A spontaneous genetically induced epiallele at a retrotransposon shapes host genome function
Figures
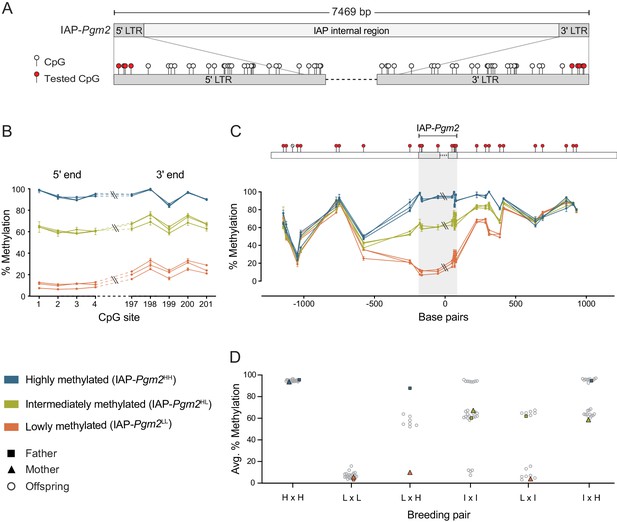
Intracisternal A-particle (IAP)-Pgm2 methylation is tri-modally distributed and stably inherited in inbred B6 mice.
(A) Map of CpG positions in the IAP-Pgm2 long terminal repeats (LTRs). CpGs assayed in panel B are shown in red. (B) IAP-Pgm2 methylation levels are consistent between the 5′ and 3′ ends. Methylation levels were quantified at the most distal CpGs of the IAP-Pgm2 5′ and 3′ LTRs (nearest to the boundary with unique DNA) using bisulphite pyrosequencing in ear DNA. Each point represents a CpG, each line represents an individual, and error bars represent standard deviations of technical triplicates. (C) Inter-individual methylation variation collapses within 500 bp on either side of IAP-Pgm2. Data presentation as in panel A. Assayed CpGs are shown in red above the graph. (D) Stable Mendelian inheritance of IAP-Pgm2 methylation reveals that high (blue, H) and low (orange, L) methylation reflect two allelic states of IAP-Pgm2, with intermediate methylation (orange, I) representing heterozygosity. Each data point represents average methylation of ear DNA across the four most distal CpGs of the 5′ LTR for one individual.
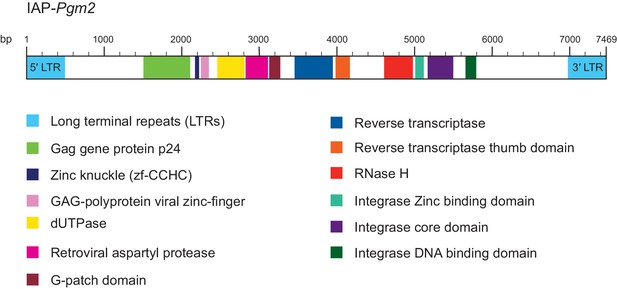
Genomic structure and coding potential of intracisternal A-particle (IAP)-Pgm2.
Details lifted from the Pfam database (https://pfam.xfam.org). Genomic distances are drawn to scale.
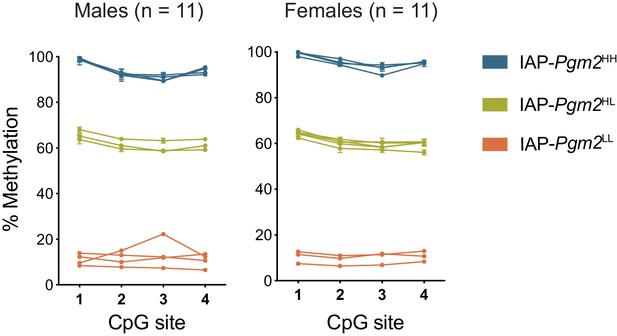
The intracisternal A-particle (IAP)-Pgm2 methylation pattern is not sex-linked.
IAP-Pgm2 methylation levels in both B6 males and females segregate into three distinct DNA methylation states: high (blue), intermediate (green), and low (orange).
-
Figure 1—figure supplement 2—source data 1
Numerical data represented in panels B-D, and Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/65233/elife-65233-fig1-figsupp2-data1-v2.xlsx
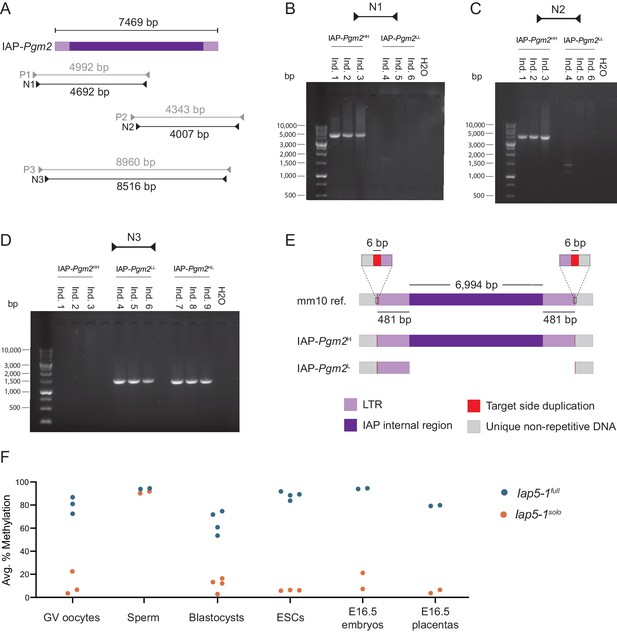
The intracisternal A-particle (IAP)-Pgm2L allele is a solo long terminal repeat (LTR) formed via inter-LTR homologous recombination.
(A) Map of PCR primer pairs P1–P3 and nested PCR primer pairs N1–N3 (not drawn to scale). (B and C) Agarose gel electrophoresis of PCR products amplified from three IAP-Pgm2HH and three IAP-Pgm2LL DNA samples using primer pairs N1 and N2. (D) As in panels B and C, but using primer pair N3 and including three IAP-Pgm2HL DNA samples. (E) Schematic representation of the alignment of the IAP-Pgm2 mm10 reference sequence and the assembled IAP-Pgm2H and IAP-Pgm2L sequences following Sanger sequencing (not drawn to scale). The IAP-Pgm2L solo LTR could have equivalently been shown aligned to the 3′ LTR because the 5′ and 3′ LTRs have identical sequences. The base-resolution sequence of the new IAP-Pgm2L allele, renamed Iap5-1solo, is available on GenBank (accession number: MW308129). (F) Both Iap5-1 variants are methylated in the male germline, with differential methylation re-established in early embryonic development. DNA methylation levels at the Iap5-1full and Iap5-1solo alleles were quantified in oocytes, sperm, blastocysts, ESC lines, and E16.5 embryonic tail and placenta samples. Respective data points represent the following: 100 pooled oocytes, sperm collected from one male, pooled littermate blastocysts, one ESC line, and individual E16.5 embryos and placentas. All data points represent average DNA methylation across the four most distal CpGs of the 5′ end of Iap5-1.
-
Figure 2—source data 1
Numerical data represented in panel F.
- https://cdn.elifesciences.org/articles/65233/elife-65233-fig2-data1-v2.xlsx
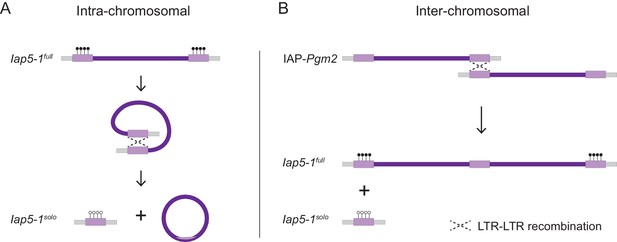
Possible mechanisms of homologous recombination between intracisternal A-particle (IAP) long terminal repeats (LTRs) leading to the formation of a solo LTR at the Iap5-1 locus.
(A) Intra-chromosomal homologous recombination between the 5′ and 3′ LTRs of the repeat element, giving rise to a solo LTR and a circularised unintegrated viral fragment. (B) Inter-chromosomal recombination between identical LTRs on sister chromatids or homologous chromosomes, producing a solo LTR on one DNA strand and a tandem duplication on the other. Diagrams adapted from Seperack et al., 1988.
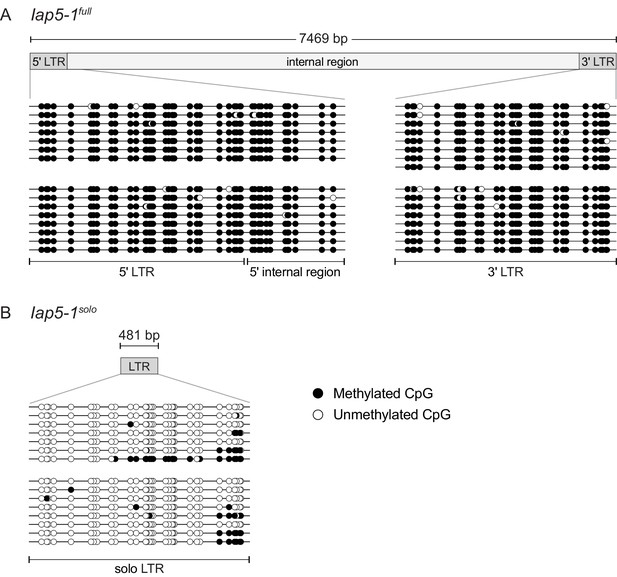
Differential DNA methylation between the Iap5-1full and Iap5-1solo variants extends over the entire long terminal repeat(s) (LTR[s]).
(A and B) Clonal bisulphite sequencing of the 5′ and 3′ ends of Iap5-1full (A) and of the Iap5-1solo LTR (B) in adult liver. Each line represents an individual PCR clone and each circle represents a single CpG (white circle, unmethylated CpG; black circle, methylated CpG). Each block of clones represents an individual mouse. Diagrams of the two variants are shown above the sequencing data.
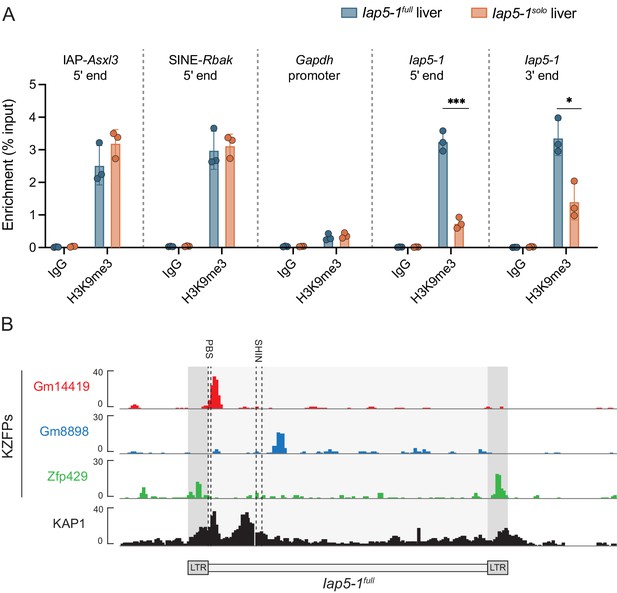
Lack of DNA methylation at Iap5-1solo is accompanied by a loss of H3K9me3 marks.
(A) H3K9me3 ChIP-qPCR on Iap5-1full and Iap5-1solo adult male livers. IAP-Asxl3 and SINE-Rbak are positive control loci, the Gapdh promoter is a negative control locus, and the Rabbit IgG antibody serves as a negative isotype control in the two B6 populations. H3K9me3 enrichment was calculated using the per cent input method and compared between genotypes using unpaired t-tests (*p<0.05; ***p<0.0005). Error bars represent standard deviations of biological replicates. (B) Iap5-1full is bound by multiple KZFPs. Publicly available ChIP-seq data sets indicate that KZFPs Gm14419 and Gm8898 are capable of binding the internal region of Iap5-1full and that KZFP Zfp429 is capable of binding the long terminal repeat (LTR) sequence shared by both Iap5-1full and Iap5-1solo. The primer binding site (PBS) and the short heterochromatin inducing sequence (SHIN), both frequently bound by KZFPs, are shown with dashed lines. ChIP-seq data sets were downloaded from the GEO database (accession numbers: Gm14419, GSM3173720; Gm8898, GSM3173728; Zfp429, GSM3173732; KAP1, sum of GSM3173661, GSM3173662, and GSM3173663).
-
Figure 3—source data 1
Numerical data represented in panel A.
- https://cdn.elifesciences.org/articles/65233/elife-65233-fig3-data1-v2.xlsx
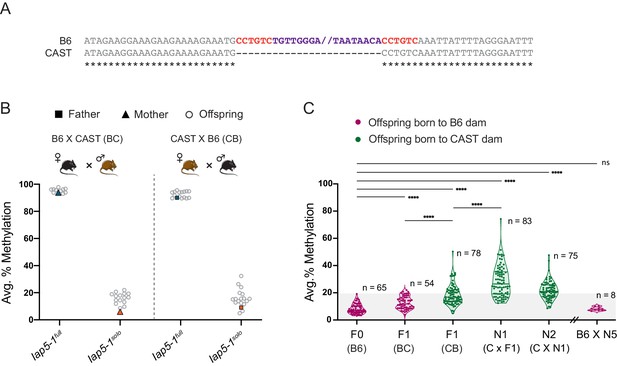
Genetic background influences Iap5-1solo methylation levels.
(A) B6 and CAST sequence alignment of the Iap5-1 insertion site reveals that the Iap5-1 element (purple) and its associated target site duplication (TSD) sequence (red) are not present in the CAST genome. Asterisks indicate nucleotide conservation. The internal portion of the B6 sequence was omitted; nothing was omitted from the CAST sequence. (B) Reciprocal F1 hybrids uncover a variant-specific genetic background effect. Iap5-1full and Iap5-1solo B6 females were crossed to CAST males (BC, left); Iap5-1full and Iap5-1solo B6 males were crossed to CAST females (CB, right). DNA methylation levels were quantified in ear samples collected from hemizygous offspring. Each data point represents average DNA methylation across the four distal CpGs of the 5′ end of Iap5-1 for one individual. (C) The genetic background-specific maternal effect at Iap5-1solo is strengthened upon backcrossing to a CAST female and lost following backcross to a B6 female. The N1 generation was produced by crossing F1 CB males to CAST females, and the N2 generation was generated by crossing N1 males harbouring the Iap5-1solo allele to CAST females. After five generations of backcrossing to CAST, N5 males were crossed to B6 females. Offspring that did not inherit the Iap5-1solo allele were not included in the analysis. Thick and thin lines in the violin plots designate the median and distribution quartiles, respectively. Sample sizes are shown on the graph. Grey shaded area represents the range of methylation levels observed in B6 individuals. Statistics: Welch’s ANOVA test followed by Games-Howell’s post hoc multiple comparison test (****p<0.0001; ns: not significant).
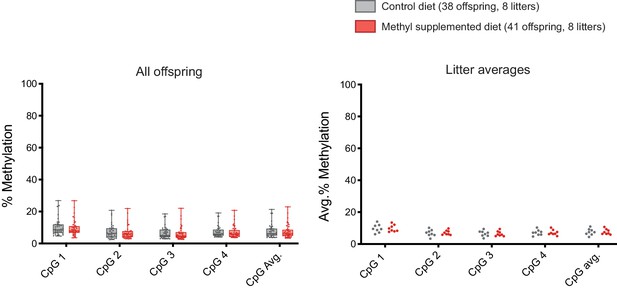
DNA methylation at Iap5-1solo is unresponsive to maternal dietary methyl supplementation.
Iap5-1solo females were fed a control or methyl supplemented diet 2 weeks prior to mating and for the duration of pregnancy and lactation. DNA methylation levels were quantified in 8-week-old offspring liver samples at the four distal CpGs of the 5′ end of Iap5-1solo. Statistics: Unpaired t-tests on litter averages for each CpG and for the CpG average.
-
Figure 4—figure supplement 1—source data 1
Numerical data represented in panels B, C, and Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/65233/elife-65233-fig4-figsupp1-data1-v2.xlsx
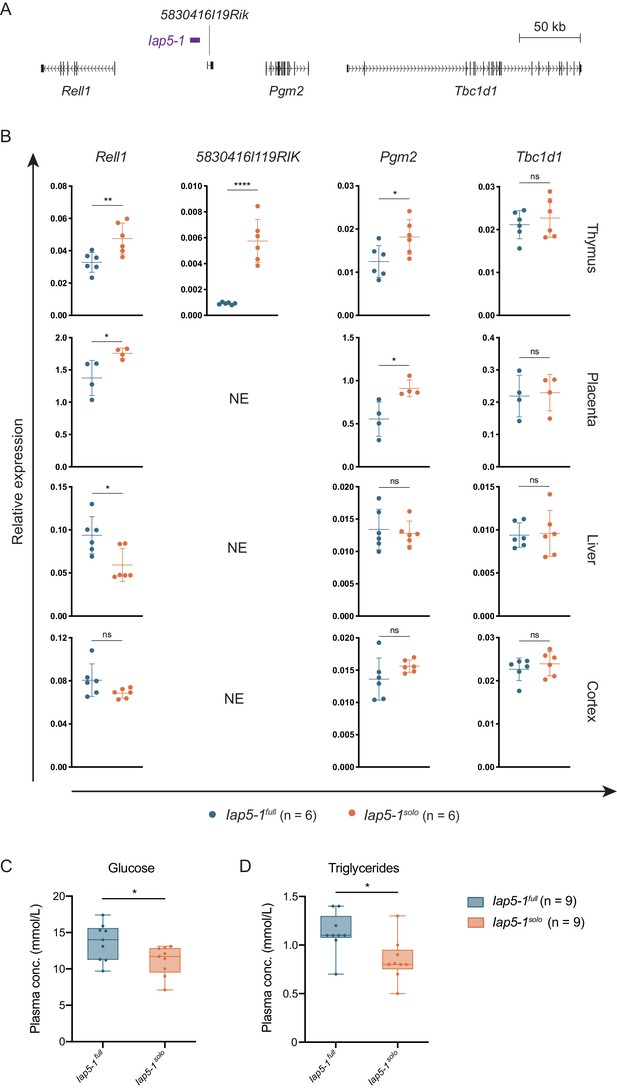
Functional consequences of the genetically induced epiallele at the Iap5-1 locus.
(A) Scaled map of the genes surrounding Iap5-1. Gene transcripts were extracted from the UCSC Genome Browser (Haeussler et al., 2019). (B) The Iap5-1 polymorphism influences neighbouring gene expression in a tissue-specific manner. Expression of Rell1 (exon 7), 5830416l19Rik (exon 3), Pgm2 (exon 9), and Tbc1d1 (exon 3) was quantified in Iap5-1full and Iap5-1solo thymus, placenta, liver, and brain tissues via RT-qPCR (NE: not expressed). Relative expression was normalised to Hprt1 and β-actin expression and calculated using the ΔCt method. Means and standard deviations of biological replicates are shown in the graphs. (C and D) Plasma glucose (C) and triglyceride (D) concentrations in Iap5-1full and Iap5-1solo adult males. Box plots show the distribution quartiles and median. Statistics: unpaired t-tests (*p<0.05; **p<0.005; ****p<0.0001; ns: not significant).
-
Figure 5—source data 1
Numerical gene expression data represented in Figure 5B and Figure 5—figure supplement 1B.
- https://cdn.elifesciences.org/articles/65233/elife-65233-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Numerical data for metabolic experiments shown in Figure 5C and D and Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/65233/elife-65233-fig5-data2-v2.xlsx
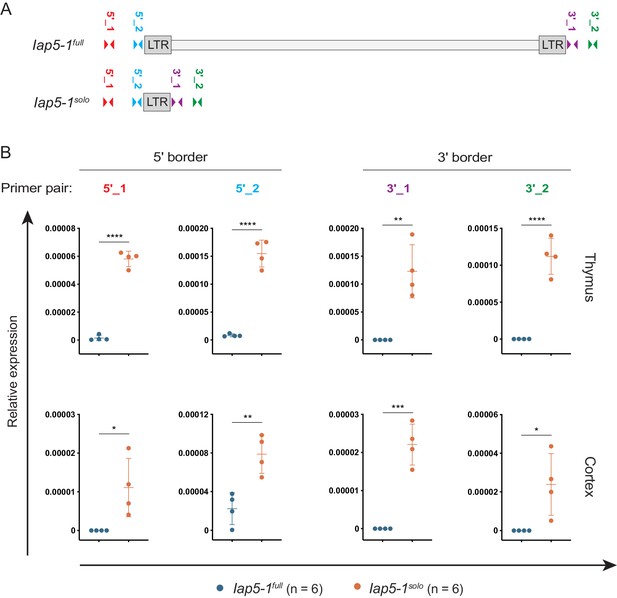
Bordering genomic regions are transcribed in Iap5-1solo tissues.
(A) Map of RT-qPCR primers (drawn to scale). (B) Transcription of bordering unique DNA was quantified in Iap5-1full and Iap5-1solo thymus and cortex tissues via RT-qPCR. Relative expression was normalised to β-actin expression and calculated using the ΔCt method. Means and standard deviations of biological replicates are indicated on the graphs. Statistics: unpaired t-tests (*p<0.05; **p<0.005; ***p<0.001; ****p<0.0001).
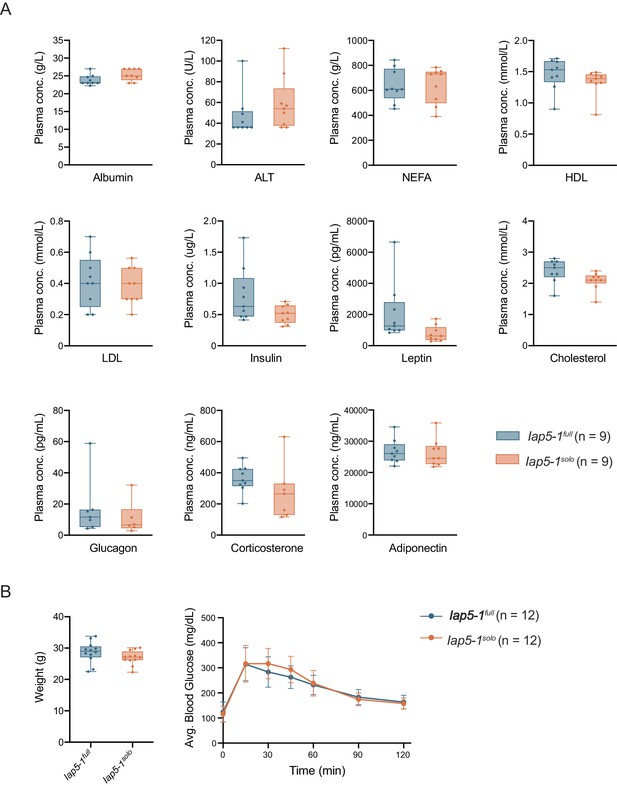
Metabolic phenotyping of Iap5-1full and Iap5-1solo individuals.
(A) Concentration of metabolic biomarkers in plasma samples collected from Iap5-1full and Iap5-1solo adult males. Box plots show the distribution quartiles and median. Statistics: unpaired t-tests. (B) Glucose tolerance test on Iap5-1full and Iap5-1solo adult males. Following an overnight fast, mice were weighed (left panel) and fasting tail blood glucose was measured prior to administering an intraperitoneal 2 g/kg glucose injection. Tail blood glucose measurements were taken 15, 30, 45, 60, 90, and 120 min from the time of injection (right panel). Error bars represent standard deviations of biological replicates. Statistics: unpaired t-test (left panel) and multiple unpaired t-tests corrected for multiple comparisons using the Holm-Šidák method (right panel).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL/6J | The Jackson Laboratory via Charles River | 000664 (JAX) 632 (Charles River) | |
| Strain, strain background (Mus musculus) | CAST/EiJ | MRC Harwell Institute | FESA:773 | |
| Cell line (Mus musculus) | C57BL6/J ES cell lines | This paper | See Materials and methods, Generation of ESC lines. | |
| Antibody | Rabbit polyclonal anti-H3K9me3 | Active Motif | RRID:AB_2532132 | ChIP (2.5 µl/30 mg liver tissue) |
Additional files
-
Supplementary file 1
Primer sequences and GEO accession numbers.
- https://cdn.elifesciences.org/articles/65233/elife-65233-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65233/elife-65233-transrepform-v2.transparentreportingform.docx





