Cytokine ranking via mutual information algorithm correlates cytokine profiles with presenting disease severity in patients infected with SARS-CoV-2
Figures
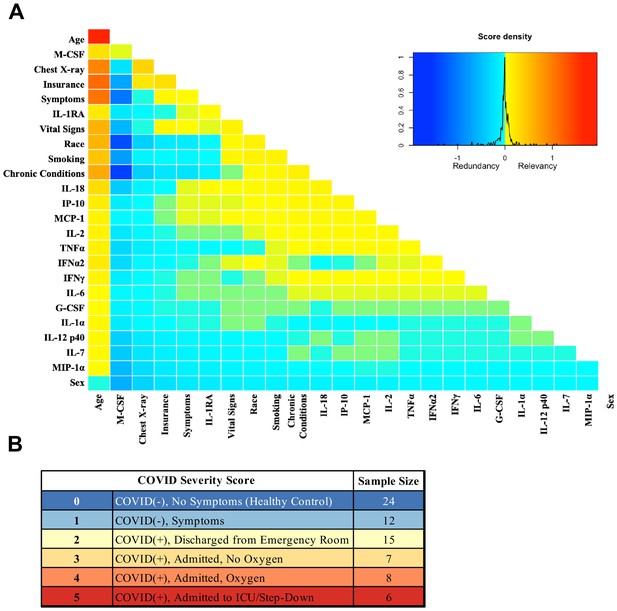
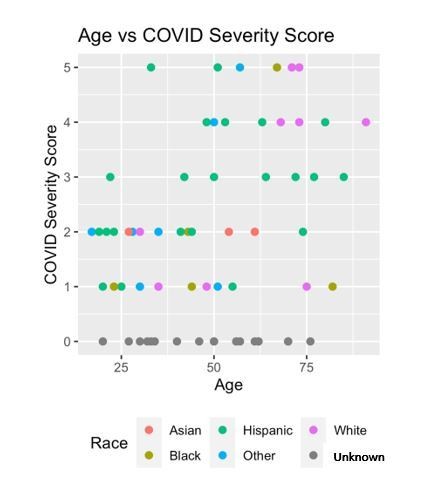
Mutual information COVID Severity Score (CSS) relevancy matrix.
(A) Comprehensive matrix of relevancy to CSS of all variables assessed by mutual information algorithm, relevancy scores computed for not-yet selected variables are shown in each column, and variables are ordered to place maximum local scores on the diagonal, yielding a list in decreasing order from the upper left of variable relevancy. Warmer colors indicate higher relevancy, while cooler colors indicate higher redundancy. (B) COVID Severity Score Table with breakdown of categories as well as sample size per category.
-
Figure 1—source data 1
Mutual algorithm criteria table.
- https://cdn.elifesciences.org/articles/64958/elife-64958-fig1-data1-v2.docx
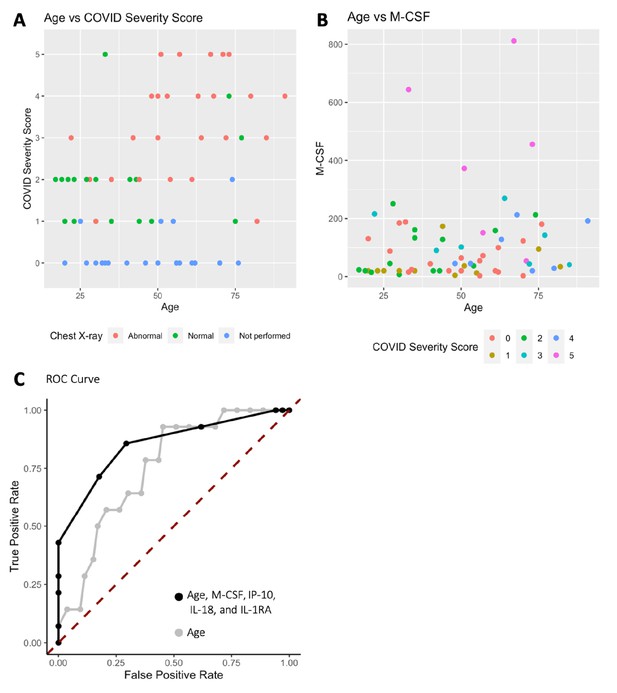
Age, macrophage colony-stimulating factor (M-CSF), and chest x-ray are the most predictive variables for COVID Severity Score (CSS).
(A) The y-axis is CSS, and the x-axis is age in years with points colored by chest x-ray status. (B) The y-axis is M-CSF concentration in pg/mL, and the x-axis is age in years, with points colored based on CSS. See individual legends below graphs. (C) Receiver operating characteristic (ROC) curve predicting CSS 4–5 using age, M-CSF, IP-10, IL-18, and IL-1RA, and ROC curve predicting CSS 4–5 using only age.
-
Figure 2—source data 1
Patient information.
- https://cdn.elifesciences.org/articles/64958/elife-64958-fig2-data1-v2.docx
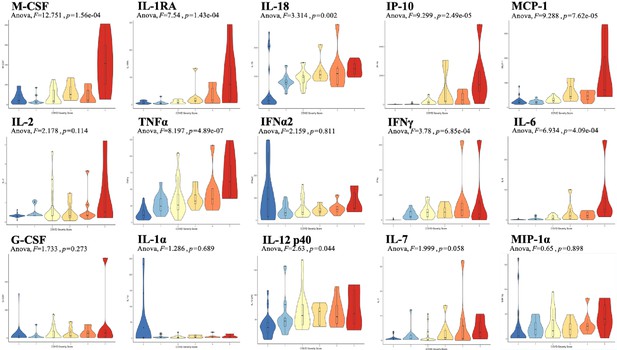
Violin plot representations of cytokine expression levels ordered by COVID Severity Score (CSS).
Cytokines ordered by row from upper left corner based on mutual information relevancy matrix (upper left being most relevant and lower right being least relevant). The x-axis is CSS, and the y-axis is analyte concentration in pg/mL. One-way ANOVA F values and p values are listed on each plot. Source code 1. R code with sections to apply the varrank package to the source data, create a receiver operating characteristic curve, and calculate analysis of variance for each cytokine.
-
Figure 3—source data 1
Raw data.
Source Data Table 1. Mutual algorithm criteria table. *Documented obesity or overweight for height < 99th percentile. Source Data Table 2. Patient information.
- https://cdn.elifesciences.org/articles/64958/elife-64958-fig3-data1-v2.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Biological samples (Human) | Human plasma | Lifespan Brown COVID-19 Biobank | See data table | 48 unique patients |
| Biological samples (Human) | Human plasma | Lee BioSolutions | 991–58-PS | 24 unique patients |
| Commercial assay or kit | MILLIPLEX MAP Human Immunology Multiplex Assay | Millipore Sigma | 15-Plex # HCYTA-60K | |
| Software, algorithm | R | R Project for Statistical Computing | RRID:SCR_001905 | |
| Other | Luminex 100/200 System assay platform | Thermo Fisher Scientific | https://www.luminexcorp.com/luminex-100200 |
Additional files
-
Source code 1
Cytokine Mutual Information R Script.
- https://cdn.elifesciences.org/articles/64958/elife-64958-code1-v2.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64958/elife-64958-transrepform-v2.docx