A common 1.6 mb Y-chromosomal inversion predisposes to subsequent deletions and severe spermatogenic failure in humans
Figures
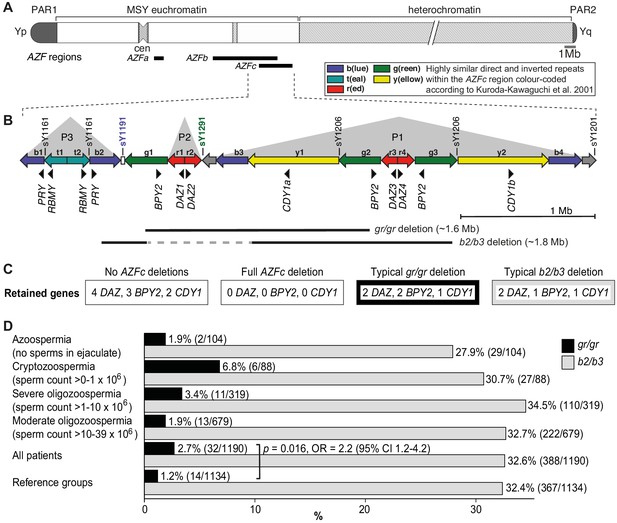
Y-chromosomal AZFc region and its partial deletions in the study group.
(A) Schematic representation of the human Y chromosome with the AZFa, AZFb, and AZFc regions shown as black bars. (B) Magnified structure of the AZFc region with approximate locations of multicopy protein-coding genes, STS (sY) markers for the detection of AZFc partial deletions and the span of typical gr/gr and b2/b3 deletions (Kuroda-Kawaguchi et al., 2001). P1–P3 (gray triangles) denote palindromic genomic segments consisting of two ‘arms’ representing highly similar inverted DNA repeats (>99.7% sequence identity) that flank a relatively short distinct ‘spacer’ sequence. Of note, the occurrence of the b2/b3 deletion requires a preceding inversion in the AZFc region and therefore its presentation on the reference sequence includes also the retained segment (gray dashed line). Full details about alternative gr/gr and b2/b3 deletion types are presented in Figure 1—figure supplement 1. (C) Dosage of multicopy genes on human Y chromosomes with or without AZFc deletions. (D) Prevalence of the gr/gr and b2/b3 deletions detected in the subgroups of this study. Fisher’s exact test was used to test the statistical significance in the deletion frequencies between the groups. PAR, pseudoautosomal region; MSY, male-specific region of the Y chromosome; cen, centromere; AZF, azoospermia factor region.
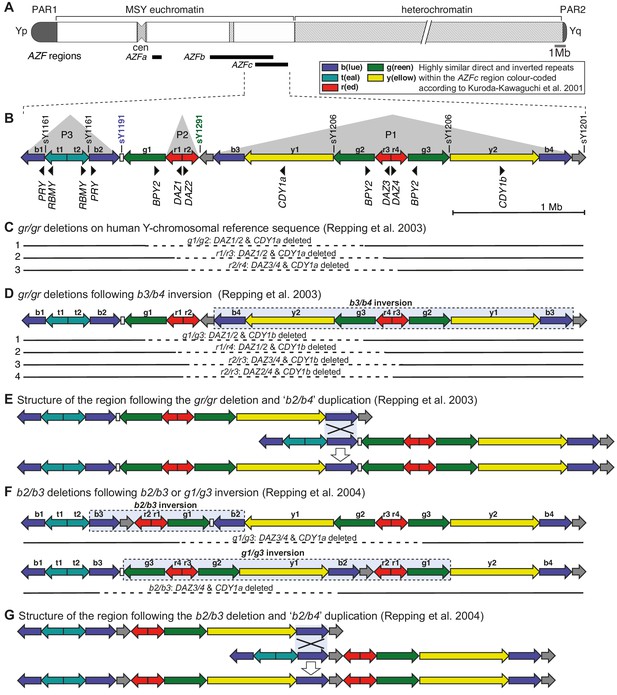
The human Y chromosome and the Azoospermia Factor (AZF) regions.
(A) The human Y chromosome drawn to approximate scale with regions of AZFa, AZFb, and AZFc deletions shown below as black bars. PAR, pseudoautosomal region; MSY, male-specific region of the Y chromosome; cen, centromere; AZF, azoospermia factor region. (B) Structure of the AZFc region on human Y-chromosomal reference sequence with approximate locations of protein-coding genes and STS (sY) markers used for detection of partial AZFc deletions (Kuroda-Kawaguchi et al., 2001). Alternative involved regions in gr/gr deletions arising on (C). The human Y-chromosomal reference sequence and (D) the Y chromosome with a preceding b3/b4 inversion. The approximate region removed by each deletion is shown as a dashed line. (E) Structure of the AZFc region undergone the gr/gr deletion and the proposed model of homologous recombination leading to the subsequent ‘b2/b4’ duplication. The light blue box denotes the recombination targets. The duplication is presumably the result of recombination between sister chromatids. (F) Reported models for b2/b3 deletions arising on the Y chromosomes with preceding b2/b3 or g1/g3 inversions. The approximate region removed by each deletion is shown as a dashed line. (G) Structure of the AZFc region undergone the b2/b3 deletion and the proposed model of homologous recombination leading to the subsequent ‘b2/b4’ duplication. The light blue box denotes the recombination targets. The duplication is presumably the result of recombination between sister chromatids.
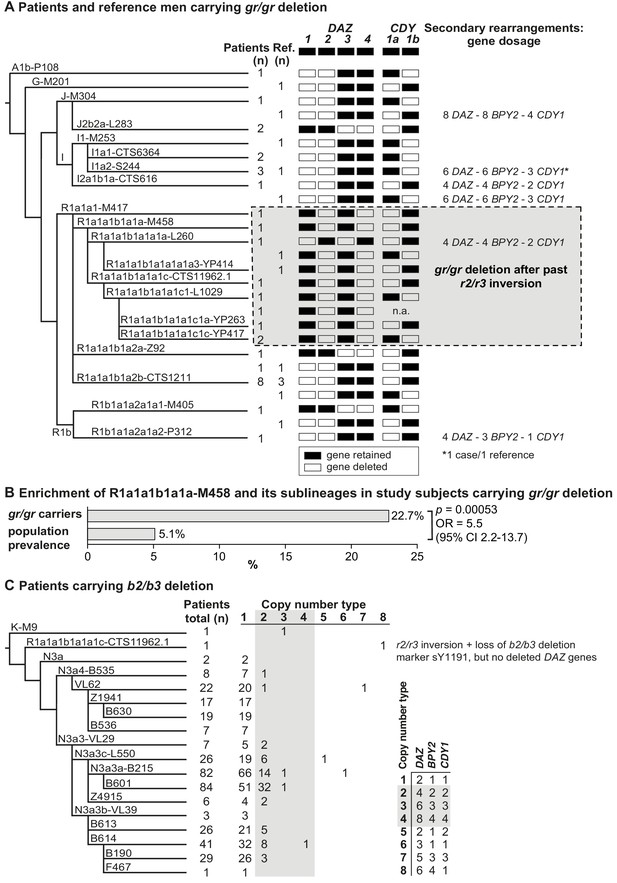
Phylogenetic relationships and gene copies in study subjects with partial AZFc deletions.
(A) Y-chromosomal lineages indicated with typed terminal markers (left), deleted (white)/retained (black) DAZ and CDY1 gene copies (middle), and secondary rearrangements in the AZFc region (right) of idiopathic male factor infertility (n = 31) and reference cases (n = 13) carrying the gr/gr deletion. The human Y-chromosomal reference sequence has four DAZ and two CDY1 copies; the retained gene copies on each Y chromosome with a gr/gr deletion are shown as filled boxes. Chromosomes carrying atypical gr/gr subtypes with the loss of either the DAZ1/DAZ3 or DAZ2/DAZ4 gene pair due to complex genomic rearrangement combining the previous r2/r3 inversion with a subsequent gr/gr deletion are highlighted with a dashed gray square. (B) Enrichment of the Y-chromosomal lineage R1a1-M458 and its sub-lineages in study subjects carrying the gr/gr deletion in comparison to the Estonian general population (data from Underhill et al., 2015). Fisher’s exact test was used to test the statistical significance between the groups. (C) Y-chromosomal lineages indicated with typed terminal markers (left) and the copy number of the DAZ, BPY2, and CDY1 gene copies (right) determined for 382 idiopathic male factor infertility cases carrying the b2/b3 deletion. The light gray box denotes DAZ, BPY2, and CDY1 gene dosage consistent with full b2/b4 duplication(s). The legend for the deletion subtype is shown in the bottom right corner. Further information on the distribution of Y-chromosomal lineages in the carriers of AZFc partial deletions are provided in Supplementary files 5 and 6, and the AZFc rearrangement types are detailed in Figure 1—figure supplement 1 and Supplementary files 7 and 9–12. n, number; n.a., not available; Ref, reference cases.
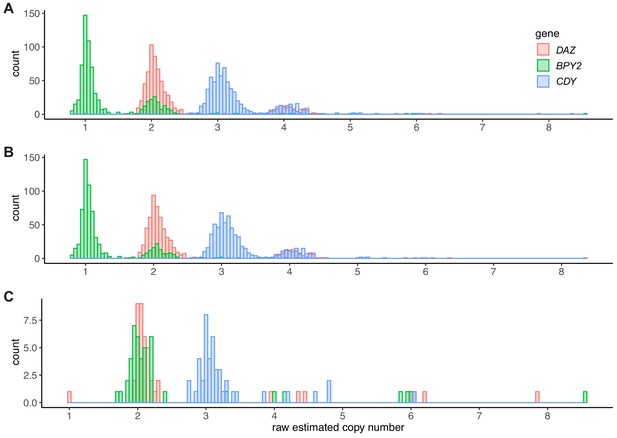
Histograms of estimated raw copy number values for DAZ, BPY2, and CDY genes by ddPCR.
(A) Distribution of copy number estimates for all typed samples (n = 675); (B) samples carrying the b2/b3 or sY1191 marker deletions (n = 631); and (C) samples carrying the gr/gr deletion (n = 44). The CDY gene copy number is a sum of CDY1 and CDY2 (assumed to be two) copy numbers. Note: one patient with a gr/gr deletion appeared to carry a single copy of DAZ gene (ddPCR repeated four times). However, both the Illumina re-sequencing data and paralogous sequence variant (PSV) typing were consistent with two copies of DAZ genes being retained, and therefore in all downstream analysis, the presence of two DAZ genes was assumed. The likely reason for the discrepancy between ddPCR and re-sequencing/PSV typing is a disruption of ddPCR primer or probe binding site.
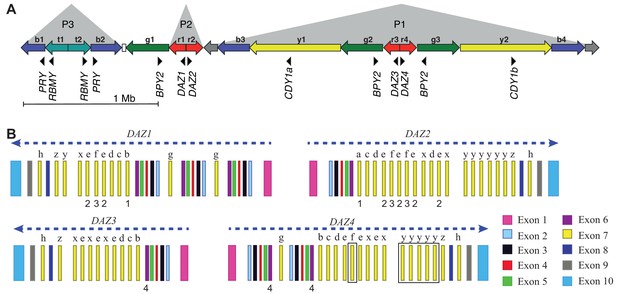
Location of the identified exonic variants in the DAZ genes.
(A) Schematic representation of the human Y-chromosomal reference sequence containing the AZFc region, with approximate locations of protein-coding genes and their direction of transcription shown as black triangles. Direct and inverted repeats with highly similar DNA sequences are denoted as coloured arrows. (B) The structure of human DAZ genes, modified from Fernandes et al., 2002 to represent the human Y-chromosomal reference sequence (GRCh38). Blue dashed arrows show the direction of transcription. Exons with high DNA sequence similarity within and between DAZ genes are denoted with the same fill colour. The number of highly similar exon seven copies (in yellow) varies between genes and copies marked with the same letter denote identical sequences. Numbers below the gene structure denote exonic variants. 1 – DAZ1 p.H173Y or DAZ2 p.H173Y. 2 – DAZ1 p.Q262E or DAZ2 p.Q262E. 3 – DAZ1 p.Y243C or DAZ2 p.Y219C. 4 – potential splicing variant in DAZ3 or DAZ4. Black squares in DAZ4 denote the deleted exons 7f and 7y in Y lineages I1 and R1a1a1b1a2, respectively.
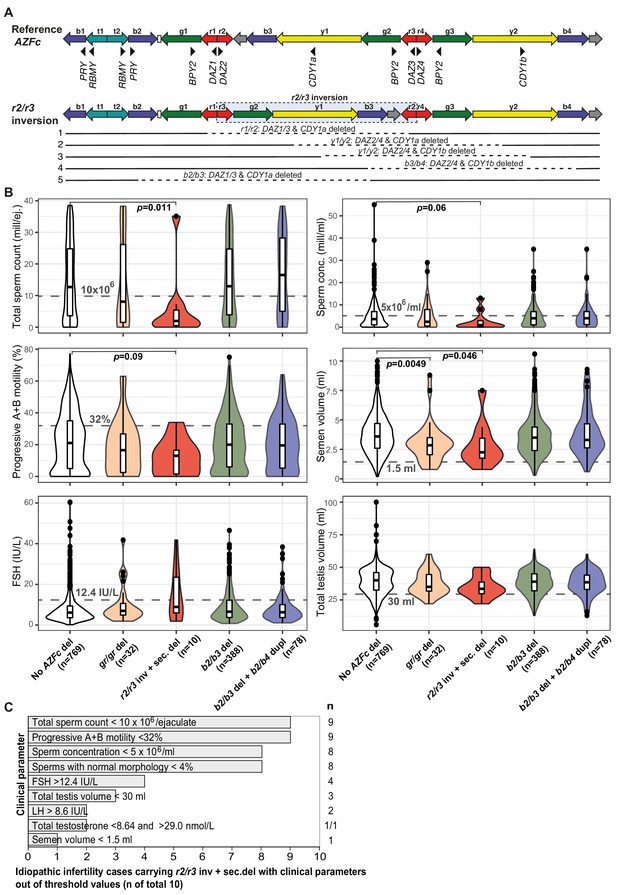
Complex structural variants at the Y-chromosomal lineage R1a1-M458 and their effect on andrological parameters.
(A) Schematic presentation of the Y chromosome with the r2/r3 inversion compared to the reference sequence. The r2/r3 inversion structure nearly destroys the large palindrome P1 and, consequently, destabilizes the AZFc region since several long DNA amplicons with highly similar DNA sequence (b2, b3, and b4; g2 and g3; y1 and y3) are positioned in the same sequence orientation. This structure promotes non-allelic homologous recombination mediating recurrent deletion and duplication events. The approximate regions removed by the identified gr/gr and b2/b3 deletions arising on the r2/r3 inverted Y chromosome are shown as dashed lines. (B) Distribution of andrological parameters in the idiopathic male factor infertility cases (total sperm counts 0–39 × 106) subgrouped based on the structure of the AZFc region. The pairwise Wilcoxon rank-sum test was applied to estimate the statistical difference between groups (Bonferroni threshold for multiple testing correction, p<1.0×10−3). Threshold values (shown in gray) for sperm parameters corresponding to severe spermatogenic failure are based on international guidelines (World Health Organization, 2010). For reproductive hormones, reference values of the laboratory service provider are shown. The empirical threshold for the total testis volume was based on routinely applied clinical criteria at the AC-TUH. For additional details, see Figure 3—figure supplements 1–3, Supplementary files 3 and 12. (C) The majority of idiopathic infertility cases carrying the r2/r3 inversion plus secondary AZFc partial deletions (total n = 10) exhibit severe oligoasthenoteratozoospermia (OAT) defined as extremely reduced sperm counts (<5 × 106/ml) and concentration (<10 × 106/ejaculate) combined with low fraction of sperms with normal morphology (<4% normal forms) and motility (<32% progressive motile spermatozoa). Reference values for andrological parameters have been applied as referred in (B). As total testis volume is mostly within the expected range, their infertility is not caused by intrinsic congenital testicular damage but rather due to severe spermatogenic failure per se. Del, deletion; inv, inversion; dupl, duplication; n, number; sec, secondary; mill, million; ej., ejaculate.
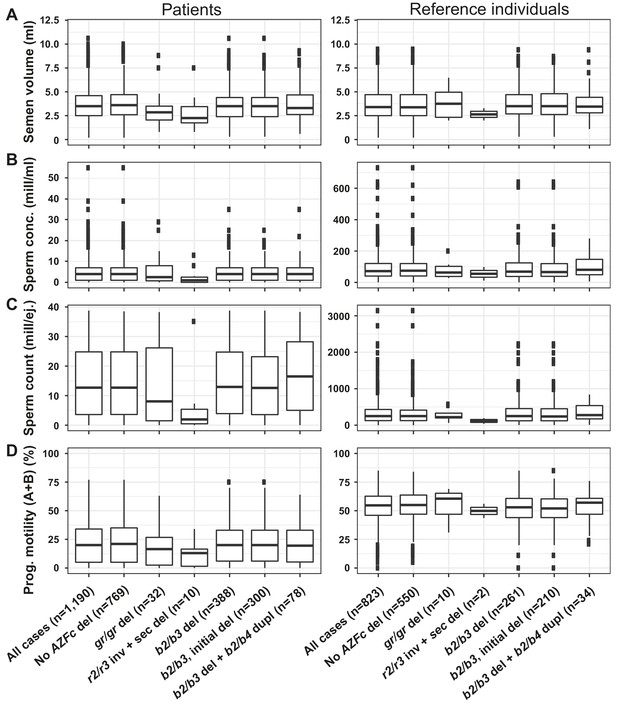
Distribution of seminal parameters in idiopathic male factor infertility cases with spermatogenic impairment and reference subjects.
(A) Semen volume, (B) sperm concentration, (C) sperm count and (D) progressive motility. Samples are grouped by genotype with the number of samples (n) shown in brackets. del, deletion; sec, secondary; inv, inversion; dupl, duplication; mill, million; ej., ejaculate. Note different scaling of the Y-axis for the two study groups in (B) and (C).
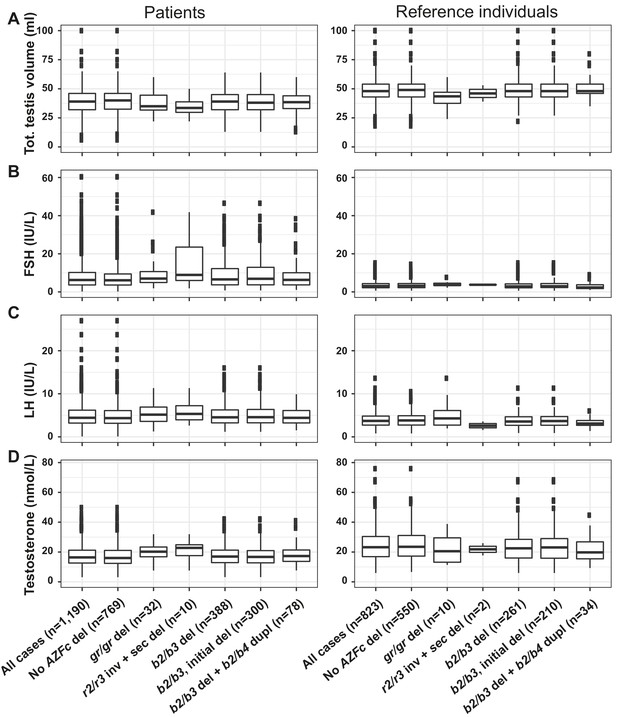
Distribution of hormonal and testicular parameters in idiopathic male factor infertility cases with spermatogenic impairment and reference individuals.
(A) Total testis volume, (B) FSH, (C) LH and (D) testosterone. Samples are grouped by genotype with the number of samples (n) shown in brackets. Del, deletion; sec, secondary; inv, inversion; dupl, duplication.
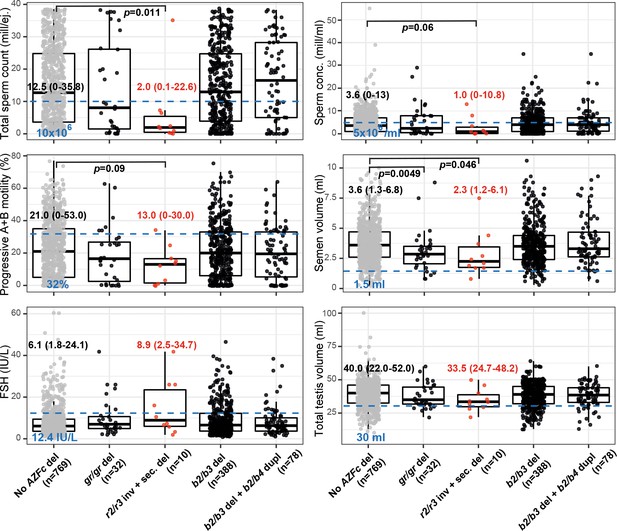
Distribution of andrological parameters in the idiopathic male factor infertility cases (total sperm counts 0–39 × 106) subgrouped based on the structure of the AZFc region.
The median (5–95% range) of each parameter is shown for cases carrying the r2/r3 inversion plus secondary deletions compared to infertile men without any AZFc deletion. The pairwise Wilcoxon rank–sum test was applied to estimate the statistical difference between groups. Statistical significance threshold after correction for multiple testing was estimated p<1.0×10−3 (a total of 5 tests × 10 independent parameters). Threshold values (shown in blue) for sperm parameters corresponding to severe spermatogenic failure are based on international guidelines (World Health Organization, 2010). For reproductive hormones, reference values of the laboratory service provider are shown. The empirical threshold for the total testis volume was based on routinely applied clinical criteria at the AC-TUH. For full details, see Supplementary files 3 and 12.
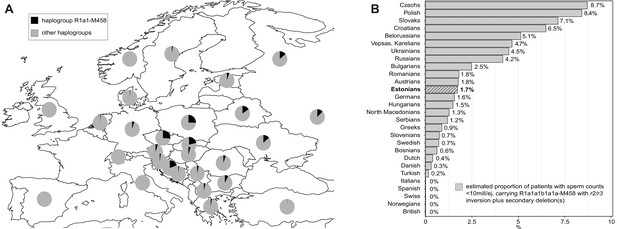
The prevalence of the Y-chromosomal haplogroup R1a1-M458 carrying a fixed r2/r3 inversion.
(A) Geographical distribution of haplogroup R1a1-M458 and its sub-lineages in Europe. Pie charts indicate populations, with the black sector showing the proportion of R1a1-M458 according to Underhill et al., 2015. (B) The estimated proportion of subjects among idiopathic cases with severe spermatogenic failure (sperm counts 0–10 × 106/ejaculate) carrying R1a1-M458 Y lineage (and its sub-lineages) chromosomes that have undergone a subsequent partial AZFc deletion. The prevalence was estimated using reported population frequencies of R1a1-M458, including Estonians (Underhill et al., 2015) and data available in the current study for Estonian men with spermatogenic failure. Estonians are shown in bold and with a striped filling. For full details, see Supplementary file 15.
Tables
Characteristics of the patients with male factor infertility and reference groups used for comparison.
| Idiopathic spermatogenic impairment (n = 1190)* | Reference groups (n = 1134) | ||||||
|---|---|---|---|---|---|---|---|
| Parameter | Unit | Azoo-/cryptozoospermia | Severe oligozoospermia | Moderate oligozoospermia | Partners of pregnant women† | Estonian young men cohort‡ | REPROMETA proven fathers§ |
| n | 104/88 | 319 | 679 | 324 | 499 | 311 | |
| Age | Years | 33.2 (23.6–51.8) | 32.2 (23.9–49.5) | 31.7 (23.0–44.6) | 31.0 (22.9–45.0) | 18.6 (17.2–22.9) | 31.0 (21.0–43.0) |
| BMI | kg/m2 | 26.0 (21.2–34.4) | 25.9 (20.2–35.5) | 25.8 (20.1–34.6) | 24.8 (20.0–32.2) | 22.0 (18.7–27.5) | 25.9 (20.2–33.1) |
| Total testis volume | ml | 33.5 (17.0–49.0) | 39.0 (22.0–50.0) | 40.0 (26.0–52.0) | 46.0 (34.0–62.4) | 50.0 (35.0–70.0) | n.d. |
| Semen volume | ml | 3.3 (0.8–6.6) | 3.3 (1.1–7.0) | 3.6 (1.6–6.9) | 3.7 (1.7–8.0) | 3.2 (1.2–6.4) | n.d. |
| Sperm concentration | × 106/ml | 0 (0–0.2) | 1.4 (0.4–5.2) | 6.0 (2.2–15.2) | 76.0 (16.7–236.0) | 66.8 (8.2–225.1) | n.d. |
| Total sperm count | × 106/ ejaculate | 0 (0–0.7) | 4.7 (1.3–9.3) | 23.1 (11.0–37.5) | 295.2 (60.0–980.1) | 221.6 (18.4–788.0) | n.d. |
| Progressive A+B motility | % | 0 (0–37.2) | 16.0 (0–47.2) | 27.0 (1.0–57.0) | 50.0 (30.0–69.0) | 57.3 (34.7–75.3) | n.d. |
| Sperms with normal morphology | % | 0 (0–1.0) | 0 (0–6.0) | 2.0 (0–9.0) | 10.0 (2.0–19.1) | 12.0 (4.0–20.0) | n.d. |
| FSH | IU/l | 13.7 (2.7–38.2) | 6.6 (1.9–22.8) | 5.2 (1.8–16.5) | 3.6 (1.5–8.3) | 2.8 (1.2–6.7) | n.d. |
| LH | IU/l | 5.7 (2.1–12.0) | 4.6 (1.9–9.9) | 4.2 (1.8–8.4) | 3.6 (1.5–6.7) | 3.8 (1.8–7.2) | n.d. |
| Total testosterone | nmol/l | 15.3 (7.7–28.4) | 16.6 (7.9–30.0) | 16.6 (8.5–30.3) | 16.5 (8.8–27.2) | 27.7 (15.4–46.3) | n.d. |
-
All study subjects were recruited in Estonia. For each parameter, median and (5th–95th) percentile values are shown. Additional details in Supplementary file 1.
*Patients were subgrouped based on total sperm counts per ejaculate: azoospermia, no sperm; cryptozoospermia, sperm counts > 0–1 × 106; severe oligozoospermia, >1–10 × 106; moderate oligozoospermia, >10–39 × 106 (Punab et al., 2017).
-
†Male partners of pregnant women (Punab et al., 2017); eight men had sperm counts < 39 × 106; for four men, sperm analysis was not available.
‡Male cohort without fatherhood data (Grigorova et al., 2008); 47 men had sperm counts < 39 × 106; for nine men, sperm analysis was not available.
-
§REPROMETA study recruited and sampled couples after delivery of their newborn; details in Kikas et al., 2020; Pilvar et al., 2019.
n.d., not determined.
Summary of the identified Y-chromosomal AZF deletion subtypes.
| Y-chromosomal rearrangements | Idiopathic male infertility patients (n) | Reference men (n) |
|---|---|---|
| All analyzed cases | 1190 | 1134 |
| Any AZFc gr/gr deletion | 32 (2.7%) | 14 (1.2%) |
| Fisher’s exact test, p=0.016; OR = 2.2 [95% CI 1.2–4.2] | ||
| Any AZFc b2/b3 deletion | 388 (32.6%) | 367 (32.4%) |
| Other type of AZF deletion | Loss of b2/b3 marker sY1191 (one case) | AZFc b1/b3 del (one case); partial AZFa del (one case) |
| No deletion | 769 (64.6%) | 751 (66.2%) |
| Simple partial AZFc deletions | ||
| Typical gr/gr deletion | 19/31 (61.3%) | 8/13 (61.5%) |
| Typical b2/b3 deletion* | 300/382 (78.5%) | 210/249 (84.3%) |
| AZFc partial deletion followed by b2/b4 duplication | ||
| gr/gr del + b2/b4 dupl† | 2/31 (6.5%) | 3/13 (23.1%) |
| Fisher’s exact test, p=0.144; OR = 0.2 [95% CI 0.0–1.6] | ||
| b2/b3 del + b2/b4 dupl*,† | 78/382 (20.4%) | 34/249 (13.7%) |
| Fisher’s exact test, p=0.026; OR = 1.6 [95% CI 1.0–2.4] | ||
| AZFc partial deletion and atypical genomic rearrangements‡ | ||
| gr/gr del + extra gene copies | 1 | 0 |
| b2/b3 del + extra gene copies | 3 | 4 |
| Complex events on the Y lineage R1a1-M458 with the preceding AZFc r2/r3 inversion | ||
| r2/r3 inv + gr/gr del | 8 | 2§ |
| r2/r3 inv + gr/gr del + b2/b4 dupl | 1 | 0 |
| r2/r3 inv + loss of marker sY1191 + secondary gene duplications¶ | 1 | 0 |
| r2/r3 inv + b2/b3 del + b2/b4 dupl | 0 | 1** |
| Carriers of any AZFc gr/gr deletion type without the preceding r2/r3 inversion | ||
| gr/gr del w/o detected r2/r3 inv | 23/1190 (1.9 %) | 12/1134 (1.1%) |
| Fisher’s exact test, p=0.090; OR = 1.8 [95% CI 0.9–3.7] | ||
-
*Deletion subtype analysis was carried out for cases with available sufficient quantities of DNA. REPROMETA subjects were excluded from the b2/b3 deletion subtype analysis and subsequent statistical testing due to missing andrological data.
†One or more amplicons of the retained ‘2xDAZ, 2xBPY2, 1xCDY1’ (gr/gr deletion) or ‘2xDAZ, 1xBPY2, 1xCDY1’ (b2/b3 deletion) genes.
-
‡Additional copies of DAZ, BPY2, and/or CDY1 genes inconsistent with the full ‘b2/b4’ duplication.
§Including one REPROMETA man without andrological data.
-
¶Detected gene copy numbers 6xDAZ, 4xBPY2, 3xCDY1; the obligate presence of r2/r3 inversion was defined based on Y-chromosomal phylogeny as the man carries Y lineage R1a1a1b1a1a1c-CTS11962.1 that was also identified in two cases with the r2/r3 inversion (Supplementary file 12).
**Man from ‘Partners of pregnant women’ cohort with sperm concentration 12 × 106/ml below normozoospermia threshold (15 × 106/ml) and sperm counts 39.4 × 106/ejaculate at the borderline of the lowest reference value (39.0 × 106/ejaculate).
Enrichment of the AZFc r2/r3 inversion followed by a partial AZFc deletion in men with severe spermatogenic failure.
| AZFc r2/r3 inversion + AZFc partial deletion | ||||
|---|---|---|---|---|
| Group | All (n) | Estimated non-carriers (n) | Detected carriers (n) | % of carriers in the (sub)group |
| a. Full study group | ||||
| All analyzed study subjects | 2324 | 2311 | 13 | 0.6% |
| Study subjects with sperm counts | 2000 | 1988 | 12 | 0.6% |
| Subjects stratified based on total sperm counts per ejaculate | ||||
| Sperm counts 0–10 × 106 | 524 | 515 | 9 | 1.7% |
| Sperm counts > 10 × 106 | 1476 | 1473 | 3 | 0.2% |
| Fisher’s exact test, p=6.0×10−4, OR = 8.6 [95% CI 2.3–31.8] | ||||
| b. Carriers of the Y lineage R1a1a-M458* | ||||
| In all analyzed study subjects | 119 | 106 | 13 | 11.0% |
| In study subjects with sperm counts | 102 | 90 | 12 | 11.8% |
| Subjects stratified based on total sperm counts per ejaculate | ||||
| Sperm counts 0–10 × 106 | 27 | 18 | 9 | 33.7% |
| Sperm counts > 10 × 106 | 75 | 72 | 3 | 4.0% |
| Fisher’s exact test, p=3.0×10−4, OR = 12.0 [95% CI 2.9–48.9] | ||||
-
*Expected number of Y lineage R1a1-M458 in each subgroup was estimated using the known Estonian population prevalence 5.1% (Underhill et al., 2015).
Additional files
-
Supplementary file 1
Characteristics of the Estonian patients with idiopathic spermatogenic impairment and the used reference groups showing mean and standard deviation values.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp1-v1.xlsx
-
Supplementary file 2
Frequencies of AZFc partial deletions identified in patients and reference groups.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp2-v1.xlsx
-
Supplementary file 3
Andrological parameters of patients and reference cases with and without the AZFc rearrangements.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp3-v1.xlsx
-
Supplementary file 4
Genetic association test with the carrier status of AZFc b2/b3 deletion results using linear regression.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp4-v1.xlsx
-
Supplementary file 5
Y haplogroup distribution and enrichment of lineage R1a1a1b1a1a-M458 among Estonian patients and reference cases carrying the gr/gr deletions.
(a) Y haplogroup distribution of the Estonian patients with idiopathic spermatogenic impairment and reference cases carrying gr/gr deletions.
(b) Enrichment of Y-chromosomal lineage R1a1a1b1a1a-M458 in men carrying gr/gr deletion.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp5-v1.xlsx
-
Supplementary file 6
Y haplogroup distribution of the Estonian patients with idiopathic spermatogenic impairment and reference cases with Y chromosomes having lost the b2/b3 deletion marker sY1191.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp6-v1.xlsx
-
Supplementary file 7
Retained DAZ, BPY2, and CDY1 copy numbers on the Y chromosomes with either gr/gr deletion or having lost the b2/b3 deletion marker sY1191.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp7-v1.xlsx
-
Supplementary file 8
Retained DAZ, BPY2, and CDY1 copy numbers on the Y chromosomes with either gr/gr deletion or having lost the b2/b3 deletion marker sY1191.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp8-v1.xlsx
-
Supplementary file 9
Retained DAZ, BPY2, and CDY1 copy numbers according to Y lineage in samples having lost the b2/b3 deletion marker sY1191.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp9-v1.xlsx
-
Supplementary file 10
Deleted DAZ and CDY1 gene types in gr/gr and b2/b3 carriers.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp10-v1.xlsx
-
Supplementary file 11
Detailed copy number, deletion type, and Y haplogroup information for samples carrying the gr/gr deletion.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp11-v1.xlsx
-
Supplementary file 12
Andrological parameters of 10 patients and two reference cases with ‘r2/r3’ inversion plus gr/gr, b2/b3, or complex deletion.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp12-v1.xlsx
-
Supplementary file 13
Summary of genetic variation identified on the Y chromosomes with AZFc region rearrangements (n = 476).
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp13-v1.xlsx
-
Supplementary file 14
Identified SNVs and indels from re-sequencing of retained DAZ, BPY2, and CDY genes on the Y chromosomes with AZFc region rearrangements.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp14-v1.xlsx
-
Supplementary file 15
Population frequencies of R1a1a1b1a1a-M458 Y lineage and expected proportion of cases with the complex rearrangement r2/r3 inversion + secondary rearrangement among men with sperm counts of <10 mill/ejaculate.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp15-v1.xlsx
-
Supplementary file 16
Y-chromosomal STS markers and PCR primers used for detection of partial AZFc deletions.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp16-v1.xlsx
-
Supplementary file 17
Genomic coordinates of regions sequenced using Illumina MiSeq.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp17-v1.xlsx
-
Supplementary file 18
PCR primers and reaction conditions to amplify regions of interest for sequencing with Illumina MiSeq.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp18-v1.xlsx
-
Supplementary file 19
PCR primers used for typing of Y phylogenetic markers.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp19-v1.xlsx
-
Supplementary file 20
PCR primers and probes used for copy number detection of DAZ, BPY2, and CDY genes using droplet digital PCR.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp20-v1.xlsx
-
Supplementary file 21
Paralogous sequence variants used to determine the retained DAZ and CDY1 genes.
- https://cdn.elifesciences.org/articles/65420/elife-65420-supp21-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65420/elife-65420-transrepform-v1.docx





