Mitochondrial respiration contributes to the interferon gamma response in antigen-presenting cells
Figures
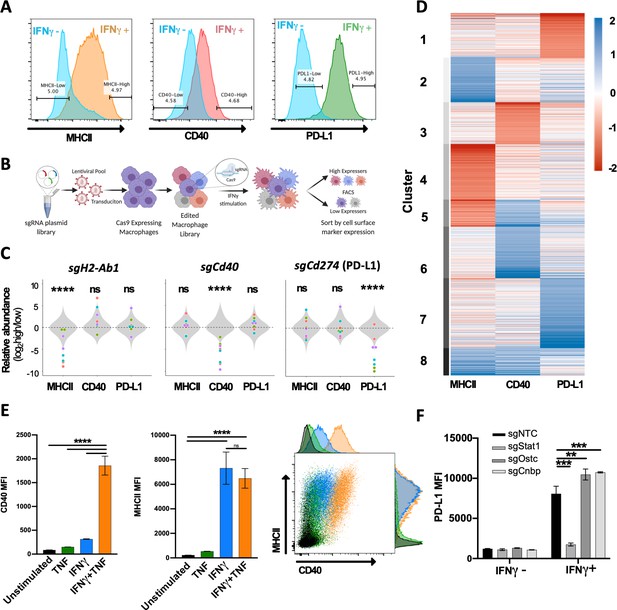
Forward genetic screen to identify regulators of the IFNγ response.
(A) Representative histograms of the three selected cell surface markers targeted in macrophage CRISPR screens: MHCII, CD40, and PD-L1. Blue histograms indicate expression of each marker in unstimulated macrophages, and alternatively colored histograms show expression following 24 hr stimulation with recombinant murine IFNγ (10 ng/mL). Gates used for sorting ‘high’ and ‘low’ populations are shown. (B) Schematic of CRISPR screens. (C) Relative enrichment of select positive control (points) and all 1000 non-targeting control (NTC) sgRNAs (gray distribution) are plotted as a function of their log2 fold enrichment (‘high’ vs. ‘low’ bins). Data are from both replicate selections for each sgRNA (sgRNA denoted by shape). (D) Heatmap of β scores from CRISPR analysis, ordered according to k-means clustering (k = 8) of the 5% most enriched or depleted genes in each screen. (E) Macrophages were stimulated for 24 hr with TNF (25 ng/mL), IFNγ (10 ng/mL), or both TNF and IFNγ. Mean fluorescence intensity (MFI) of CD40 and MHCII was quantified by flow cytometry. Data are mean ± standard deviation for three biological replicates. Representative scatter plot from two independent experiments is provided. (F) Macrophages transduced with sgRNA targeting Stat1, Ostc, Cnbp, or a NTC were cultured with or without IFNγ for 24 hr, and cell surface expression of PD-L1 (MFI) was quantified by flow cytometry. For each genotype, data are the mean of cell lines with two independent sgRNAs ± standard deviation. Data are representative of three independent experiments. Statistical testing in panel (C) was performed with Tukey’s multiple comparisons test. Within each screen, the sgRNA effects for each gene were compared to the distribution of NTC sgRNAs. Statistical testing in panels (E) and (F) was performed by one-way ANOVA with Holm–Sidak multiple comparisons correction. p-Values of 0.05, 0.01, 0.001, and 0.001 are indicated by *, **, *** and ****, respectively.
-
Figure 1—source data 1
Whole-genome profiling of macrophage MHCII, CD40, and PD-L1 expression.
- https://cdn.elifesciences.org/articles/65109/elife-65109-fig1-data1-v2.xlsx
-
Figure 1—source data 2
k-means clustering of CRISPR-KO beta scores for regulators of the IFNγ response.
- https://cdn.elifesciences.org/articles/65109/elife-65109-fig1-data2-v2.xlsx
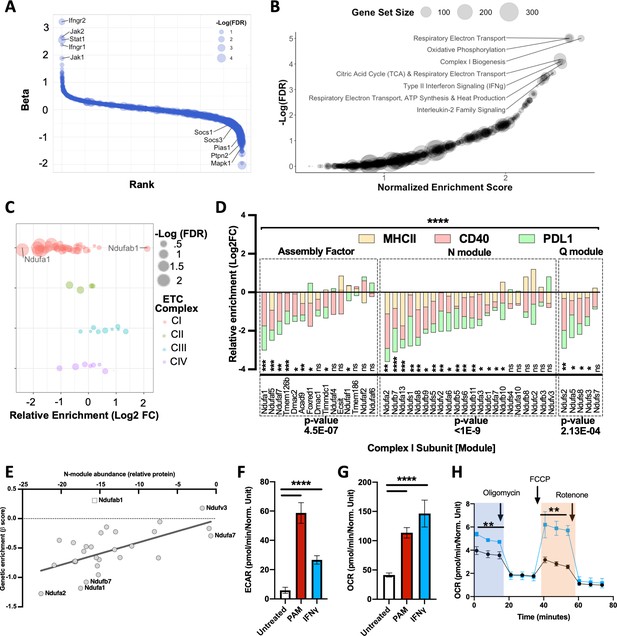
Global analysis of knockout (KO) libraries implicates mitochondrial complex I is a positive regulator of the IFNγ response.
(A) Rank plot of the combined analysis for all genome-wide KO screens. Gene ranks (x-axis) were determined by maximal likelihood estimation (MLE). Known positive (left) and negative (right) regulators of IFNγ-mediated signaling are highlighted. The q-value (false discovery rate [FDR]) for each gene is indicated by dot size (-log10 FDR). (B) Gene set enrichment analysis (GSEA) is based on the ranked list of positive regulators. Non-redundant pathways with a normalized enrichment score (NES) exceeding 2.0 and an FDR below 0.025 are labeled. (C) Relative enrichment (log2 fold change between ‘high’ and “low” bins) of genes that comprise the mitochondrial respirasome (GeneOntology 0005746) and were targeted in the CRISPR KO library. Respirasome components are grouped by electron transport chain (ETC) complex. FDR is based on MAGeCK-MLE. (D) Screen-specific enrichment score is plotted for complex I structural subunits and assembly factors. The statistical enrichment of a gene (e.g., Ndufa1) or module (e.g., N) was calculated using a binomial distribution function to calculate the probability that observed sgRNAs under examination would be depleted or enriched given the expected median probability. p-Values of 0.05, 0.01, 0.001, and 0.001 are indicated by *, **, ***, and ****, respectively. (E) Correlation between the relative effect of each complex I subunit on the structural integrity of the N-module (x-axis) with the relative requirement of each complex I subunit for the IFNγ response (y-axis; β score, as in panel D). The Pearson correlation coefficient (r) was calculated to be 0.6452 (95% confidence interval 0.3584–0.8207); p-value=0.0002. As Ndufab1 (empty square) is an essential gene, its detection in the library indicates editing did not eliminate function; therefore, it was excluded from correlation analysis. (F) Following stimulation with IFNγ or PAM, extracellular acidification rate (ECAR)and (G) Oxygen consumption rate (OCR) values were measured by Seahorse in primary bone marrow-derived macrophages (BMDMs). Basal OCR and ECAR were determined 24 hr after stimulation with 10 ng/mL IFNγ or 200 ng/mL PAM. ****p<0.0001 by one-way ANOVA. (H) In a parallel experiment, the indicated chemical modulators were added to resting (Black) or IFNγ-activated (Blue) BMDMs at the indicated time points after initiating metabolic monitoring and the OCR response was monitored. Basal OCR (blue box) and maximal OCR (red box) are highlighted (right panel). **p<0.01 by two-tailed t-test.
-
Figure 2—source data 1
Whole-genome profiling of the IFNγ response in macrophages.
- https://cdn.elifesciences.org/articles/65109/elife-65109-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Gene set enrichment analysis (GSEA) to identify pathways that regulate the IFNγ response in macrophages.
- https://cdn.elifesciences.org/articles/65109/elife-65109-fig2-data2-v2.xlsx
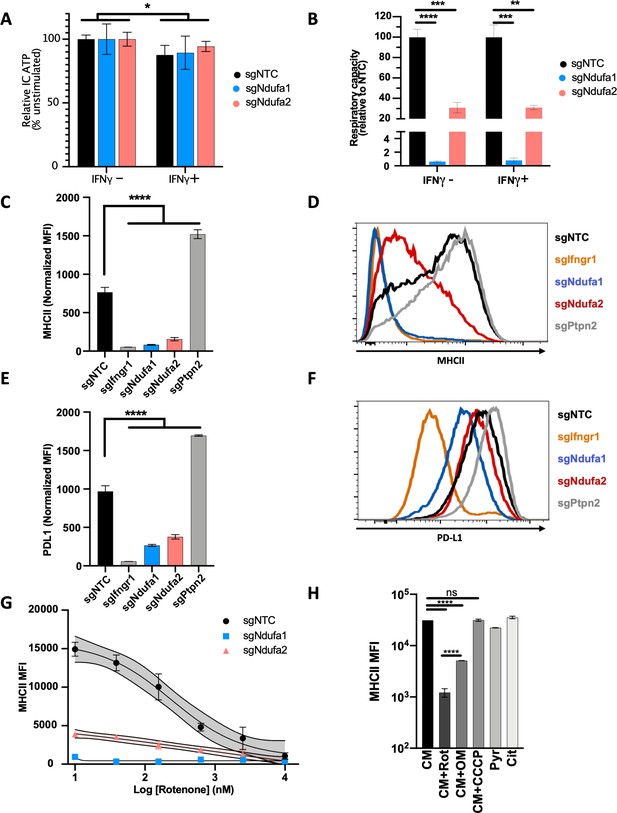
Complex I is necessary for IFNγ-induced MHCII and PD-L1 expression.
Metabolic phenotypes in macrophage mutants were confirmed by measuring intracellular (IC) ATP abundance following culture in media containing only (A) glucose or (B) pyruvate. Values are normalized to the average respiratory capacity of non-targeting control macrophages (NTC) and are the mean ± standard deviation for four biological replicates. Statistical testing within each condition (with or without IFNγ for 24 hr) was performed by one-way ANOVA with Dunnett’s multiple comparisons correction. (C–F) NTC, positive control (sgIfngr1 and sgPtpn2), and complex I mutant (sgNdufa1 and sgNdufa2) macrophages were stimulated for 24 hr with recombinant murine IFNγ. Plotted values in (C) and (E) are the geometric mean fluorescence intensity (MFI) for a given mutant normalized to an internal control present in each well; for each gene, the data are the mean for two independent sgRNAs ± standard deviation. Representative histograms are provided in (D) and (F). Data are representative of >5 independent experiments. (G) MHCII MFI of macrophages stimulated with IFNγ and treated with rotenone at the indicated concentrations for 24 hr. Mean ± standard deviation for two biological replicates are shown. Data are representative of four independent experiments. (H) Left: MHCII MFI on macrophages cultured in complete media (CM) and stimulated with IFNγ and the indicated inhibitors for 24 hr. Right: MHCII MFI on macrophages cultured in CM or media containing only pyruvate (Pyr) or citrate (Cit) stimulated with IFNγ for 24 hr. Mean ± standard deviation for two or three biological replicates is indicated. Data are representative of four independent experiments. Statistical testing was performed by one-way ANOVA with Tukey’s correction for multiple hypothesis testing. p-Values of 0.05, 0.01, 0.001, and 0.001 are indicated by *, **, ***, and ****, respectively.
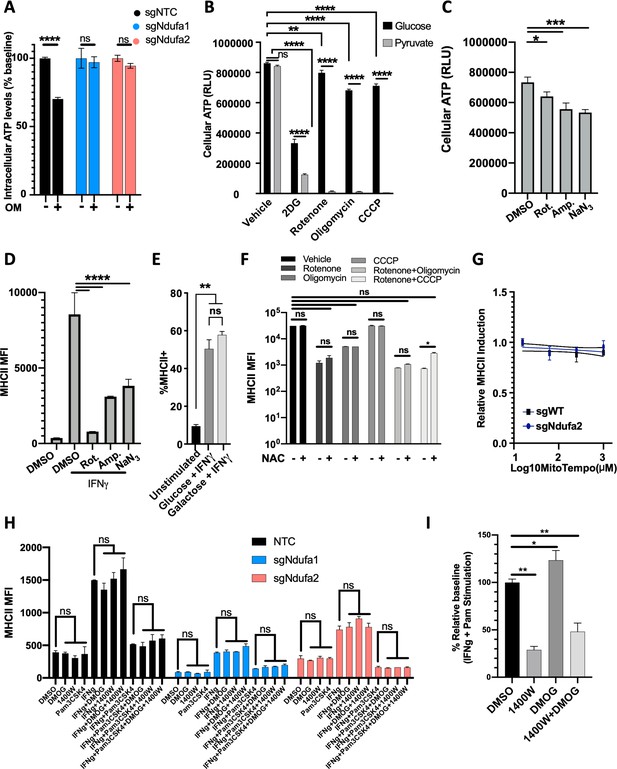
The complex I dependency of IFNγ signaling is independent of reactive oxygen and nitrogen radicals, and Hif-1α.
(A) sgNTC, sgNdufa1, and sgNdufa2 cells cultured in complete media and treated with or without oligomycin (2.5 μM) for 4 hr. Relative ATP levels were determined as in Figure 2A. (B) Intracellular ATP levels quantified as relative light units (RLUs) using CellTiter-Glo2.0 (Promega) for macrophages in specified growth conditions for 4 hr. Concentrations of carbon source and inhibitors are indicated in Materials and methods. (C) sgNTC macrophages were grown in glucose then treated with DMSO, rotenone (10 µM), antimycin A (10 µM), and sodium azide (10 mM) for 24 hr, and the reduction in ATP was quantified using CellTiter-Glo2.0. (D) In parallel, sample cells were left resting or were stimulated with IFNγ and the surface expression of MHCII was quantified by flow cytometry. (E) Macrophages were cultured in either glucose or galactose and stimulated with IFNγ for 24 hr. Following stimulation, the proportion of cells with MHCII expression was determined by flow cytometry. (F) Macrophages were cultured in conditions as described in Figure 4H. For each condition, cells were stimulated with IFNγ or IFNγ and N-acetyl cysteine (NAC) for 24 hr after which cell surface levels of MHCII were quantified. (G) Control or complex I mutant (sgNdufa2) macrophages were stimulated with IFNγ for 24 hr with increasing doses of mitochondrial reactive oxygen species scavenger MitoTEMPO. For each concentration, values are plotted as a fold change relative to no scavenger; Mean ± standard deviation for two biological replicates of each condition. (H) Control or complex I-deficient macrophages were stimulated with IFNγ for 24 hr with or without the addition of dimethyloxalylglycine (DMOG) or 1400W. Following stimulation, the proportion of cells with MHCII expression was determined by flow cytometry. (I) Nitric oxide was measured using Griess Reagent System (Promega) from cell supernatants following stimulation with IFNγ and Pam3CSK4 for 24 hr with or without the addition of DMOG or 1400W. Relative nitric oxide levels were calculated as a percent relative to control (IFNγ and Pam3CSK4 with DMSO). All data are representative of at least two independent experiments. Statistical testing was performed using one-way ANOVA with Holm–Sidak multiple comparison correction. p-Values of 0.05, 0.01, 0.001, and 0.001 are indicated by *, **, ***, and ****, respectively.
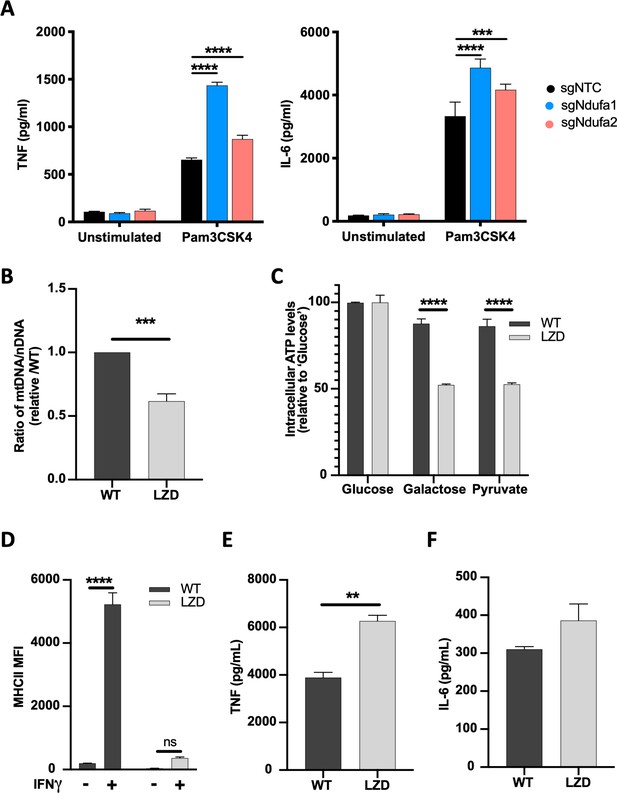
Diminished mitochondrial function specifically limits IFNγ-dependent responses.
(A) TNF and IL-6 production by non-targeting control (NTC) or complex I mutant macrophages stimulated with Pam3CSK4 for 24 hr was determined by ELISA. Statistical testing between mutant and NTC macrophages from triplicate samples was performed by ANOVA with Dunnett’s correction for multiple comparisons. Data are representative of two independent experiments. (B) qPCR determination of relative mitochondrial genomes present per nuclear genome in macrophages cultured in vehicle (WT) or 50 µg/mL linezolid (LZD). Ct values were normalized to reference nuclear gene hexokinase 2 (Hk2) and plotted as abundance relative to WT. Data were analyzed by two-way unpaired t-test. (C) ATP abundance in control or LZD-conditioned macrophages cultured in 10 mM glucose, galactose, or pyruvate. ATP values normalized to mean of 10 mM glucose and plotted as percent. Mean ± standard deviation for two biological replicates of each condition. Differences were tested by two-way ANOVA using the Sidak method to correct for multiple hypothesis testing. (D) Mean fluorescence intensity (MFI) of MHCII was determined by flow cytometry on control or LZD-conditioned macrophages following 24 hr stimulation with IFNγ. Mean ± standard deviation for two biological replicates of each condition and representative of two independent experiments. Differences were tested by two-way ANOVA using Tukey’s method to correct for multiple hypothesis testing. (E, F) Secretion of TNF and IL-6 in WT and LZD-conditioned macrophages following Pam3CSK4 stimulation for 6 hr was quantified by ELISA. Mean ± standard deviation for three biological replicates of each condition and two independent experiments. Data were analyzed by two-way unpaired t-test. p-Values of 0.05, 0.01, 0.001, and 0.001 are indicated by *, **, ***, and ****, respectively.
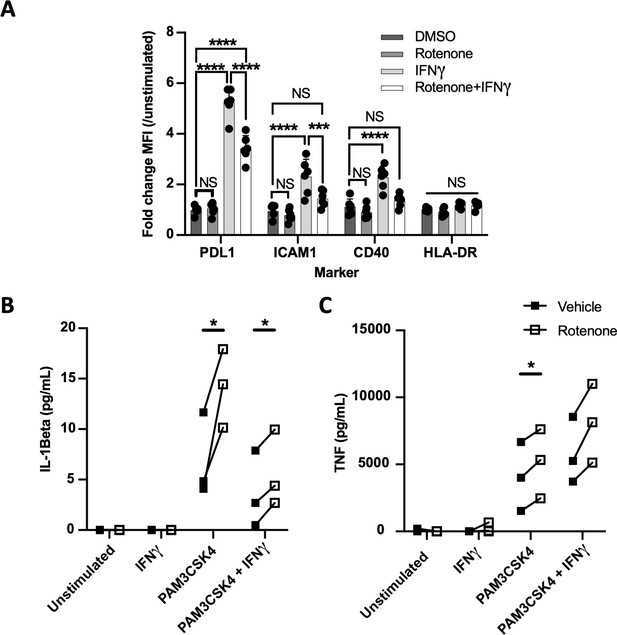
Complex I is specifically required for IFNγ signaling in human cells.
(A) CD14+ monocytes from healthy human donors were differentiated into macrophages with GM-CSF. Mean fluorescence intensity (MFI) of cell surface markers PD-L1, ICAM1, CD40, and HLA-DR was determined by flow cytometry following stimulation with IFNγ and/or inhibition of complex I with rotenone (10 µM) for 24 hr. Data are representative of two independent experiments, and values are normalized to donor-specific unstimulated/vehicle control. Mean ± standard deviation for six biological replicates of each condition. Differences were tested by two-way ANOVA using the Sidak–Holm method to correct for multiple hypothesis testing. (B, C) Quantification of IL-1β and TNF production from primary human macrophages, measured by ELISA from cell supernatants following stimulation. Lines connect values for individual donors treated with vehicle (DMSO, black squares) or rotenone (empty squares). Differences were tested by repeated measures two-way ANOVA using the Sidak–Holm method to correct for multiple hypothesis testing. p-Values of 0.05, 0.01, 0.001, and 0.001 are indicated by *, **, ***, and ****, respectively.
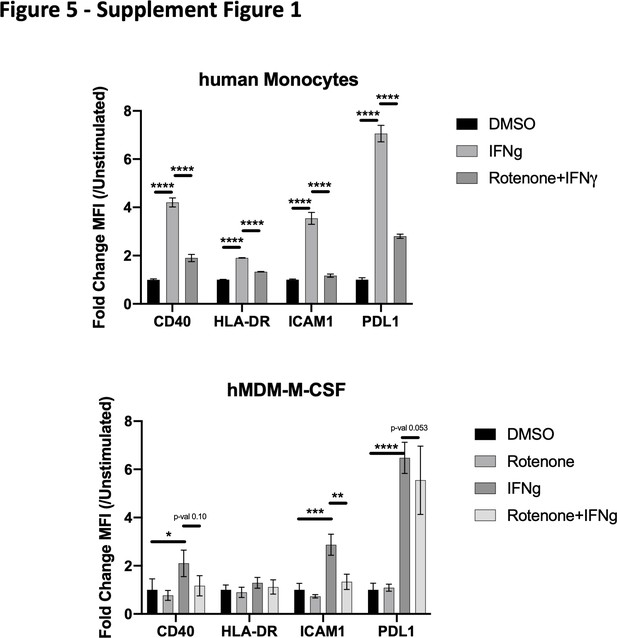
Complex I is specifically required for IFNγ signaling in diverse human myeloid cells.
CD14+ monocytes were isolated from healthy human donors and stimulated directly (upper panel) or subsequent to differentiation into macrophages with M-CSF (lower panel). Mean fluorescence intensity (MFI) of cell surface markers PD-L1, ICAM1, CD40, and HLA-DR was determined by flow cytometry following stimulation with IFNγ and/or inhibition of complex I with rotenone (10 µM) for 24 hr. Data are representative of two independent experiments, and values are normalized to donor-specific unstimulated/vehicle control. Mean ± standard deviation for six biological replicates of each condition. Differences were tested by two-way ANOVA using the Sidak–Holm method to correct for multiple hypothesis testing. p-Values of 0.05, 0.01, 0.001, and 0.001 are indicated by *, **, ***, and ****, respectively.
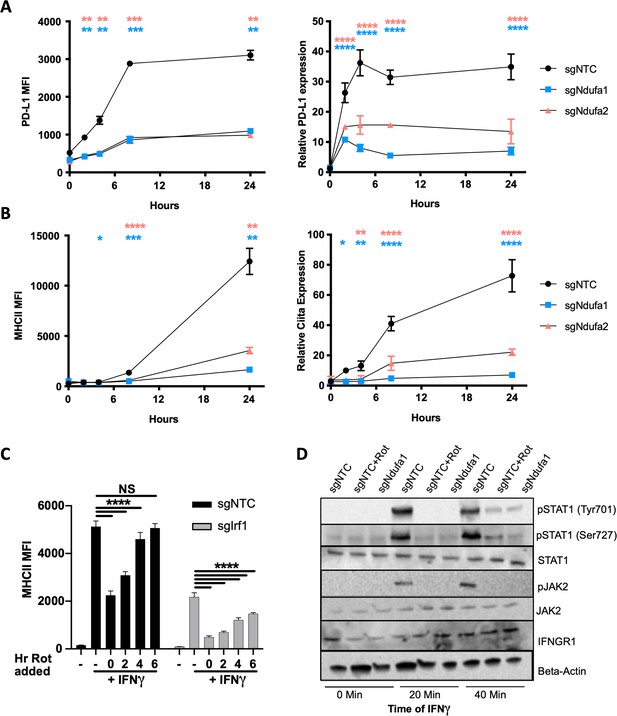
Complex I inhibition reduces IFNγ receptor activity.
(A) PD-L1 transcript was quantified by qRT-PCR using ΔΔCt relative to β-actin in macrophages of the indicated genotype after stimulation with 10 ng/mL IFNγ. PD-L1 mean fluorescence intensity (MFI) was determined at the same time points by flow cytometry. (B) Ciita transcript was quantified by qRT-PCR using ΔΔCt relative to β-actin Gapdh in macrophages of the indicated genotype after stimulation with 10 ng/mL IFNγ. MHCII MFI was determined at the same time points by flow cytometry. Data shown are from biological triplicate samples with technical replicates for RT-PCR experiments and are representative of two independent experiments. (C) sgNTC (left) or sgIrf1 (right) macrophages were cultured for 24 hr with or without IFNγ stimulation. At 2 hr intervals post-IFNγ stimulation, rotenone was added. After 24 hr of stimulation, cells were harvested and surface expression of MHCII (MFI) was quantified by flow cytometry. Data are mean ± standard deviation for three biological replicates and are representative of two independent experiments. Statistical testing was performed by one-way ANOVA with Tukey’s correction for multiple hypothesis testing. (D) Control (non-targeting control [NTC]) or sgNdufa1 macrophages were stimulated with IFNγ for the indicated times while NTC macrophages were pretreated with 10 μM rotenone for 2 hr prior to IFNγ stimulation. Cell lysates analyzed by immunoblot for STAT1 abundance and phosphorylation (Y701 and S727), JAK2 abundance and phosphorylation (Y1007/8), and IFNGR1. β-Actin was used as a loading control. Data are representative of three independent experiments. Results shown are from a single experiment analyzed on three parallel blots. p-Values of 0.05, 0.01, 0.001, and 0.001 are indicated by *, **, ***, and ****, respectively.
-
Figure 6—source data 1
Raw blots.
- https://cdn.elifesciences.org/articles/65109/elife-65109-fig6-data1-v2.zip
-
Figure 6—source data 2
Labeled raw blots.
- https://cdn.elifesciences.org/articles/65109/elife-65109-fig6-data2-v2.zip
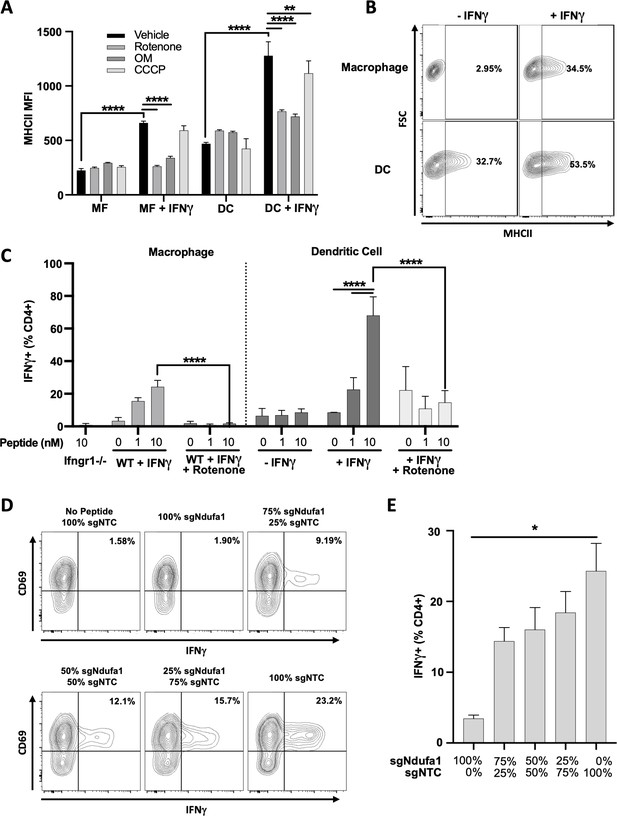
Mitochondrial respiration in antigen-presenting cells (APCs) is required for IFNγ-dependent T cell activation.
(A) Cell surface expression of MHCII (mean fluorescence intensity [MFI]) in macrophages (MF) or dendritic cells (DCs) derived from conditionally immortalized progenitor lines. IFNγ was added for 24 hr where indicated. Cells were treated with vehicle (DMSO), rotenone (10 µM), oligomycin (OM, 2.5 µM), or carbonyl cyanide m-chlorophenyl hydrazone (CCCP) concurrent with IFNγ. Data are three biological replicates and are representative of at least two independent experiments. (B) Contour plot of macrophage (top row) or DC (bottom row) MHCII expression in the absence of (left column) or following (right column) stimulation with IFNγ for 24 hr. Representative samples were selected from (A). The percent MHCII positive are indicated for each of the conditions. (C) CD4+ T cell activation as measured by the percent of live cells positive for IFNγ by intracellular cytokine staining. Prior to co-culture with T cells, APCs were stimulated with the indicated combinations of IFNγ (10 ng/mL), and/or rotenone (10 µM) for 24 hr. After washing and pulsing with ESAT-61–15 at the indicated concentrations (nm), T cells were added to APCs at an effector to target (E:T) ratio of 1:1 and co-cultured for a total of 5 hr. Data are representative of two independent experiments. Data are mean ± standard deviation for three biological replicates. Statistical testing was performed by one-way ANOVA with Tukey’s correction for multiple hypothesis testing. (D, E) sgNdufa1 or non-targeting control (NTC) macrophages were differentiated from immortalized progenitors and mixed at the ratios indicated (labeled as percent of knockout [KO] cells). Mixed cultures were stimulated with IFNγ for 24 hr, peptide loaded, and co-cultured with CD4+ T cells (E:T 1:1). Production of IFNγ was measured by ICS and quantified as the percent of cells positive for staining by flow cytometry. Representative contour plots (D) and quantification (E) of the experiment are shown. Data shown are for biological triplicate samples and are representative of two independent experiments. p-Values of 0.05, 0.01, 0.001, and 0.001 are indicated by *, **, ***, and ****, respectively.
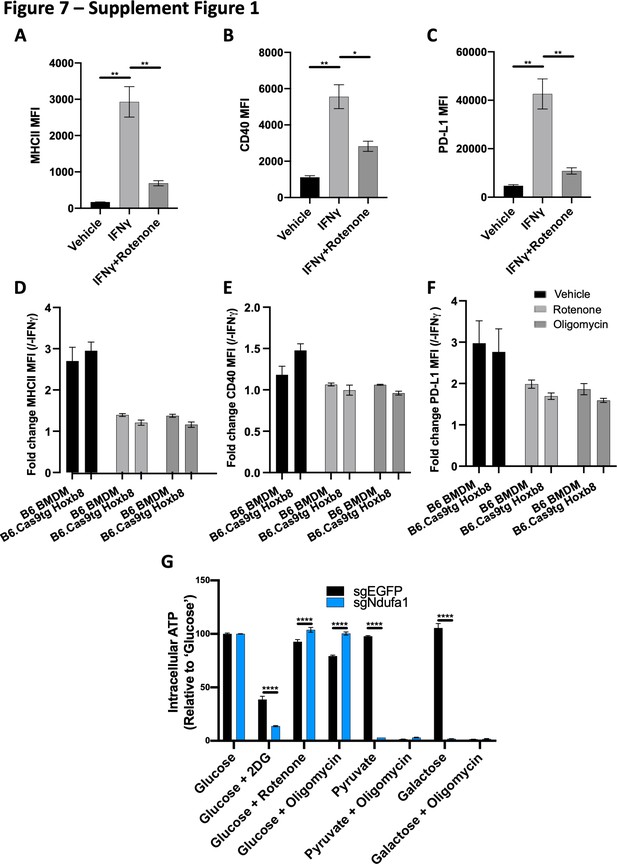
The metabolic dependencies for IFNγ signaling are preserved in macrophages derived from myeloid progenitor lines.
(A–C) Myeloid progenitors cells were conditionally immortalized by transducing murine bone marrow with an estrogen-dependent Hoxb8 transgene that maintains stem-like properties. Following differentiation of progenitors into macrophages using M-CSF-enriched conditioned media, macrophages were stimulated with IFNγ with or without rotenone. 24 hr after stimulation, cell surface levels of (A) MHCII, (B) CD40, and (C) PD-L1 were quantified by flow cytometry. Data are representative of three independent experiments and are the mean ± standard deviation for two biological replicates. Statistical testing was performed by one-way ANOVA with Tukey’s correction for multiple hypothesis testing. (D–F) As in panels (A–C), macrophages from either immortalized macrophage progenitors or primary bone marrow were stimulated with IFNγ with or without rotenone or oligomycin. 24 hr after stimulation, cell surface levels of (D) MHCII, (E) CD40, and (F) PD-L1 were quantified by flow cytometry. (G) Wild-type or ΔNdufa1 macrophages derived from Hoxb8-immortalized bone marrow progenitors were cultured in the specified media and inhibitor condition before total intracellular ATP was quantified by CellTiter-Glo2.0. For each genotype, values are relative to ‘glucose’ control. Mean ± standard deviation for two biological replicates of each condition. p-Values of 0.05, 0.01, 0.001, and 0.001 are indicated by *, **, ***, and ****, respectively.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Mus musculus) | L3-Cas9+ | Kiritsy and Ankley et al. (co-submitted) | Primary BMDMs immortalized with J2 virus were transduced with Cas9 and single cell cloned | |
| Cell line (M. musculus) | EGFP-Cas9 iBMDMs | This paper | Primary BMDMs from Jackson Stock 026179 were immortalized with J2 virus | |
| Cell line (M. musculus) | sgNdufa1 EGFP-Cas9 iBMDMs | This paper | Cas9+ iBMDMs were transduced with Ndufa1 sgRNA | |
| Cell line (M. musculus) | sgNdufa2 EGFP-Cas9 iBMDMs | This paper | Cas9+ iBMDMs were transduced with Ndufa2 sgRNA | |
| Cell line (M. musculus) | Cas9+ C57BL/6J estradiol-inducible HoxB8 progenitors | This paper | Myeloid progenitors from Jackson stock 026179 were immortalized with HoxB8 retrovirus and maintained with 10 µM estradiol | |
| Cell line (M. musculus) | sgNdufa1 C57BL/6J estradiol-inducible HoxB8 progenitors | This study | Cas9+ HoxB8 cells were transduced with Ndufa1 sgRNA | |
| Cell line (M. musculus) | Cas9+ C57BL/6J estradiol-inducible HoxB8 progenitors | This paper | Myeloid progenitors from Jackson stock 003288 were immortalized with HoxB8 retrovirus and maintained with 10 µM estradiol | |
| Strain, strain background (M. musculus) | C57BL6J | Jackson Laboratories | Stock 000664 | |
| Strain, strain background (M. musculus) | C7 TCR-transgenic mice (specific for ESAT-6 antigen) | PMID:18779346 | Mice were donated to and maintained by the Behar laboratory | |
| Peptide, recombinant protein | ESAT-6 peptide (MTEQQW NFAGIEAAA) | New England Peptide | ||
| Recombinant DNA reagent | sgOpti | Addgene | RRID:Addgene_ 85681 | |
| Recombinant DNA reagent | sgOpti with blasticidin and zeocyin selection | Kiritsy and Ankley et al. (co- submitted) | sgOpti (RRID 85681) was modified with bacterial selection replaced with zeocyin and mammalian selection replaced with blasticidin | |
| Recombinant DNA reagent | sgOpti with hygromycin and kanamycin selection | Kiritsy and Ankley et al. (co-submitted) | sgOpti (RRID 85681) was modified with bacterial selection replaced with kanamycin and mammalian selection replaced with hygromycin | |
| Recombinant DNA reagent | VSVG | Addgene | RRID:Addgene_8454 | |
| Recombinant DNA reagent | psPax2 | Addgene | RRID:Addgene_12260 | |
| Antibody | MHCII-PE, clone M5/114.15.2 (rat monoclonal) | BioLegend | RRID:AB_313323 | FC (1:800) |
| Recombinant DNA reagent | Mouse CRISPR KO pooled library (BRIE) | Addgene | RRID:Addgene_7363 | |
| Chemical compound, drug | Rotenone | Sigma | Cat# R8875 | |
| Chemical compound, drug | Oligomycin | Cayman | Cat# 11342 | |
| Chemical compound, drug | CCCP | Cayman | Cat# 25458 | |
| Chemical compound, drug | 1400W | Cayman | Cat# 81520 | |
| Chemical compound, drug | N-Acetyl cysteine | Cayman | Cat# 20261 | |
| Chemical compound, drug | Dimethyloxallyl glycine (DMOG) | Cayman | Cat# 71210 | |
| Chemical compound, drug | UK5099 | Cayman | Cat# 16,980 | |
| Chemical compound, drug | 2-Deoxyglucose (2-DG) | Cayman | Cat# 14325 | |
| Chemical compound, drug | MitoTEMPO hydrate | Cayman | Cat# 16621 | |
| Chemical compound, drug | Sodium azide | Sigma | Cat# S2002 | |
| Chemical compound, drug | Antimycin A | Sigma | Cat# A8674 | |
| Chemical compound, drug | Pam3CSK4 | Invivogen | Cat# tlrl-pms | |
| Chemical compound, drug | Linezolid (LZD) | Gift from Clifton Barry (used here PMID:32477361) | ||
| Antibody | Purified anti-STAT1 antibody clone A15158C (mouse monoclonal) | BioLegend | Cat# 603701; RRID:AB_2749867 | WB (1:1000) |
| Antibody | Phospho-Stat1 Tyr701, clone 58D6 (rabbit monoclonal) | Cell Signaling Technology | Cat# 5375; RRID:AB_10860071 | WB (1:1000) |
| Antibody | Purified anti-STAT1 Phospho Ser727 antibody, clone A15158B (mouse monoclonal) | BioLegend | Cat# 686408; RRID:AB_2650782 | WB (1:1000) |
| Antibody | Jak2 XP, clone D2E12 (rabbit monoclonal) | Cell Signaling Technology | Cat# 4040; RRID:AB_10691469 | WB (1:500) |
| Antibody | Phospho-Jak2 (Tyr1007/1008) antibody (unknown) | Cell Signaling Technology | Cat# 3771; RRID:AB_330403 | WB (1:500) |
| Antibody | Biotin anti-mouse CD119, IFNγ Rα chain antibody clone 2E2 (Armenian hamster monoclonal) | BioLegend | Cat# 112803; RRID:AB_2123476 | WB (1:1000) |
| Antibody | Anti-mouse β-actin antibody, clone C4 (mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-51850; RRID:AB_629337 | WB (1:2000) |
| Antibody | CD274-Bv421 clone 10F.9G2 (rat monoclonal) | BioLegend | RRID:AB_10897097 | FC (1:400) |
| Commercial assay or kit | Zombie Aqua Fixable Viability Kit | BioLegend | Cat# 423101 | FC (1:100) |
| Commercial assay or kit | DNeasy Blood and Tissue Kit | Qiagen | Cat# 69504 | |
| Antibody | CD40 APC anti-mouse CD40 antibody, clone 3/23 (rat monoclonal) | BioLegend | Cat# 124611;RRID:AB_1134081 | FC (1:200) |
| Antibody | Human anti-CD54, clone HCD54 (mouse monoclonal) | BioLegend | Cat# 322718; RRID:AB_2248731 | FC (1:400) |
| Antibody | Human anti-CD40 clone 5C3 (mouse monoclonal) | BioLegend | Cat# 334307; RRID:AB_1186060 | FC (1:400) |
| Antibody | Human anti-CD274, B7-H1, PD-L1, clone 29E (mouse monoclonal) | BioLegend | Cat# 329713; RRID:AB_10901164 | FC (1:400) |
| Antibody | Human anti-HLA-DR antibody, clone L243 (mouse monoclonal) | BioLegend | Cat# 307657; RRID:AB_2572100 | FC (1:400) |
| Antibody | Anti-mouse IFNγ antibody (rat monoclonal) | BioLegend | Cat# 505807; RRID:AB_315401 | FC (1:200) |
| Peptide, recombinant protein | Murine IL-12 | PeproTech | Cat# 210-12 | |
| Peptide, recombinant protein | Human GM-CSF | PeproTech | Cat# 300-03 | |
| Peptide, recombinant protein | Human IFN gamma | PeproTech | Cat# 300-02 | |
| Peptide, recombinant protein | Murine TNF | PeproTech | Cat# 315-01A | |
| Antibody | Anti-IL4 clone: 11B11 (rat monoclonal) | BioLegend | RRID:AB_2750407 | Neutralization (1:500) |
| Antibody | Goat anti-rabbit HRP (goat polyclonal) | Invitrogen | Cat# 31460 | WB (1:1000) |
| Antibody | Goat anti-mouse HRP (goat polyclonal) | Invitrogen | Cat# 31430 | WB (1:1000) |
| Commercial assay or kit | One-Step RT PCR Kit | Qiagen | Cat# 210215 | |
| Commercial assay or kit | Luna Universal One-Step RT-qPCR Kit | NEB | Cat# E3005 | |
| Commercial assay or kit | Trizol | Thermo Fisher Scientific | Cat# 15596026 | |
| Commercial assay or kit | CellTiter-Glo 2.0 Cell Viability Assay | Promega | Cat# G9241 | |
| Commercial assay or kit | Seahorse assay media | Agilent | Cat# 103575-100 | |
| Software, algorithm | MAGECK | PMID:25476604 | ||
| Peptide, recombinant protein | Interferon gamma | BioLegend | Cat# 575308 | |
| Commercial assay or kit | MojoSORT – Mouse CD4 Naïve T cell Isolation Kit | BioLegend | Cat# 480040 | |
| Commercial assay or kit | MojoSort Human CD14 Nanobeads | BioLegend | Cat# BioLegend 480093 | |
| Commercial assay or kit | IL6 ELISA-max | BioLegend | Cat# 431301 | |
| Commercial assay or kit | TNF ELISA-max | BioLegend | Cat# 430901 | |
| Commercial assay or kit | Human IL1b | R&D Systems | Cat# DY201 | |
| Commercial assay or kit | Human TNF-alpha DuoSet ELISA | R&D Systems | Cat# DY210 | |
| Commercial assay or kit | Greiss reagent | Promega | G2930 | |
| Sequence-based reagent | All oligonucleotide sequences are contained in Supplementary file 1. | This paper | All oligonucleotide sequences are contained in Supplementary file 1 |
Additional files
-
Supplementary file 1
Oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/65109/elife-65109-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65109/elife-65109-transrepform1-v2.docx





