | Gibco | Cat#14175-079 | |
| Chemical compound, drug | Trypsin | Gibco | Cat#25300054 | |
| Chemical compound, drug | DNAse I | New England Biolabs | Cat#M0303L | |
| Chemical compound, drug | MgSO4 | Thermo Fisher | Cat#7487-88-9 | |
| Chemical compound, drug | Neurobasal | Gibco | Cat#21103-049 | |
| Chemical compound, drug | FBS | Gibco | Cat#10437-028 | |
| Chemical compound, drug | B-27 | Thermo Fisher | Cat#A3582801 | |
| Chemical compound, drug | Effectene Transfection Reagent | Qiagen | Cat#301425 | |
| Chemical compound, drug | Rat Tail Collagen | Corning | Cat#354249 | |
| Chemical compound, drug | Paraformaldehyde 16% solution, EM grade | Electron Microscopy Services | Cat#15710 | |
| Chemical compound, drug | Amicon Ultracel 30K filters | Millipore | Cat#UFC903096 | |
| Chemical compound, drug | Drosophila Schneider’s Media | Life Technologies | Cat#21720024 | |
| Chemical compound, drug | Surfact-AMPS NP40 | Thermo Fisher | Cat#85124 | |
| Chemical compound, drug | Protease Inhibitor (Complete) | Roche | Cat#11697498001 | |
| Chemical compound, drug | 2x Laemmli Sample Buffer | Bio-Rad | Cat#1610737 | |
| Chemical compound, drug | 4x Laemmli Sample Buffer | Bio-Rad | Cat#1610747 | |
| Chemical compound, drug | Clarity Western ECL Substrate | Bio-Rad | Cat#1705061 | |
| Chemical compound, drug | NotI | New England Biolabs | Cat#R3189S | |
| Chemical compound, drug | XbaI | New England Biolabs | Cat#R0145S | |
| Chemical compound, drug | BglII | New England Biolabs | Cat#R0144S | |
| Chemical compound, drug | XhoI | New England Biolabs | Cat#R0146S | |
| Chemical compound, drug | BbsI | New England Biolabs | Cat#R3539S | |
| Chemical compound, drug | MfeI | New England Biolabs | Cat#R3589S | |
| Chemical compound, drug | Proteinase K | Roche Diagnostics | Cat#03115828001 | |
| Chemical compound, drug | Protein A Agarose beads | Invitrogen | Cat#15918-014 | |
| Chemical compound, drug | rProteinG Agarose beads | Invitrogen | Cat#15920–010 | |
| Commercial assay or kit | Quikchange II site-directed mutagenesis kit | Agilent | Cat#200523 | |
| Software, algorithm | Image J Fiji | Fiji | https://imagej.net/Fiji | RRID:SCR_002285 |
| Software, algorithm | Adobe Photoshop | Adobe | N/A | RRID:SCR_014199 |
| Software, algorithm | Bio-Rad Image Lab | Bio-Rad | http://www.bio-rad.com/zh-cn/product/image-lab-software | RRID:SCR_014210 |
| Software, algorithm | GraphPad Prism 9 | GraphPad software | https://www.graphpad.com/ | RRID:SCR_002798 |
| Software, algorithm | ChemiDoc Imaging System | Bio-Rad | Cat#171001401 | |
| Software, algorithm | Volocity Software | Perkin Elmer | http://cellularimaging.perkinelmer.com/downloads/ | RRID:SCR_002668 |
| Other | BTX Electroporator | BTX Harvard Apparatus | Cat#45-0662 | |
| Other | Nikon Ti-U microscope | Nikon | N/A |





