Evolution of cytokine production capacity in ancient and modern European populations
Figures
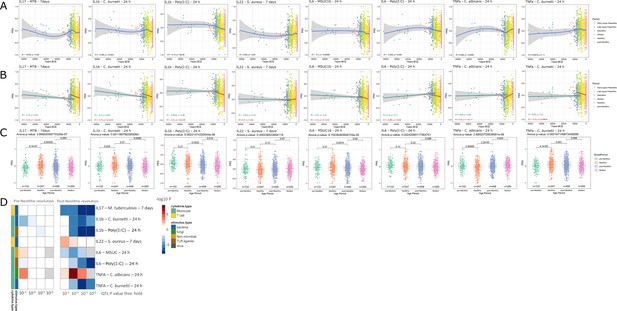
Distribution of PRS of cytokine response QTLs across time.
(A–C) Polygenic risk score (PRS) models based on quantitative trait loci (QTLs) at a p-value threshold of 1e-5. Gray area around regression lines represents the 95% confidence interval. (A) LOESS regression models showing the changing differences in PRS across time. (B) Dual linear models showing the difference in trends before and after the Neolithic revolution. (C) Boxplots showing the difference in mean PRS between adjacent broad time periods using a t-test and the overall difference in means using analysis of variance (ANOVA). (D) Heatmap showing the consistency in regression trends before and after the Neolithic revolution using different QTL inclusion thresholds (1e-3 to 1e-6). Intracellular organisms that can cause zoonotic disease: Mycobacterium tuberculosis, Coxiella burnetti. Extracellular organisms: Staphylococcus aureus, Candida albicans. MSUC: monosodium urate crystals. PolyIC: polyinosinic:polycytidylic acid.
Correlation between cytokine PRS and time.
Missing genotype rates ranged through 0.96, 0.9, 0.8, and 0.7. Quantitative trait locus (QTL) p-values for variants included in our polygenic risk score (PRS) models ranged through 10−3, 10−4, 10−5, and 10−6. The color key indicates the range of -log10 p-values of the Pearson correlation between PRS and time. Red and blue indicate positive and negative association, respectively.
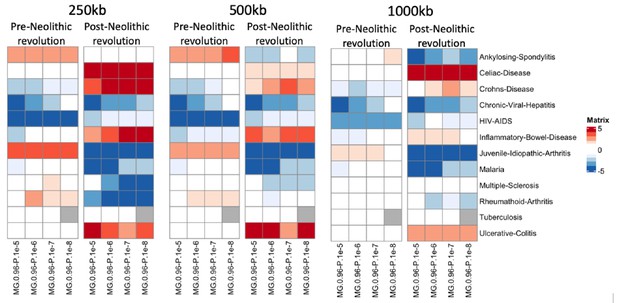
PRS correlation pre- and post-Neolithic revolution using polygenic scores calculated at various window sizes.
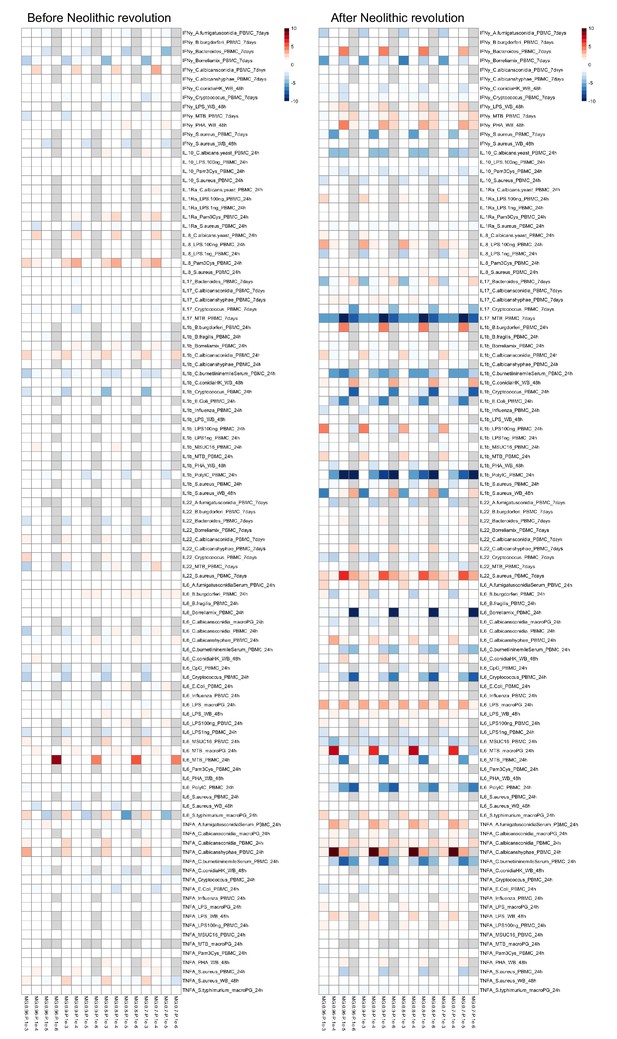
Heatmaps showing changes in polygenic risk scores (PRS) for immune-related complex traits before and after the Neolithic revolution are shown using window sizes 250 kb, 500 kb, and 1000 kb around lead single-nucleotide polymorphisms (SNPs).
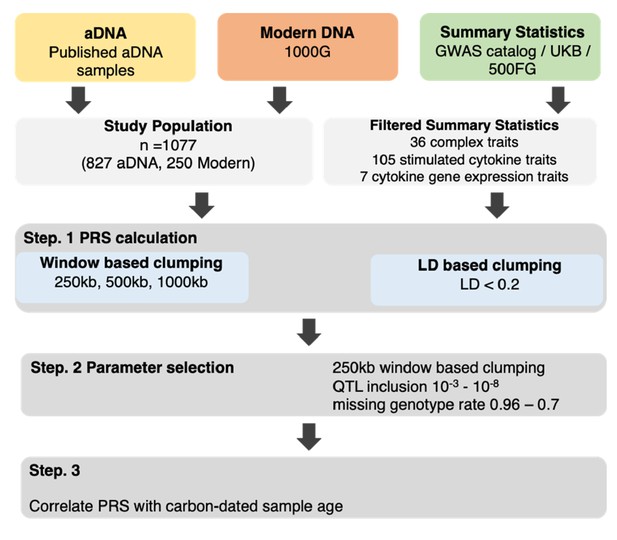
aDNA and modern DNA samples of European individuals were used in combination with summary statistics from predominantly.
European populations to calculate PRS of immune-related traits. This was done at various threshold combinations before correlating the scores with the sample age.
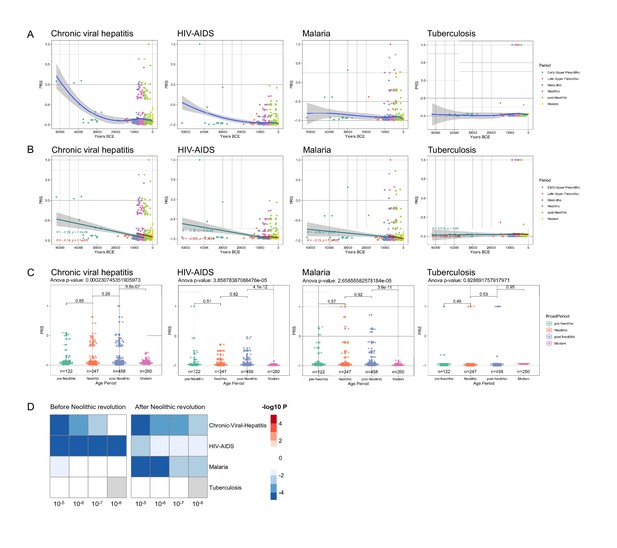
Distribution of PRS of infectious diseases across time.
(A–C) Polygenic risk score (PRS) models based on a p-value threshold of 1e-5. Gray area around regression lines represents the 95% confidence interval. (A) LOESS regression models showing the changing differences in PRS across time. (B) Dual linear models showing the difference in trends before and after the Neolithic revolution. (C) Boxplots showing the difference in mean PRS between adjacent broad time periods using a t-test and the overall difference in means using analysis of variance (ANOVA). (D) Heatmap showing the consistency in regression trends before and after the Neolithic revolution using different quantitative trait locus (QTL) inclusion thresholds (1e-5 to 1e-8).
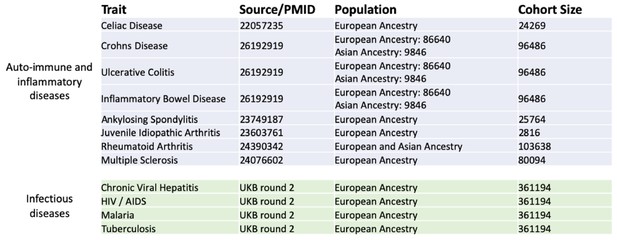
GWAS summary statistics and cohorts used for PRS calculation.
Traits were separated into two categories: auto-immune and inflammatory disease-related traits, and infectious disease-related traits. Genome-wide association studies (GWAS) summary statistics from predominantly European populations were selected.
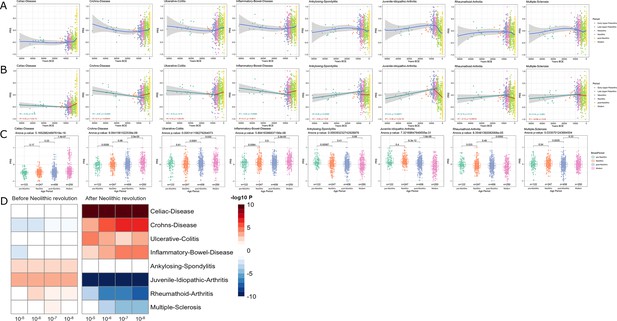
Distribution of PRS of inflammatory diseases across time.
(A–C) Polygenic risk score (PRS) models based on a p-value threshold of 1e-5. Gray area around regression lines represents the 95% confidence interval. (A) LOESS regression models showing the changing differences in PRS across time. (B) Dual linear models showing the difference in trends before and after the Neolithic revolution. (C) Boxplots showing the difference in mean PRS between adjacent broad time periods using a t-test and the overall difference in means using analysis of variance (ANOVA). (D) Heatmap showing the consistency in regression trends before and after the Neolithic revolution using different quantitative trait locus (QTL) inclusion thresholds (1e-5 to 1e-8).
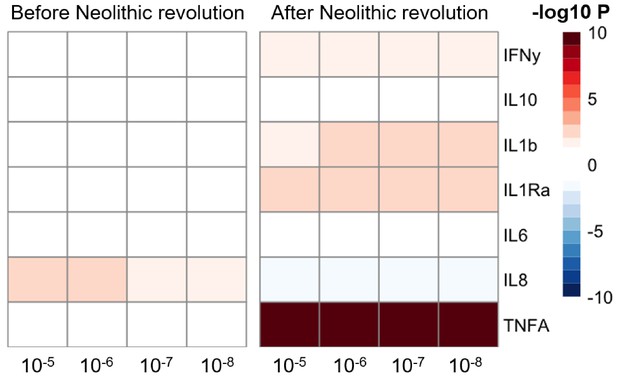
Changes in cytokine gene expression PRS across time before and after the Neolithic revolution using different QTL thresholds (1e-5 to 1e-8).
Missing genotype rates ranged through 0.96, 0.9, 0.8, and 0.7. Quantitative trait locus (QTL) p-values for variants included in our polygenic risk score (PRS) models ranged through 10−3, 10−4, 10−5, 10−6, 10−7, and 10−8. The color key indicates the range of -log10 p-values of the Pearson correlation between PRS and time. Red and blue indicate positive and negative association, respectively.
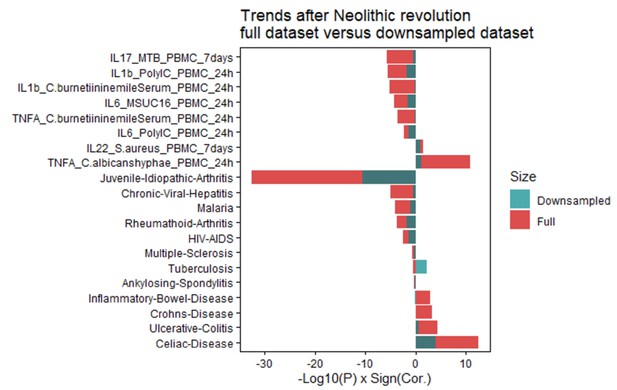
Robustness of correlation coefficients post-Neolithic is independent of the sample size.
Changes in polygenic risk scores (PRS) following the Neolithic revolution remain consistent after down-sampling samples from after the start of the Neolithic period to the same amount as samples before the Neolithic period. The -log10 p-value multiplied by the sign of the correlation coefficient is shown; the results for the down-sampled set are shown in blue and those for the full dataset are shown in red.
Additional files
-
Supplementary file 1
Stimulated cytokine expression combinations at different timepoints.
(a) Overview of the stimulus, cytokine, and time-point combinations. In total, 105 unique stimulated cytokine traits were available using various types of stimuli measuring both the innate and the adaptive immune response. (b) Tests for selection as opposed to the genetic drift were performed between populations from adjacent time periods. A two-tailed test was used to determine whether the mean trait Fst between pre-Neolithic–Neolithic, Neolithic–post-Neolithic, and post-Neolithic–Modern samples was significantly different compared to 10,000 random LD- and MAF-matched mean Fsts calculated using the same amount of single-nucleotide polymorphisms (SNPs).
- https://cdn.elifesciences.org/articles/64971/elife-64971-supp1-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64971/elife-64971-transrepform-v1.docx





