SARS-CoV-2 entry into human airway organoids is serine protease-mediated and facilitated by the multibasic cleavage site
Figures
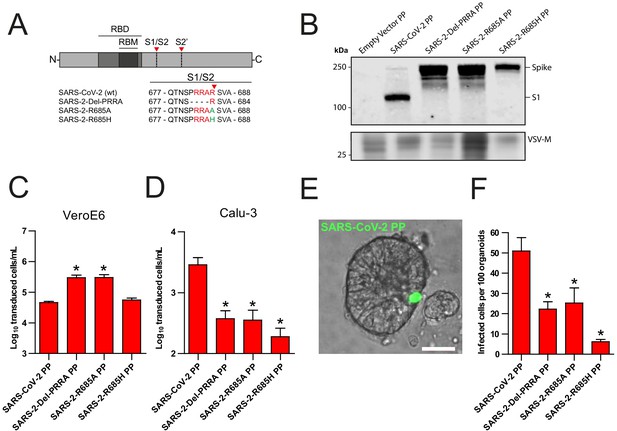
The SARS-CoV-2 S multibasic cleavage site mediates entry into human airway organoids.
(A) Schematic overview of SARS-CoV-2 S protein mutants. MBCS residues are indicated in red; amino acid substitutions are indicated in green. Red arrows indicate cleavage sites. The SARS-CoV-2 S MBCS was mutated to either remove the PRRA motif (SARS-2-Del-PRRA) or substitute the R685 site (SARS-2-R685A and R685H). (B) Comparison of S cleavage of SARS-CoV-2 PPs and the MBCS mutants. Western blots were performed against S1 with VSV-M silver stains as a production control. (C and D) PP infectivity of SARS-CoV-2 S and MBCS mutants on VeroE6 (C) and Calu-3 (D) cells. (E) Differentiated hAO cultures were infected with concentrated SARS-CoV-2 PPs containing a GFP reporter, indicated in green. Scale bar indicates 20 μm. (F) SARS-CoV-2 PP and MBCS mutant infectivity on bronchiolar hAO cultures. One-way ANOVA was performed for statistical analysis comparing all groups with SARS-CoV-2 PPs. Experiments were performed in triplicate (B and D, F). Representative experiments from at least two independent experiments are shown (C and D). Combined data from three independent experiments is shown (F). Error bars indicate SEM. *p<0.05. GFP, green fluorescent protein; hAO, human airway organoid; MBCS, multibasic cleavage site; PP, pseudoparticles; RBD, receptor binding domain; RBM, receptor binding motif.
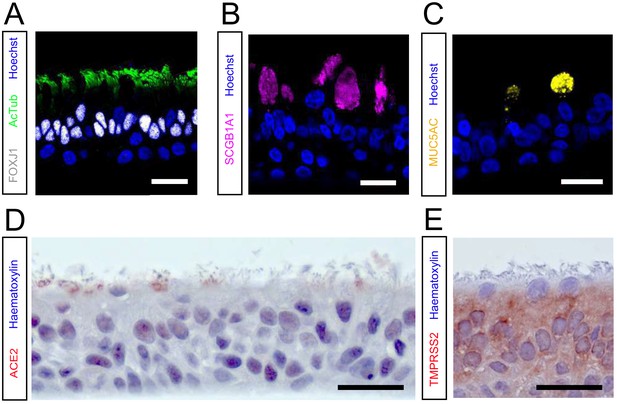
hAO cultures grown at 2D air–liquid interface are well-differentiated and express ACE2 and TMPRSS2.
(A, B, and C) Immunofluorescent or immunohistochemistry staining of differentiated airway cultures. Anti-AcTub (green) and anti-FOXJ1 (white) stains ciliated cells (A), anti-SCGB1A1 (magenta) stains club cells (B), and anti-MUC5AC (yellow) stains goblet cells (C). Nuclei are stained with hoechst (blue). (D and E) hAO cultures also expressed the SARS-CoV-2 entry receptor ACE2 (D) and TMPRSS2 (E). Hematoxylin was used as a counterstain in (D) and (E). Scale bars indicate 20 μm. Representative images are shown from a bronchiolar culture. ACE2, angiotensin-converting enzyme 2; hAO, human airway organoid; TMPRSS2, transmembrane protease serine 2.
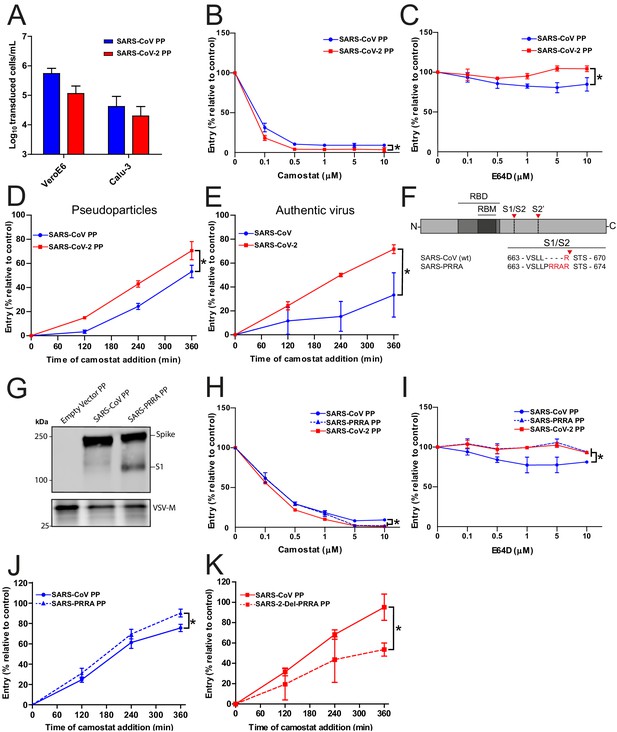
SARS-CoV-2 enters faster on Calu-3 cells than SARS-CoV and entry speed is increased by the multibasic cleavage site.
(A) SARS-CoV PP and SARS-CoV-2 PP infectivity on VeroE6 and Calu-3 cells. (B and C) SARS-CoV PP and SARS-CoV-2 PP entry route on Calu-3 cells. Cells were pretreated with a concentration range of camostat (B) or E64D (C) to inhibit serine proteases and cathepsins, respectively. T-test was performed for statistical analysis at the highest concentration. *p<0.05. (D and E) SARS-CoV PP, SARS-CoV-2 PP (D) and authentic virus (E) entry speed on Calu-3 cells. T-test was performed for statistical analysis at the latest time point. *p<0.05. (F) Schematic overview of SARS-CoV S protein mutants. MBCS residues are indicated in red. The SARS-CoV-2 PRRA motif was inserted into SARS-CoV PPs (SARS-PRRA). (G) Comparison of S1 cleavage of SARS-CoV PP and the MBCS mutant. VSV-M silver stains are shown as a production control. (H and I) SARS-CoV PP, SARS-PRRA PP, and SARS-CoV-2 PP entry route on Calu-3 cells. Cells were pretreated with a concentration range of camostat (H) or E64D (I) to inhibit plasma membrane and endosomal entry, respectively. One-way ANOVA was performed for statistical analysis comparing all groups with SARS-CoV PPs at the highest concentration. *p<0.05. (J and K) Entry speed on Calu-3 cells of SARS-CoV PPs compared with SARS-PRRA PPs (J) and SARS-CoV-2 PPs compared with SARS-2-Del-PRRA PPs (K). T-test was performed for statistical analysis at the latest time point. *p<0.05. Experiments were performed in triplicate (A–E, H–K). Representative experiments from at least two independent experiments are shown. Error bars indicate SD. ANOVA, analysis of variance; MBCS, multibasic cleavage site; PP, pseudoparticles.
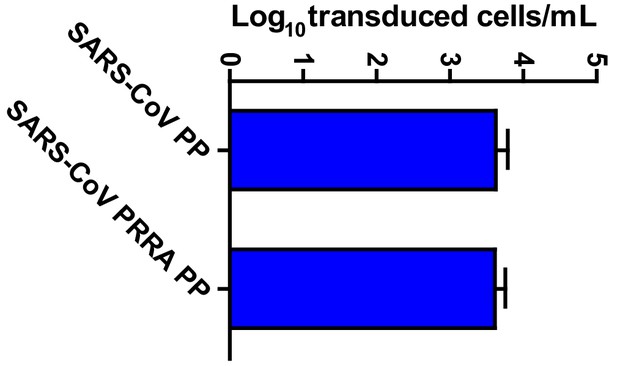
SARS-CoV PP infectivity into Calu-3 cells is not altered by the insertion of the multibasic cleavage site.
Titrations of SARS-CoV PPs and SARS-PRRA PPs on Calu-3 cells. Error bars indicate SEM. A representative experiment in triplicate from three independent experiments is shown. PP, pseudoparticles.
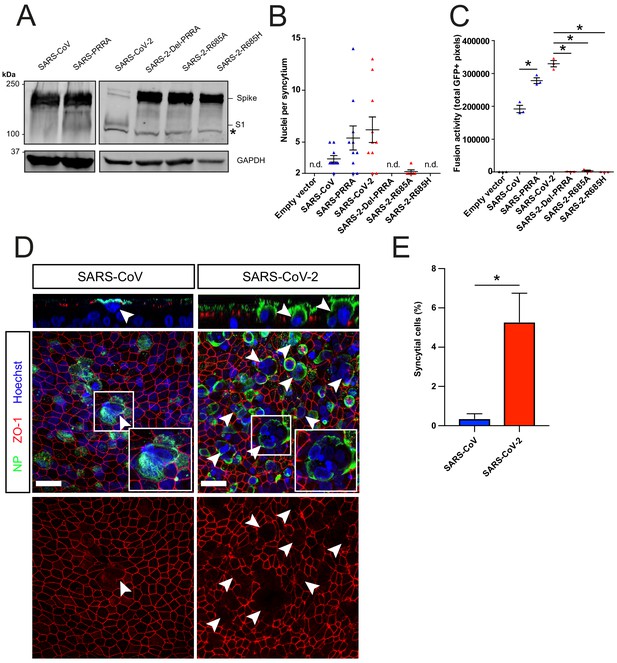
The SARS-CoV-2 multibasic cleavage site facilitates cell–cell fusion and SARS-CoV-2 is more fusogenic than SARS-CoV on human airway organoids.
(A) Proteolytic cleavage of SARS-CoV-2 S, SARS-CoV S, and S mutants was assessed by overexpression in HEK-293T cells and subsequent western blots for S1. GAPDH was used as a loading control. Asterisk indicates an unspecific band. (B and C) Fusogenicity of SARS-CoV-2 S, SARS-CoV S, and S mutants was assessed after 18 hr by counting the number of nuclei per syncytium (B) and by measuring the sum of all GFP+ pixels per well (C). Statistical analysis was performed by one-way ANOVA on SARS-CoV or SARS-CoV-2 S-mediated fusion compared with its respective mutants. *p<0.05 (C). (D) Differentiated bronchiolar hAO cultures were infected at an MOI of 1 with SARS-CoV or SARS-CoV-2. Seventy-two hours postinfection they were fixed and stained for nucleoprotein (NP; green) and tight junctions (ZO1; red) to image syncytia. Nuclei were stained with hoechst (blue). Scale bars indicate 20 μm. Arrows indicate syncytial cells. (E) Percentage of syncytial cells of total number of infected cells per field of 0.1 mm2. Five fields were counted. T-test was performed for statistical analysis. *p<0.05. Experiments were performed in triplicate (C). Representative experiments from at least two independent experiments are shown. Error bars indicate SEM. hAO, human airway organoids; H p.i., hours postinfection; MOI, multiplicity of infection; n.d., not detected.
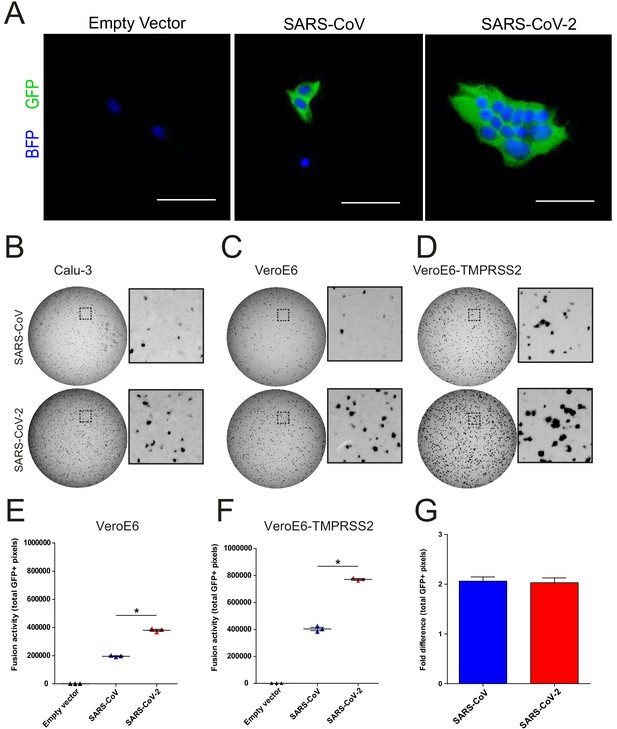
A GFP complementation based assay for assessing coronavirus fusogenicity.
(A) HEK-293T cells expressing an empty vector or S protein together with GFP-11-tagged beta-actin and a BFP containing a nuclear localization signal were added to cells stably expressing GFP1-10. Fusion of these two cell types allowed GFP complementation in cells expressing a nuclear BFP, facilitating easy quantification of nuclei per syncytial cell. Unfused cells only expressed BFP in the nucleus. Fusion with VeroE6 GFP1-10 cells 18 hr after addition of the fusogenic HEK-293T is shown as an example. (B–D) Full well scans of the complemented GFP signal 18 hr after addition of the fusogenic HEK-293T cells to Calu-3 GFP1-10 (B), VeroE6 GFP1-10 (C), and VeroE6-TMPRSS2 GFP1-10 (D) cells are shown. Dashed areas are enlarged next to each well. Scale bars indicate 50 μm. (E and F) Fusogenicity of SARS-CoV-2 S and SARS-CoV S was assessed after 18 hr by measuring the sum of all GFP+ pixels per well in VeroE6 cells (E) and VeroE6 TMPRSS2 cells (F). Statistical analysis was performed by one-way ANOVA on SARS-CoV-2 S-mediated fusion compared with SARS-CoV S. *p<0.05. (G) Fold change in total GFP+ pixels by TMPRSS2 overexpression in VeroE6 cells. ANOVA, analysis of variance; BFP, blue fluorescent protein; GFP, green fluorescent protein.
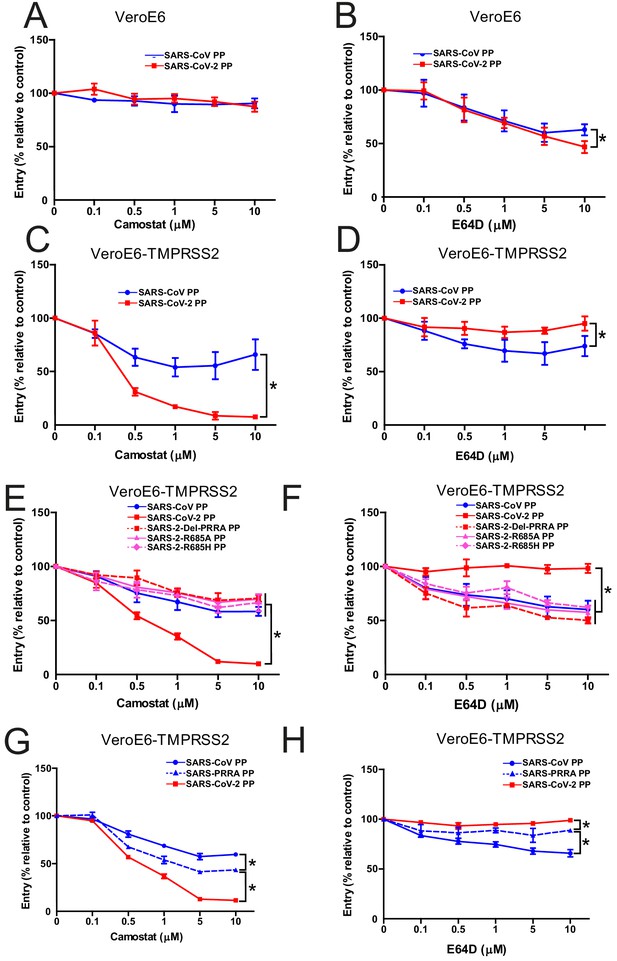
The SARS-CoV-2 multibasic cleavage site increases serine protease usage.
(A and B) SARS-CoV PP and SARS-CoV-2 PP entry route on VeroE6 cells pretreated with a concentration range of camostat (A) or E64D (B) to inhibit serine proteases and cathepsins, respectively. (C and D) SARS-CoV PP and SARS-CoV-2 PP entry route on VeroE6-TMPRSS2 cells pretreated with a concentration range of camostat (C) or E64D (D) to inhibit serine proteases and cathepsins, respectively. T-test was performed for statistical analysis at the highest concentration. *p<0.05. (E and F) Entry route of SARS-CoV-2 PP and MBCS mutants on VeroE6-TMPRSS2 cells pretreated with a concentration range of camostat (E) or E64D (F) to inhibit serine proteases and cathepsins, respectively. One-way ANOVA was performed for statistical analysis comparing all groups to SARS-CoV-2 PPs at the highest concentration. *p<0.05. (G and H) Entry route of SARS-CoV PPs and SARS-PRRA PPs on VeroE6-TMPRSS2 cells pretreated with a concentration range of camostat (G) or E64D (H) to inhibit serine proteases and cathepsins, respectively. One-way ANOVA was performed for statistical analysis comparing all groups to SARS-PRRA PPs at the highest concentration. *p<0.05. ANOVA, analysis of variance; MBCS, multibasic cleavage site; PP, pseudoparticles. Representative experiments in triplicate from at least two independent experiments are shown. Error bars indicate SD.
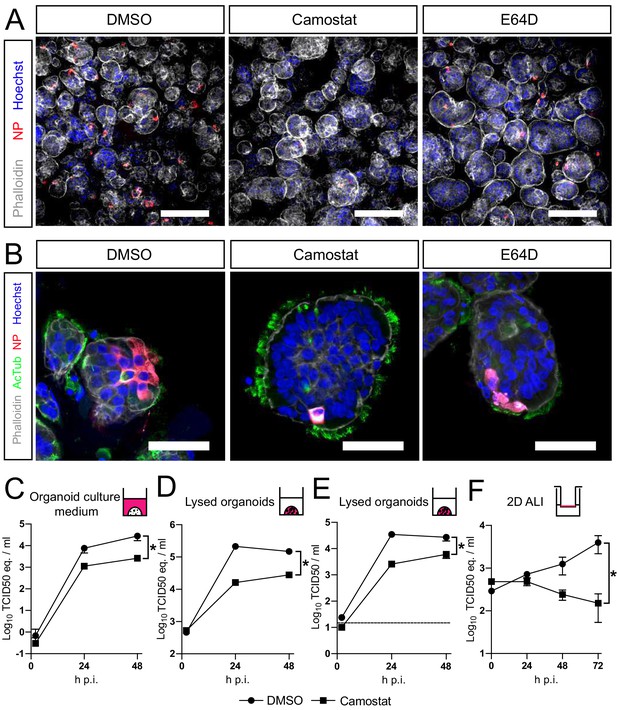
SARS-CoV-2 entry and replication are dependent on serine proteases in human airway organoids.
(A and B) Differentiated bronchiolar (A) or bronchial (B) hAO cultures were infected at an MOI of 2. Sixteen hours (A) or 24 hr (B) postinfection they were fixed and stained for viral nucleoprotein (red). Nuclei were stained with hoechst (blue) and actin was stained using phalloidin (white). AcTub stains ciliated cells (green). Scale bars indicate 200 μm in (A) and 50 μm in (B). Representative images are shown from two independent experiments. (C–E) Replication kinetics of SARS-CoV-2 in bronchiolar hAO cultures pretreated with camostat or carrier (DMSO). (C and D) TCID50 equivalents (eq.) per mL are shown in culture medium (C) and lysed organoids (D). Circles indicate DMSO-treated organoids, whereas squares indicate camostat-treated organoids. (E) Live virus titers (TCID50/mL) in lysed organoids. Dotted line indicates limit of detection. (F) Replication kinetics of SARS-CoV-2 in 2D tracheal air–liquid interface airway cultures pretreated with camostat or carrier (DMSO). TCID50 eq./mL in apical washes are shown. Two-way ANOVA was performed for statistical analysis. Error bars indicate SEM. *p<0.05. DMSO, dimethyl sulfoxide; hAO, human airway organoid; H p.i., hours postinfection; MOI, multiplicity of infection.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| VeroE6 cells (Cercopithecus aethiops) | Monkey kidney cell line | ATCC | CRL 1586TM | |
| Vero cells (Cercopithecus aethiops) | Monkey kidney cell line | WHO | RCB 10-87 | |
| Calu-3 (Homo sapiens) | Lung adenocarcinoma cell line | ATCC | HTB 55 | |
| SARS-CoV-2 BavPat1 | SARS-CoV-2 | Dr. Christian Drosten | European Virus Archive Global #026V-03883 | |
| SARS-CoV HKU39849 | SARS-CoV | Dr. Malik Peiris | N/A | |
| Airway tissue for organoids (Homo sapiens) | Airway organoids | This study | This study | |
| Aloxistatin | E64D | MedChemExpress | Cat# HY-100229 | |
| Camostat mesylate | Camostat | Sigma | Cat# SML0057 | |
| Hexadimethrine bromide | Polybrene | Sigma | 107689-10G | |
| Polyethylenimine linear | Polyethylenimine | Polysciences | Cat# 23966 | |
| Hygromycin B | Hygromycin B | Invitrogen | Cat# 10843555001 | |
| G418, Geneticin | Geneticin | Invitrogen | Cat# 10131035 | |
| Opti-MEM I (1×) + GlutaMAX | Opti-MEM I (1×) + GlutaMAX | Gibco | Cat# 51985-042 | |
| Advanced DMEM/F12 | Advanced DMEM/F12 | Thermo Fisher scientific | Cat# 12634-010 | |
| AO medium | AO medium | Sachs et al., 2019 | N/A | |
| Pneumacult ALI medium | Pneumacult ALI medium | Stemcell | Cat # 05001 | |
| TryplE | TryplE | Thermo Fisher Scientific | Cat# 12605010 | |
| Cultrex Basement Membrane Extract, Type 2 | Basement membrane extract | R&D Systems | Cat# 3533-005-02 | |
| 12 mm Transwell with 0.4 µm Pore Polyester Membrane Insert, Sterile | Transwell inserts | Corning | Cat# 3460 | |
| Collagen Type I, High concentration Rat tail | Collagen | Corning | Cat# 354249 | |
| 0.45 μm low protein binding filter | 0.45 μm low protein binding filter | Millipore | Cat# SLHV033RS | |
| Recombinant DNA reagent | pCMV-S (CUHK-W1) | Sino Biological | Cat# VG40150-G-N | Encoding S of isolate CUHK-W1; |
| Recombinant DNA reagent | pCAGGS-S (CUHK-W1) | This study | This study | Encoding S of isolate CUHK-W1; |
| Recombinant DNA reagent | pCAGGS-S (Wuhan-Hu-1) | This study | This study | Encoding S of isolate Wuhan-Hu-1 |
| Recombinant DNA reagent | pQXCIN | Clontech | Cat# 631516 | Retro-X Q vector set |
| Recombinant DNA reagent | pQXCIH | Clontech | Cat# 631516 | Retro-X Q vector set |
| Recombinant DNA reagent | pQXCIP-GFP1-10 | Addgene | Cat# 68715 | GFP1-10 |
| Recombinant DNA reagent | pCDNA-TMPRSS2-FLAG | Genscript | OHu13675D | Human TMPRSS2 cDNA |
| Recombinant DNA reagent | pCAGGS-β-Actin-7xGFP11-P2A-BFP | This study | This study | β-Actin-7xGFP11-P2A-BFP |
| Recombinant DNA reagent | pBS-gag-pol | Addgene | Cat# 35614 | gag-pol |
| Recombinant DNA reagent | pMD2.G | Addgene | Cat# 12259 | VSV-G |
| Recombinant DNA reagent | pVSV-eGFP-dG | Addgene | Cat# 31842 | VSV delta G genomic plasmid |
| Recombinant DNA reagent | pCAG-VSV-P | Addgene | Cat# 64088 | P protein |
| Recombinant DNA reagent | pCAG-VSV-L | Addgene | Cat# 64085 | L protein |
| Recombinant DNA reagent | pCAG-VSV-N | Addgene | Cat# 64087 | N protein |
| Recombinant DNA reagent | pCAGGS-T7Opt | Addgene | Cat# 65974 | T7 polymerase |
| Antibody | Mouse-anti-SARS-CoV NP (monoclonal) | Sino Biological | Cat# 40143-MM05 | IF (1:400) |
| Antibody | Rabbit-anti-SARS-CoV NP (polyclonal) | Sino Biological | Cat# 40143-T62 | IF (1:400) |
| Antibody | Goat anti-ACE2 (polyclonal) | R&D Systems | Cat# AF933 | IF (1:200) |
| Antibody | Mouse anti-TMPRSS2 (monoclonal) | Santa Cruz | Cat# sc-515727 | IF (1:200) |
| Antibody | Rabbit-anti-goat | Dako | Cat# P0160 | IF (1:400) |
| Antibody | Goat-anti-mouse | Dako | Cat# P0260 | IF (1:400) |
| Antibody | Mouse anti-VSV (monoclonal) | Absolute Antibody | Cat# EB0010 | Pseudoparticle production (1:50,000) |
| Antibody | Goat anti-rabbit IgG (H+L) Alexa Fluor Plus 488 | Invitrogen | Cat# A32731 | IF (1:400) |
| Antibody | Goat anti-mouse IgG (H+L) Alexa Fluor Plus 488 | Invitrogen | Cat# A11029 | IF (1:400) |
| Antibody | Goat anti-mouse IgG (H+L) Alexa Fluor Plus 594 | Invitrogen | Cat# A21125 | IF (1:400) |
| Antibody | Mouse anti-double stranded RNA IgG2A (monoclonal) | Scicons | J2 clone | IF (1:500) |
| Antibody | Mouse-anti-ZO1 IgG1 (monoclonal) | Invitrogen | Cat# 33-9100 | IF (1:200) |
| Antibody | Mouse-anti-CC10 IgG1 Alexa Fluor 594 (monoclonal) | Santa Cruz Biotechnology | Cat# sc-390313 AF594 | IF (1:100) |
| Antibody | Mouse-anti-AcTub IgG2A Alexa Fluor 488 (monoclonal) | Santa Cruz Biotechnology | Cat# sc-23950 AF488 | IF (1:100) |
| Antibody | Mouse anti-MUC5AC (monoclonal) | Invitrogen | Cat# MA5-12178 | IF (1:100) |
| Antibody | Mouse-anti-FOXJ1 IgG1 (monoclonal) | eBioscience | Cat# 14-9965-82 | IF (1:200) |
| Antibody | Rabbit anti-SARS-CoV S1 (polyclonal) | Sino Biological | Cat# 40150-T62 | WB (1:1000) |
| Antibody | Mouse anti-GAPDH (monoclonal) | Santa Cruz Biotechnology | Cat# sc-32233 | WB (1:1000) |
| Pierce Silver Stain for Mass Spectrometry | Silver stain | Pierce | Cat# 24600 | |
| TO-PRO-3 Iodide | TO-PRO3 | ThermoFisher | Cat# T3605 | |
| Phalloidin CruzFluor 647 Conjugate | Phalloidin | Santa Cruz | Cat# ec-363796 | |
| Hoechst 33342, Trihydrochloride, Trihydrate | Hoechst | ThermoFisher | Cat# H1399 | |
| 4× Laemmli Sample Buffer | Laemmli | Bio-Rad | Cat# 1610747 | |
| Odyssey CLx | Odyssey CLx | Licor | ||
| Amersham Typhoon Biomolecular Imager | Amersham Typhoon Biomolecular Image | GE Healthcare | ||
| Amersham Imager 600 | Amersham Imager 600 | GE Healthcare | ||
| LSM700 confocal microscope | LSM700 confocal microscope | Zeiss | ||
| Carl ZEISS Vert.A1 | Carl ZEISS Vert.A1 | Zeiss | ||
| ZEN software | ZEN | Zeiss | ||
| ImageQuant TL 8.2 | ImageQuant TL 8.2 | GE Healthcare | ||
| Studio Lite Ver 5.2 | Studio Lite Ver 5.2 | Licor | ||
| GraphPad PRISM 8 | GraphPad PRISM 8 | GraphPad | ||
| Adobe Illustrator | Illustrator | Adobe Inc |





