High-fidelity, efficient, and reversible labeling of endogenous proteins using CRISPR-based designer exon insertion
Figures
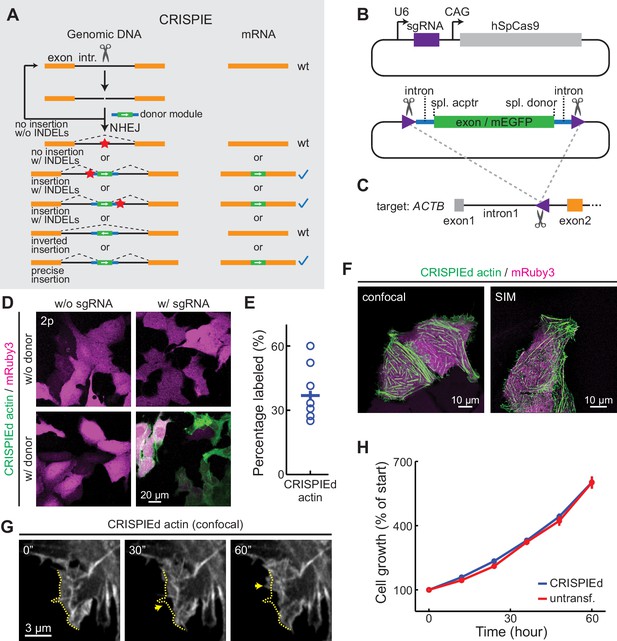
The CRISPR-mediated insertion of exon (CRISPIE) strategy and its application to label human β-actin.
(A) The conceptual design of the CRISPIE strategy showing that, although insertions/deletions (INDELs) and inverted insertion events may occur at the DNA level during editing, only wild-type mRNAs or mRNAs with the desired precise insertion are produced. Orange boxes: endogenous exon sequences; black lines: endogenous intron sequences; blue lines and green boxes: intron and exon sequences, respectively, of the designer donor module with white arrows showing the correct orientation of the reading frame; red stars: INDELs. (B) Schematics of the single guide RNA/Streptococcus pyogenes Cas9 (sgRNA/SpCas9) plasmid (upper) and the donor plasmid (lower) that are used for panels D–G. Purple triangles: the sgRNA-targeted location and orientation, spl.: splicing; acptr: acceptor. See Figure 1—figure supplement 3 for design details. (C) Schematic of the targeted intron of ACTB with the purple triangle showing the sgRNA targeting site and orientation. Orange and gray boxes: exonic sequences that do and do not encode protein sequences, respectively. (D) Representative two-photon (2 p) images of live U2OS cells with the indicated transfection. Note that the mRuby3 (magenta) expression levels were highly variable across cells due to the variability in plasmid transfection; however, the green label intensities were comparable across cells, as expected for the expression from an endogenous locus. (E) Labeling efficiency (green cell counts over red cell counts) for ACTB in U2OS cells. n = 8 field of views (FOVs), two independent transfections. (F) Representative confocal (Airyscan) and super-resolution structured illumination microscopy (SIM) images of live U2OS cells expressing CRISPIEd β-actin. (G) Representative time-lapse confocal images of live U2OS cells showing the dynamics of actin ruffles (arrows). The dashed yellow line outlines the cell morphology at time zero. (H) Cell growth curves of β-actin-CRISPIEd cells and untransfected cells. n = 6 FOVs.
-
Figure 1—source data 1
Numeric data for Figure 1E.
- https://cdn.elifesciences.org/articles/64911/elife-64911-fig1-data1-v3.xlsx
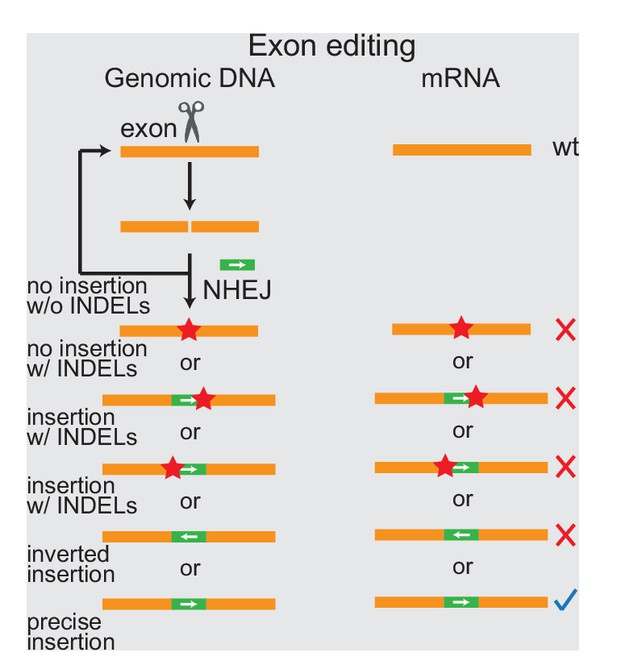
DNA insertion targeting coding sequences result in many possible modes of unwanted mutations.
Schematic of the design and consequences of DNA insertions targeting coding sequences. Orange boxes: endogenous exons; green boxes: coding sequences to be inserted, with arrows indicating the orientation of the reading frame; red stars: insertions/deletions (INDELs). The blue check mark indicates desired precise insertion, while red X’s indicate unwanted mutant mRNAs.
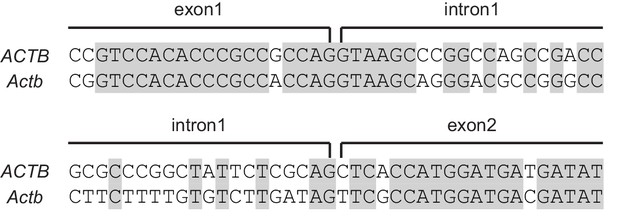
Introns are much less conserved than exons.
Representative alignment of human (ACTB) and mouse (Actb) β-actin genomic sequences at the junctions of the two ends of its first intron. Identical base pairs are highlighted in gray. .
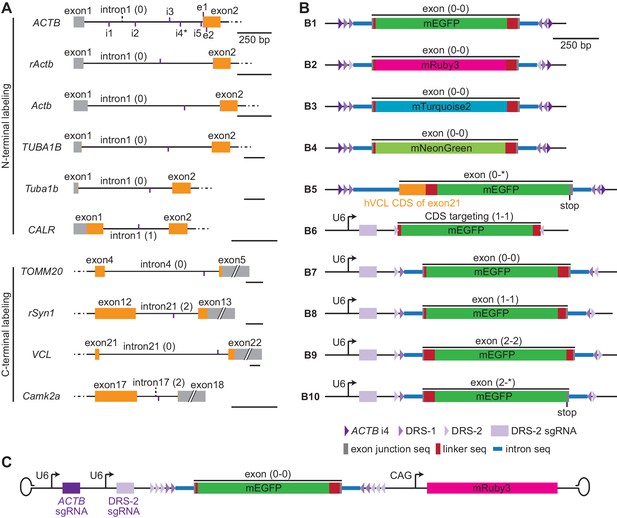
Schematic of targeting sites and donor designs.
(A) Schematic of the genomic structure around the targeting sites. Short vertical purple lines indicate the site positions and whether the targeted sequences are at the upper (purple lines above the DNA schematic) or lower (purple lines below the DNA schematic) DNA strand. All scale bars are 250 bp. (B) Schematic donor plasmid designs used in this manuscript. Triangles illustrate the indicated single guide RNA (sgRNA) targeting sites with orientation. Note that CRISPR-mediated insertion of exon (CRISPIE) allows for the inclusion of multiple cutting sites, as indicated, which provides flexibility for excising the donor DNA or the possibility to erase the inserted module at a later time. The phases of the exon modules (i.e., the reading frames) are labeled. The junctional intron (100 bp) and exon (~10 bp) sequences are taken from intron 17 of mouse Camk2a and its surrounding exons, with the exception that the 5’ intron (250 bp)/exon junction of b5 is taken from intron 21 and exon 22 of VCL, b6 does not involve any intron/exon junction (targeting coding sequences, or CDS), and the 3’ intron (250 bp)/exon junction of b8 is taken from exon 18 and intron 19 of mouse PSD-95. The donors of b1–b5 must be released from the parental plasmid by an additional sgRNA/Streptococcus pyogenes Cas9 (SpCas9) (not depicted here) that cuts one of the indicated flanking sgRNA targeting sites. b6–b10 can be excised using the DRS-2 sgRNA from the same plasmid in the presence of SpCas9. (C) Schematic adeno-associated virus (AAV) for use with SpCas9-expressing cells/animals.
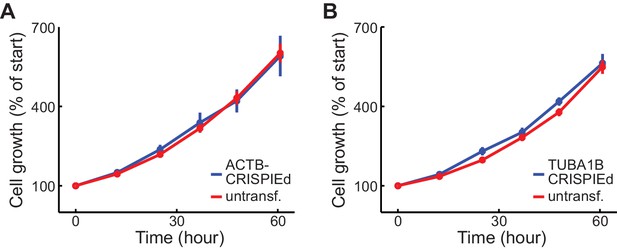
Cell growth curve of ACTB CRISPIEd U2OS cells inserted at a intronic location different from Figure 1H and TUBA1B CRISPIEd U2OS cells.
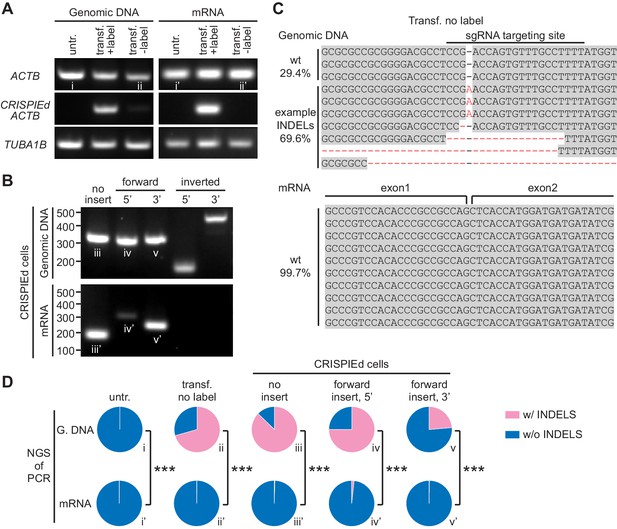
CRISPR-mediated insertion of exon (CRISPIE)-mediated β-actin labeling is resistant to inverted insertion events and insertions/deletions (INDELs).
(A) Gel images of PCR analyses for genomic DNA and RT-PCR analyses for mRNA for untransfected (untr.) and transfected U2OS cells with (transf. +label) and without (transf. -label) successful actin labeling. As detailed in Figure 2—figure supplement 1, specific primers were used for the targeting site at the ACTB gene and mRNA with and without the desired label insertion, and for a control site (TUBA1B) that is not edited in this experiment. (B) Gel images of PCR and RT-PCR analyses for the genomic DNA and mRNA, respectively, of successfully labeled cells to detect non-labeled events, and both 5’ and 3’ junctions of forward and inverted label insertion events. (C) Representative next-generation sequencing (NGS) results of the genomic DNA and mRNA prepared from transfected U2OS cells without successful actin labeling corresponding to bands ii and ii’, respectively. INDELs are marked in red. (D) Pie graphs of the relative proportions of INDEL events based on NGS results of the PCR/RT-PCR products from panels A and B, as marked by the Roman numerals. Statistical tests were performed using a χ2 test. From top to bottom, n = 54216, 1181, 1589, 2767, and 6036 for genomic DNA, and 5771, 2936, 4166, 3613, and 5949 for mRNA.
-
Figure 2—source data 1
Numeric data for Figure 2D.
- https://cdn.elifesciences.org/articles/64911/elife-64911-fig2-data1-v3.xlsx
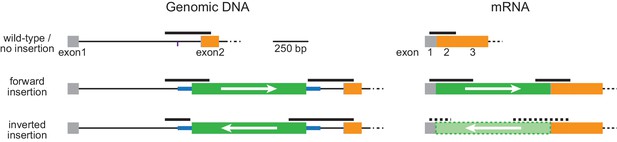
Schematic of PCR and RT-PCR products.
The thick black lines illustrate the PCR and RT-PCR products corresponding to those in Figure 2. Orange and gray boxes: exonic sequences that do and do not encode protein sequences, respectively; thin black lines: endogenous intron sequences; thick blue lines and green boxes: intron and exon sequences, respectively, of the designer donor module with white arrows indicating the correct orientation of the reading frame. Thick black dashed line and the light green box enclosed by dashed lines illustrate the hypothetical PCR products and the inverted EGFP exon, respectively, for inversely inserted mRNA. Note that this condition is only hypothetical and should not exist because there is no appropriate splicing acceptor and donor sites when the donor module is inserted in the inverted orientation.
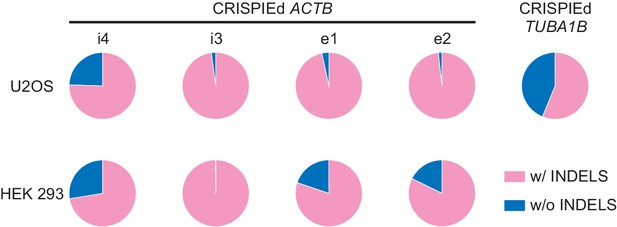
Next-generation sequencing (NGS) analysis of the genomic DNA of different insertion events.
Pie graphs of the relative proportions of insertion/deletion (INDEL) events based on NGS results of PCR amplicons of the 5’ ends of successful insertions into indicated intronic or exonic locations of ACTB or into TUBA1B in different cell lines. PCR was carried out on the genomic DNA of all cells in a dish (i.e., without fluorescence-activated cell sorting (FACS) enrichment of transfected or successfully CRISPIEd cells). The insertion locations for ACTB correspond to those shown in Figure 3. The insertion into TUBA1B is the same as that described in Figure 4B.
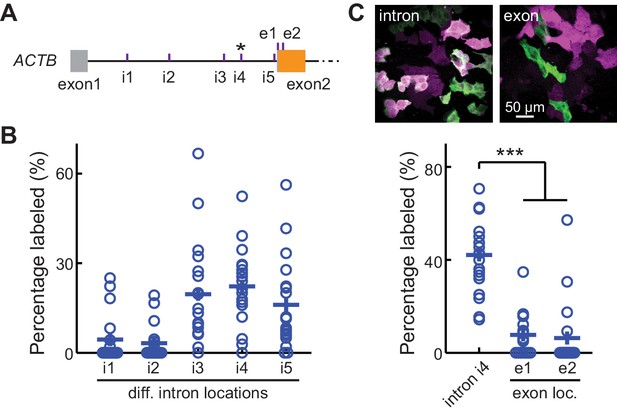
CRISPR-mediated insertion of exon (CRISPIE) is optimizable and achieves higher efficiency than exon labeling.
(A) Schematic positions of the tested editing sites in the intron 1 and exon 2 of ACTB that are targeted in panels B and C. The asterisk marks the site used for ACTB in subsequent experiments. (B) Comparison of the editing efficiency of five different intronic locations. For comparison under identical conditions, a generic donor excised using an independent single guide RNA (sgRNA) (DRS-2) was used (Figure 1—figure supplement 3B7). n = 18 FOVs (field of views) from two independent transfections. (C) Representative images and quantification of successful editing rates at the intronic location i4 vs. the only two possible exonic locations for N-terminally labeled ACTB, each using their specifically designed donors. n = 18 FOVs from two independent transfections.
-
Figure 3—source data 1
Numeric data for Figure 3B and C.
- https://cdn.elifesciences.org/articles/64911/elife-64911-fig3-data1-v3.xlsx
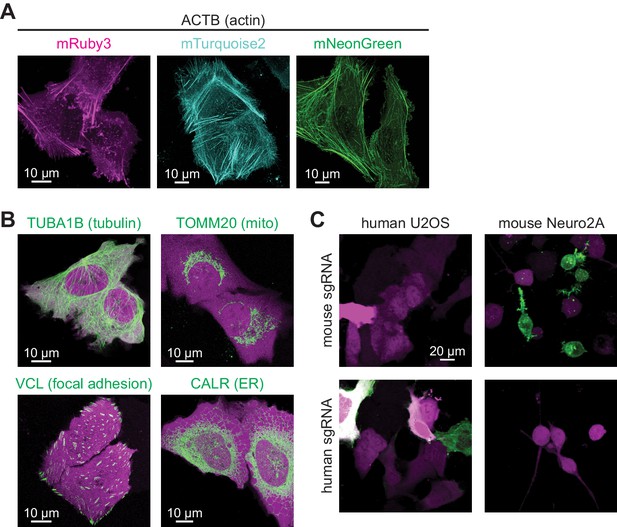
CRISPR-mediated insertion of exon (CRISPIE) is applicable to labeling with different colors, different proteins, and different animal species.
(A) Representative images of ACTB labeled with each of the three indicated fluorescent proteins (FPs) of different colors in U2OS cells. (B) Representative images of four different proteins, as indicated, labeled with monomeric EGFP (mEGFP) in U2OS cells. (C) Representative images of human and mouse β-actin labeling in human U2OS cells and mouse Neuro 2A cells, respectively, under the indicated conditions.
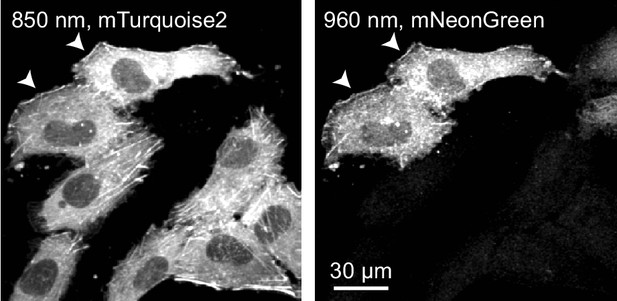
Dual color labeling within the same cells.
CRISPR-mediated insertion of exon (CRISPIE) labeling of ACTB was carried out in the presence of both mTurquoise2 and mNeonGreen templates (Figure 1—figure supplement 3B3 and 3B4). Actin was labeled with both colors in a small fraction of the cells (arrowhead), suggesting that both alleles of ACTB are labeled in these cells, each with a different color. At the majority of places where such double-labeled cells are observed, multiples of the cells are immediately adjacent to each other, suggesting that these cells are the divided daughter cells from the same parental cell.
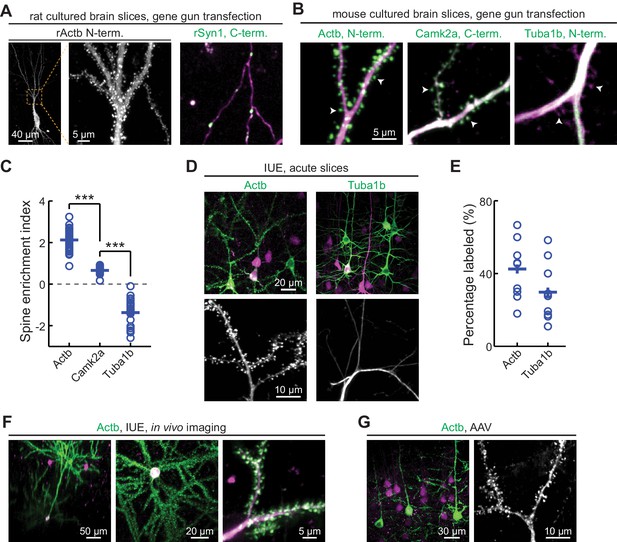
CRISPR-mediated insertion of exon (CRISPIE) is applicable to different proteins in postmitotic neurons using various transfection methods.
(A) Representative images of the indicated proteins labeled in rat cultured hippocampal slices transfected using the biolistic method. (B) Representative images of the indicated proteins labeled in cultured hippocampal slices from SpCas9 ± mice transfected using the biolistic method. Arrowheads indicate spines where each of the three proteins is differentially enriched or excluded. (C) Spine enrichment index (SEI) of the three proteins in panel B. From left to right, n (spines/neurons)=55/5, 26/3, and 22/2. (D, E) Representative images (D) and labeling efficiency quantifications (E) of β-actin or α-tubulin 1B-labeled neurons that are labeled in vivo via in utero electroporation (IUE) in the mouse somatosensory cortex and imaged in acute brain slices. n = 10 FOVs (field of views) from three mice for the quantification of both proteins. (F) Representative side view (x-z, left), top view (x-y, middle), and zoomed-in (x-y, right) images of labeled Actb (green) in cortical neurons that were transfected via IUE and were imaged in living mice via a cranial window. (G) Representative images of β-actin-CRISPIEd neurons in acute brain slices labeled via adeno-associated virus (AAV) injections into the cortex of SpCas9 ± mice.
-
Figure 5—source data 1
Numeric data for Figure 5C and E.
- https://cdn.elifesciences.org/articles/64911/elife-64911-fig5-data1-v3.xlsx
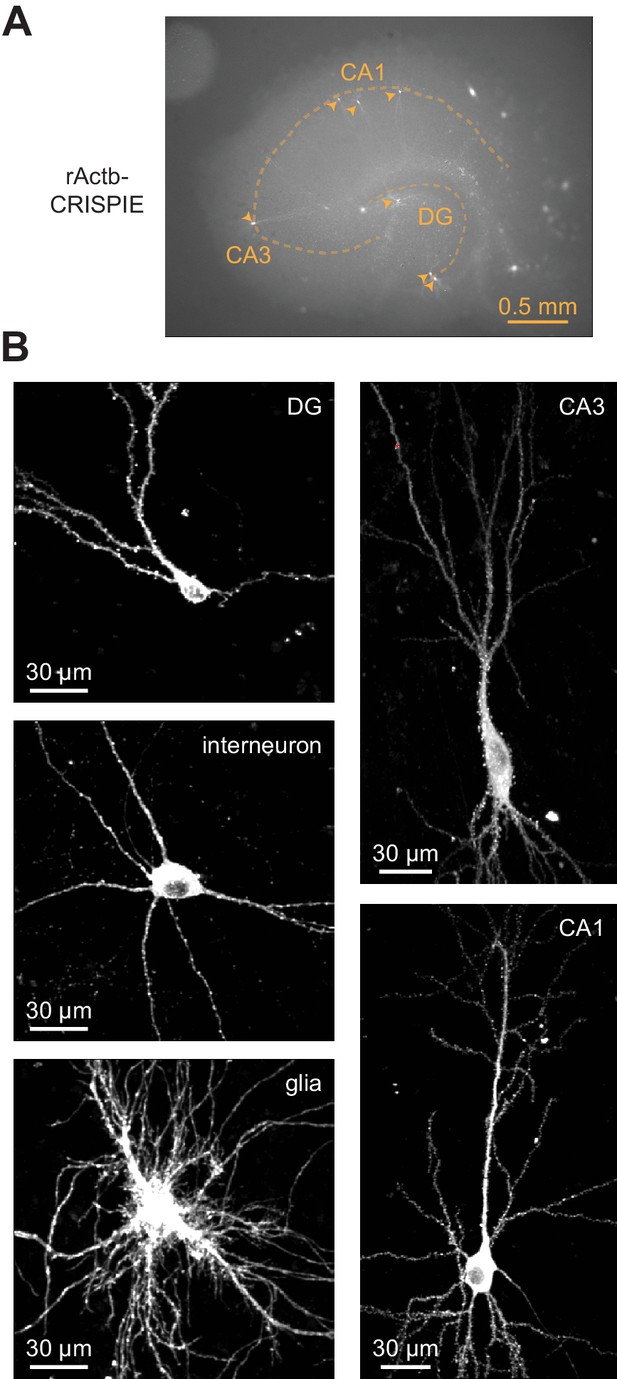
The CRISPR-mediated insertion of exon (CRISPIE) method is suitable for labeling neurons in cultured hippocampal slices transfected using the biolistic (gene gun) method.
(A) Representative macroscopic image showing multiple CRISPIEd neurons (monomeric EGFP [mEGFP]-labeled rActb) in one cultured rat hippocampal slice. (B) Representative two-photon images of CRISPIEd rActb of various cell types in cultured rat hippocampal slices. The CA1 and CA3 neurons are composites of multiple field of views (FOVs). The CA3 neuron is the same as the one presented in the left panel of Figure 5A.
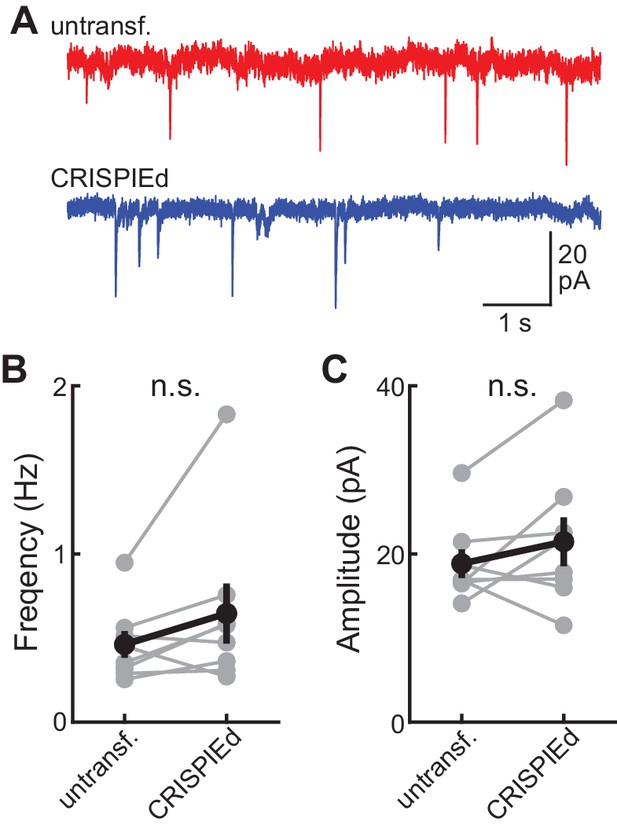
Neurons with CRISPIEd rActb exhibited normal miniature excitatory postsynaptic current (mEPSC) frequencies and amplitudes.
Example traces (A), and quantification of mEPSC frequencies (B) and amplitudes (C) of rActb-CRISPIEd neurons, as transfected by the biolistic method, compared with paired adjacent untransfected neurons in cultured rat hippocampal slices. n = 8 pairs.
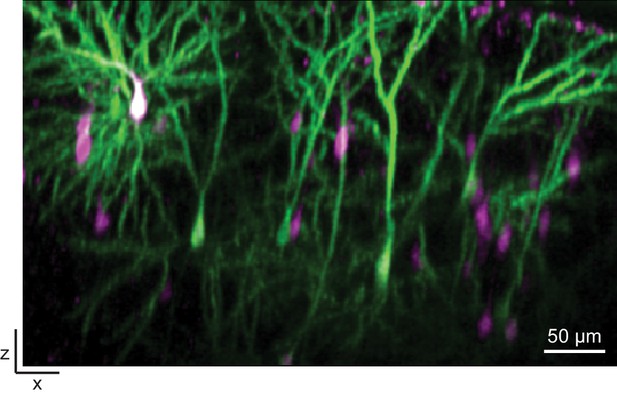
CRISPIEd Actb transfected using in utero electroporation is suitable for in vivo imaging.
Representative x-z view of a two-photon image taken through a cranial window in a living mouse transfected using in utero electroporation at E16.
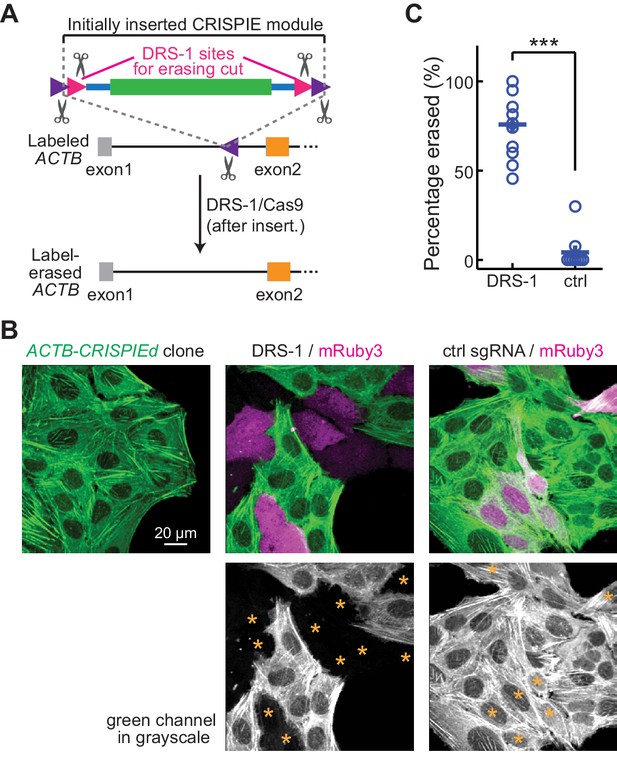
Labeling with CRISPR-mediated insertion of exon (CRISPIE) can be erased.
(A) Schematic of the experimental design for erasing CRISPIE labels. DRS-1 single guide RNA (sgRNA) targeting sequences were used to flank the exon/intron module inside the other two sgRNA sites for excising the donor from the initial insertion. Later introduction of DRS-1 sgRNA and Streptococcus pyogenes Cas9 (SpCas9) can excise the inserted CRISPIE module. Note that after the insertion and erasure, both the original sgRNA targeting site and the DRS-1 site will be destroyed. (B) Representative images of a β-actin-CRISPIEd U2OS cell clone (left), which was transfected with the transfection marker mRuby3 (magenta in upper panels) and SpCas9 together with DRS-1 (middle), or with a control sgRNA that targeted mouse, but not human β-actin (right). Asterisks in the lower panel mark the transfected (i.e., mRuby-positive) cells. (C) Quantification of the erasure efficiency among the transfected cells, as identified by mRuby3 expression. n (field of views [FOVs]/transfections)=11/3 for DRS-1, and 9/3 for control sgRNA.
-
Figure 6—source data 1
Numeric data for Figure 6C.
- https://cdn.elifesciences.org/articles/64911/elife-64911-fig6-data1-v3.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | ACTB | NA | NCBI Gene ID 60 | |
| Gene (Homo sapiens) | TUBA1B | NA | NCBI Gene ID 10376 | |
| Gene (Homo sapiens) | TOMM20 | NA | NCBI Gene ID 9804 | |
| Gene (Homo sapiens) | VCL | NA | NCBI Gene ID 7414 | |
| Gene (Homo sapiens) | CALR | NA | NCBI Gene ID 811 | |
| Gene (Mus musculus) | Actb | NA | NCBI Gene ID 11461 | |
| Gene (Mus musculus) | Tuba1b | NA | NCBI Gene ID 22143 | |
| Gene (Mus musculus) | Camk2a | NA | NCBI Gene ID 12322 | |
| Gene (Rattus norvegicus) | Actb | NA | NCBI Gene ID 81822 | |
| Gene (Rattus norvegicus) | Syn1 | NA | NCBI Gene ID 24949 | |
| Strain, strain background (Rattus norvegicus, Sprague Dawley) | Sprague Dawley rat | Charles River | Strain Code 001; RRID:RGD_734476 | |
| Genetic reagent (Mus musculus) | Cas9 mouse (B6J.129(Cg)-Igs2tm1.1(CAG-cas9*)Mmw/J) | Jax | Stock No: 028239; RRID:IMSR_JAX:028239 | |
| Cell line (Homo sapiens) | U2OS | ATCC | Cat# HTB-96; RRID:CVCL_0042 | |
| Cell line (Mus musculus) | Neuro2A | ATCC | Cat# CCL-131; RRID:CVCL_0470 | |
| Recombinant DNA reagent | px330 | Addgene | Plasmid #42230 | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting ACTB i1 | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting ACTB i2 | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting ACTB i3 | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting ACTB i4 | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting ACTB i5 | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting ACTB e1 | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting ACTB e2 | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting TUBA1B | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting TOMM20 | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting VCL | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting CALR | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting Actb | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting Tuba1b | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting Camk2a | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting rActb | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | sgRNA/Cas9 targeting rSyn1 | This paper | Progenitors: synthesized oligos; pX330; will be deposited to Addgene | |
| Recombinant DNA reagent | pUC57-Amp | Genewiz | Genewiz ID: pUC57-Amp | |
| Recombinant DNA reagent | Insertion template B1 (Figure 1—figure supplement 3) | This paper | Progenitors: synthesized DNA; pUC57-Amp; will be deposited to Addgene | |
| Recombinant DNA reagent | Insertion template B2 (Figure 1—figure supplement 3) | This paper | Progenitors: synthesized DNA; pUC57-Amp; will be deposited to Addgene | |
| Recombinant DNA reagent | Insertion template B3 (Figure 1—figure supplement 3) | This paper | Progenitors: synthesized DNA; pUC57-Amp; will be deposited to Addgene | |
| Recombinant DNA reagent | Insertion template B4 (Figure 1—figure supplement 3) | This paper | Progenitors: synthesized DNA; pUC57-Amp; will be deposited to Addgene | |
| Recombinant DNA reagent | Insertion template B5 (Figure 1—figure supplement 3) | This paper | Progenitors: synthesized DNA; pUC57-Amp; will be deposited to Addgene | |
| Recombinant DNA reagent | Insertion template B6 (Figure 1—figure supplement 3) | This paper | Progenitors: synthesized DNA; pUC57-Amp; will be deposited to Addgene | |
| Recombinant DNA reagent | Insertion template B7 (Figure 1—figure supplement 3) | This paper | Progenitors: synthesized DNA; pUC57-Amp; will be deposited to Addgene | |
| Recombinant DNA reagent | Insertion template B8 (Figure 1—figure supplement 3) | This paper | Progenitors: synthesized DNA; pUC57-Amp; will be deposited to Addgene | |
| Recombinant DNA reagent | Insertion template B9 (Figure 1—figure supplement 3) | This paper | Progenitors: synthesized DNA; pUC57-Amp; will be deposited to Addgene | |
| Recombinant DNA reagent | Insertion template B10 (Figure 1—figure supplement 3) | This paper | Progenitors: synthesized DNA; pUC57-Amp; will be deposited to Addgene | |
| Recombinant DNA reagent | pAAV-CW3SL-EGFP | Addgene | Plasmid # 61463 | |
| Recombinant DNA reagent | pKanCMV-mRuby3-18aa-actin | Addgene | Plasmid # 74255 | |
| Recombinant DNA reagent | pKanCMV-mRuby3 | This paper | Progenitors: oligos for site-directed deletion; pKanCMV-mRuby3-18aa-actin; will be deposited to Addgene | |
| Recombinant DNA reagent | Plasmid for β-actin CRISPIE (Figure 1—figure supplement 3C) | This paper | Progenitors: synthesized DNA; pKanCMV-mRuby3; pAAV-CW3SL-EGFP; will be deposited to Addgene | |
| Sequence-based reagent | Oligonucleotides used for PCR and RT-PCR analysis | Genewiz | Please see Supplementary file 1 | |
| Commercial assay or kit | NGS sequencing of PCR amplicons | Genewiz | Genewiz ID: Amplicon-EZ | |
| Chemical compound, drug | Lipofectamine 2000 | Thermo Fisher | Cat# 11668019 | |
| Chemical compound, drug | McCoy’s 5A Medium (for U2OS cell culture) | ATCC | Cat# 30–2007 | |
| Chemical compound, drug | MEM (for Neuro2A cell culture) | Thermo Fisher | Cat# 11095–080 | |
| Chemical compound, drug | MEM powder (for slice culture) | Thermo Fisher | Cat# 11700–077 | |
| Chemical compound, drug | Cell culture insert (for slice cultures) | Millipore | Cat# PICM0RG50 | |
| Chemical compound, drug | GoScript Reverse Transcriptase | Promega | Cat# A5001 | |
| Chemical compound, drug | OneTaq Hot Start 2X Master Mix with GC Buffer | NEB | Cat# M0485S | |
| Chemical compound, drug | Q5 Hot Start High-Fidelity 2X Master Mix | NEB | Cat# M0494S | |
| Chemical compound, drug | Gold particles, 1.6 µm (for gene gun bullet) | Bio-Rad | Cat# 165–2264 | |
| Chemical compound, drug | Tefzel tubing | Bio-Rad | Cat# 165–2441 | |
| Software, algorithm | MATLAB | MathWorks | RRID:SCR_001622;https://www.mathworks.com/ | |
| Software, algorithm | GPP sgRNA designer | BROAD Institute | https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design | |
| Other | β-Actin CRISPIE AAV9 (Figure 1—figure supplement 3C) | Vigene | Custom production |
Additional files
-
Supplementary file 1
PCR primer list.
- https://cdn.elifesciences.org/articles/64911/elife-64911-supp1-v3.docx
-
Supplementary file 2
PCR primer combinations.
- https://cdn.elifesciences.org/articles/64911/elife-64911-supp2-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64911/elife-64911-transrepform-v3.docx





