Scaled, high fidelity electrophysiological, morphological, and transcriptomic cell characterization
Figures
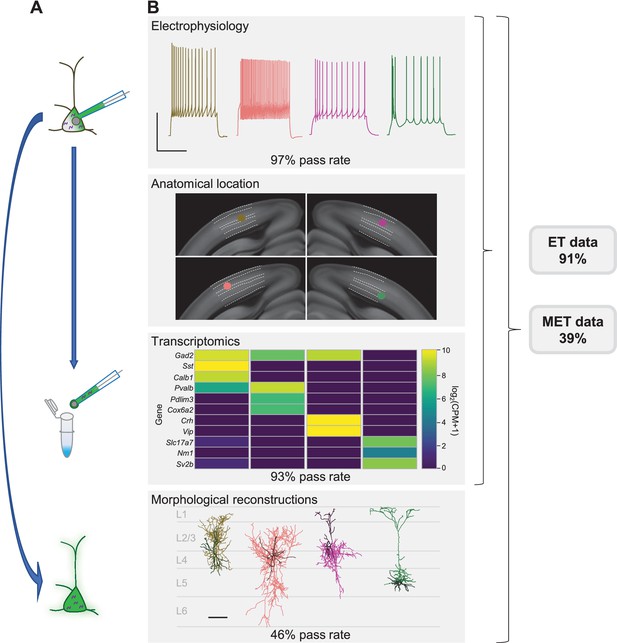
Patch-seq is a powerful technique that allows for the characterization of a neuron based on electrophysiology (E), transcriptomics (T), and morphology (M).
(A) A neuron is patched to characterize the intrinsic properties of a neuron while biocytin and Alexa dye diffuses throughout the soma, dendrites, and axon. At the conclusion of the recording the nucleus is extracted and submitted for RNA-sequencing while the slice is fixed, stained, mounted, and assessed for morphological recovery. (B) Exemplar single neuron examples: from left to right are inhibitory neurons from Sst-Cre, Pvalb-Cre, Vip-Cre mice and an excitatory neuron from an Rbp4-Cre mouse. Electrophysiology data shown are the voltage responses to a suprathreshold stimulus to assess a neuron’s intrinsic firing properties. Scale bar is 50 mV and 500 ms. The patched neuron was mapped to a common coordinate framework to define anatomical location. Transcriptomic data are shown as a heat map of selected ‘on’ marker genes detected from the patched neuron. Morphological reconstructions from the patched neurons are shown, as well as their placement within the cortical layer. For inhibitory neurons the darker line represents the dendrite and the lighter line represents the axon. For the excitatory neuron the darker line represents the basal dendrite and lighter line represents the apical dendrite. Data and rates represented here are from the steady-state, fully functional pipeline. Scale bar is 50 mV/500 ms for electrophysiology and 100 µm for reconstructions. Neurons that have a failing morphology outcome but pass for electrophysiology and transcriptomics are salvageable for ET characterization, while neurons that pass all data modalities proceed for MET characterization. Cell IDs are: Sst-Cre – 851016910, Pvalb-Cre – 837594867, Vip-Cre – 755592855, Rbp4-Cre – 841854478.
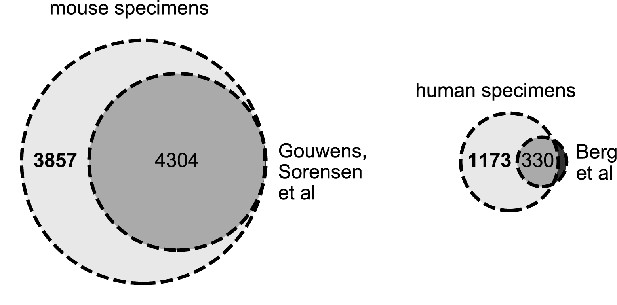
Distribution of data.
The Venn diagrams represent the distribution of mouse and human data and the amount of data that was either included or not included in the respective papers – Gouwens et al., 2020 and Berg et al., 2020.
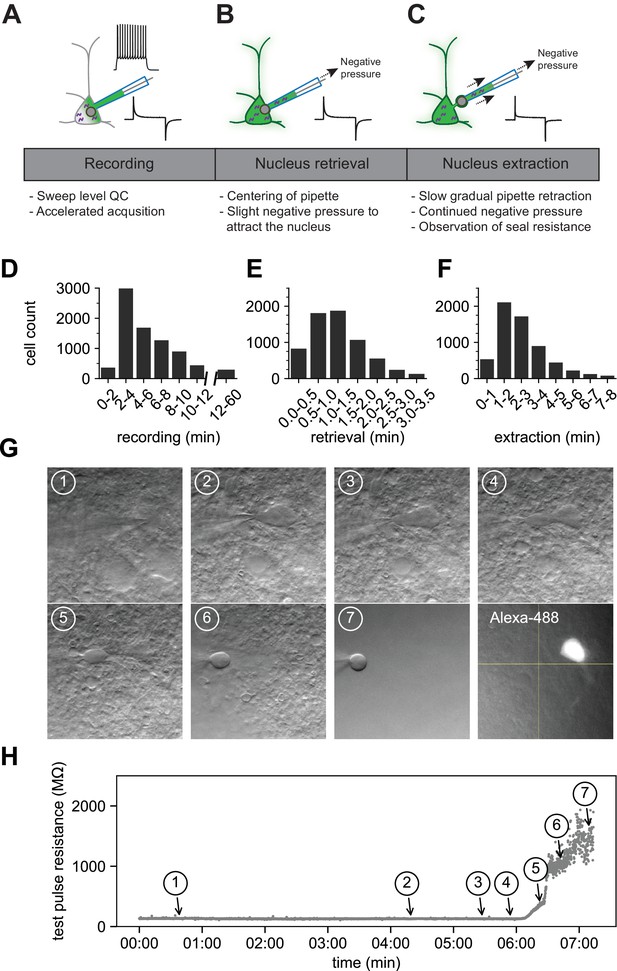
The optimized Patch-seq protocol consists of electrophysiological recording followed by nucleus retrieval and extraction.
A schematic of the three major stages of the Patch-seq protocol: (A) recording, (B) nucleus retrieval, and (C) nucleus extraction. Histograms represent the binned time spent for (D) recording (N = 7950), (E) nucleus retrieval (N = 6750), and (F) nucleus extraction (N = 6281). (G) Time series of high-resolution images showing the gradual pipette retraction with subsequent nucleus extraction. In (G1), the red line denotes outline of soma and the last panel is the fill of the soma and processes visualized by Alexa-488. Yellow asterisk identifies the nucleus as it is extracted from the soma. Blue caret identifies the somatic membrane as it is stretched with the extraction of the nucleus. (H) is the time plot of steady state resistance, as measured from the test pulse, during the nucleus extraction phase for the neuron in (G) with numbers corresponding to brightfield image.
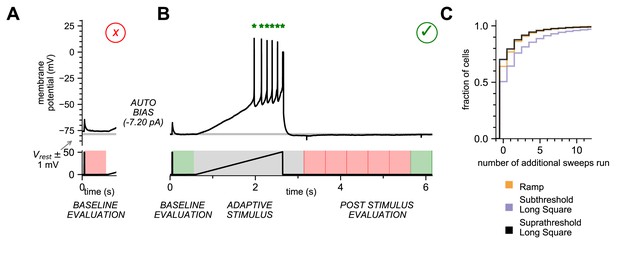
Online analysis during electrophysiology recording allows for fast, high-quality data generation.
(A) Baseline Evaluation failed. Resting Membrane Potential (RMP) was not maintained at ±1 mV (Allen Institute criteria) of the established RMP (gray line at –78 mV); sweep failed and was terminated. Additional autobias current of –7.20 pA was applied during the inter-sweep interval to bring the RMP to the target value of –78 mV. (B) Baseline Evaluation passed and the ramp current stimulus was applied (Adaptive Stimulus). Current injection rose at 25 pA/s until five action potentials were detected, at which point current injection was automatically terminated. There was a 500 ms minimal recovery period upon conclusion of stimulus, followed by an assay of RMP every 500 ms (Post Stimulus Evaluation) to test for return to the initial RMP. Once the RMP recovered to within ±1 mV (Allen Institute criteria) acquisition was terminated and sweep was considered as ‘passed.’ (C) Cumulative probability plot showing the number of additional sweeps needed to qualify as passed for each of the three stimulus sets (N = 4488 recordings).
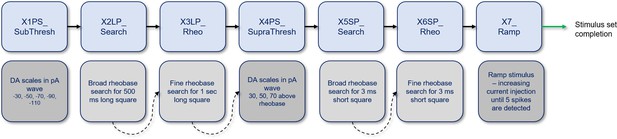
MIES analysis functions for stimulus sets.
Workflow describing the sequence of Patch-seq stimulus sets (in blue) and the associated analysis functions (in gray) used to identify and/or implement in the corresponding stimulus set. Dashed line indicates that data are used from one stimulus set to the next.
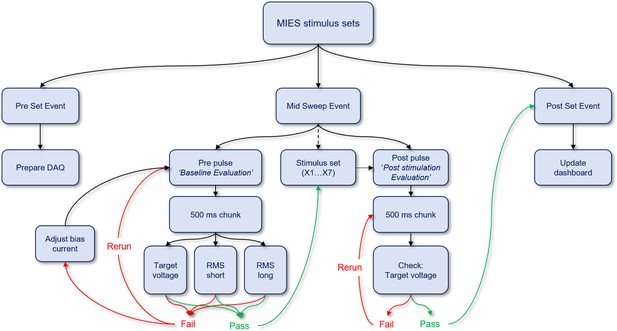
MIES analysis functions for quality control.
Workflow describing how various quality control analysis functions are performed for each stimulus set.
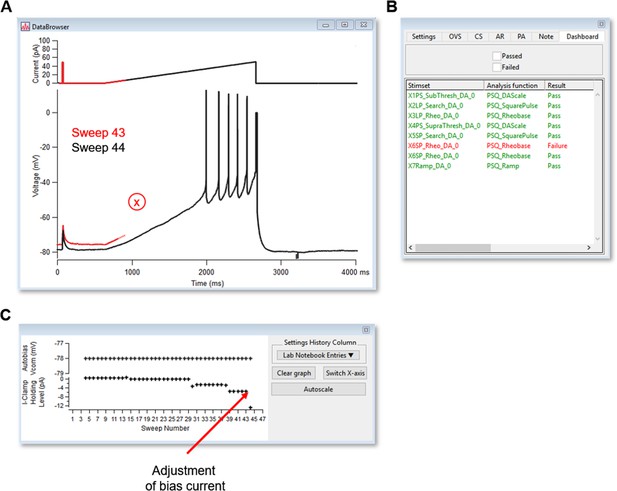
MIES dashboard sweep viewer.
(A) Data browser/sweep viewer used to display most recent sweep and/or previous sweeps. Example shown is Sweep 43, in red, and its failure to maintain Allen Institute criteria of ±1 mV of designated RMP. Adjustment of Bias current was applied to maintain designated RMP, as noted in (B) and sweep 44 was rerun, passing all QC criteria. (B) Lab notebook. A variety of features can be plotted during the course of recording. (C) Dashboard displaying results of each stimulus set.
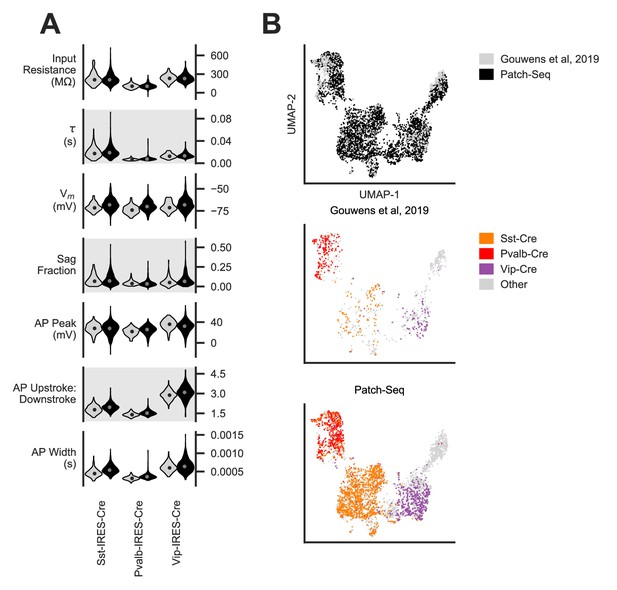
Consistency of electrophysiological features in Patch-seq and traditional acquisition protocols.
(A) Electrophysiological features collected from neurons in three major inhibitory subclass Cre-lines using the Patch-seq protocol match those from previously published neurons (Gouwens et al., 2019). Sst (N = 112 Gouwens et al., 2019, 265 Patch-seq); Pvalb (N = 185 Gouwens et al., 2019, 441 Patch-seq); Vip (N = 90 Gouwens et al., 2019, 514 Patch-seq). (B) UMAP embedded from z-scored electrophysiology sparse principal components (sPCs) collected from the combined inhibitory neuron electrophysiology data sets from Gouwens et al., 2019 and Patch-seq datasets shows that neurons from the two data sets reside in overlapping electrophysiological space. UMAP contains N = 4266 from Gouwens et al., 2020 previously unreported cells.
-
Figure 3—figure supplement 4—source data 1
UMAP coordinates for ME and MET specimens.
- https://cdn.elifesciences.org/articles/65482/elife-65482-fig3-figsupp4-data1-v2.csv
-
Figure 3—figure supplement 4—source data 2
Select electrophysiological features for ME and MET specimens.
- https://cdn.elifesciences.org/articles/65482/elife-65482-fig3-figsupp4-data2-v2.csv
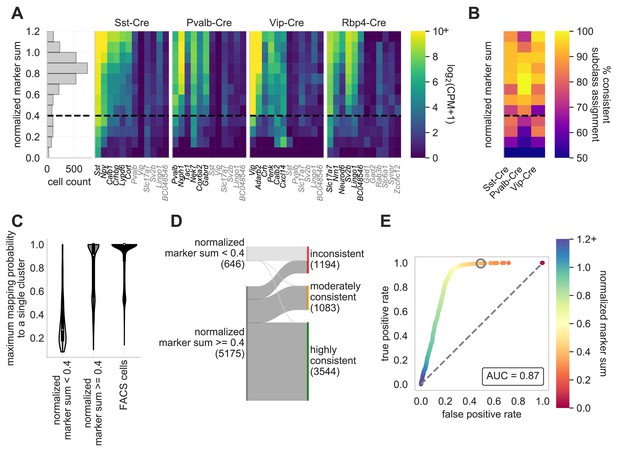
Normalized marker sum is a measure for transcriptomic quality.
To examine the quality of Patch-seq samples we used NMS. The first panel (A) is a histogram that displays the range of scores from all neurons from three of the major inhibitory Cre lines (Sst-Cre, Pvalb-Cre, and Vip-Cre) and one excitatory Cre line (Rbp4-Cre) with the dotted line, at 0.4, indicating the separation between pass and fail (N = 2994). The heat maps display the average gene expression of ‘on-marker’ (in black) or ‘off-marker’ (in gray) genes for these neurons. Gene expression values are log2(CPM + 1). (B) A heatmap showing how the NMS score relates to the % of cells assigned to a specific subclass and the corresponding Cre line. (C) Patch-seq samples above the NMS threshold map with a higher probability to a single reference transcriptomic type (t-type) derived from dissociated (FACS) cells (Tasic et al., 2018; mapping procedure described in Gouwens et al., 2020). FACS cells also map with high probability to single reference t-types when subjected to the same mapping procedure (N = 646 NMS < 0.4 samples; N = 5175 NMS ≥ 0.4 samples; N = 13464 neuronal FACS samples). (D) Sankey plot showing the confidence with which Patch-seq transcriptomes with NMS above or below 0.4 mapped to one or more reference t-types derived from dissociated cells (mapping confidence measure described in Gouwens et al., 2020). (E) ROC analysis comparing N = 4627 highly and moderately consistently mapping versus N = 1194 inconsistently mapping patched neurons. Heat map of the ROC curve is the NMS. Gray circle is at NMS = 0.4 and gray dashed line is the line of identity.
-
Figure 4—source data 1
Select genes and corresponding values for cell specimens obtained from Pvalb-IRES-Cre;Ai14 mice.
- https://cdn.elifesciences.org/articles/65482/elife-65482-fig4-data1-v2.csv
-
Figure 4—source data 2
Select genes and corresponding values for cell specimens obtained from Rbp4_KL100-Cre;Ai14 mice.
- https://cdn.elifesciences.org/articles/65482/elife-65482-fig4-data2-v2.csv
-
Figure 4—source data 3
Select genes and corresponding values for cell specimens obtained from Sst-IRES-Cre;Ai14 mice.
- https://cdn.elifesciences.org/articles/65482/elife-65482-fig4-data3-v2.csv
-
Figure 4—source data 4
Select genes and corresponding values for cell specimens obtained from Vip-IRES-Cre;Ai14 mice.
- https://cdn.elifesciences.org/articles/65482/elife-65482-fig4-data4-v2.csv
-
Figure 4—source data 5
Percentage values of cells assigned to a specific subclass and NMS score for Sst-Cre, Pvalb-Cre and Vip-Cre mice.
- https://cdn.elifesciences.org/articles/65482/elife-65482-fig4-data5-v2.csv
-
Figure 4—source data 6
Probability of single cell’s ability to map to a single cluster.
- https://cdn.elifesciences.org/articles/65482/elife-65482-fig4-data6-v2.csv
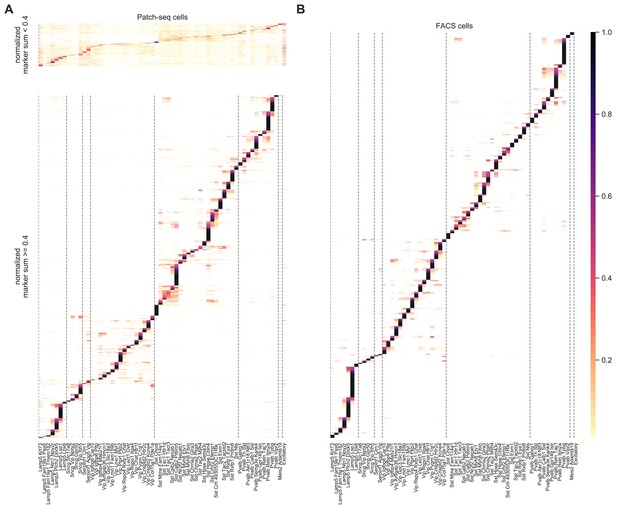
Mapping probabilities for neurons with low/high normalized marker sum.
(A) Mapping probability matrix between cell types for Patch-seq data. Neurons with an NMS < 0.4 (top panel; N = 646) map to multiple types, while the majority of neurons with an NMS ≥ 0.4 map to a single type, or types within the same subtype (bottom panel; N = 5175). (B) Applying the same mapping method to map data from dissociated FACS-sorted neurons (Tasic et al., 2018, N = 6125) onto the same taxonomy results in a reference probability matrix that demonstrates the level of expected uncertainty in mapping. Color bar indicates mapping probability.
-
Figure 4—figure supplement 1—source data 1
Probability of mapping to transcriptomic cell types for FACS specimens.
- https://cdn.elifesciences.org/articles/65482/elife-65482-fig4-figsupp1-data1-v2.csv
-
Figure 4—figure supplement 1—source data 2
Probability of mapping to transcriptomic cell types for NMS-fail Patch-seq specimens.
- https://cdn.elifesciences.org/articles/65482/elife-65482-fig4-figsupp1-data2-v2.csv
-
Figure 4—figure supplement 1—source data 3
Probability of mapping to transcriptomic cell types for NMS-pass Patch-seq specimens.
- https://cdn.elifesciences.org/articles/65482/elife-65482-fig4-figsupp1-data3-v2.csv
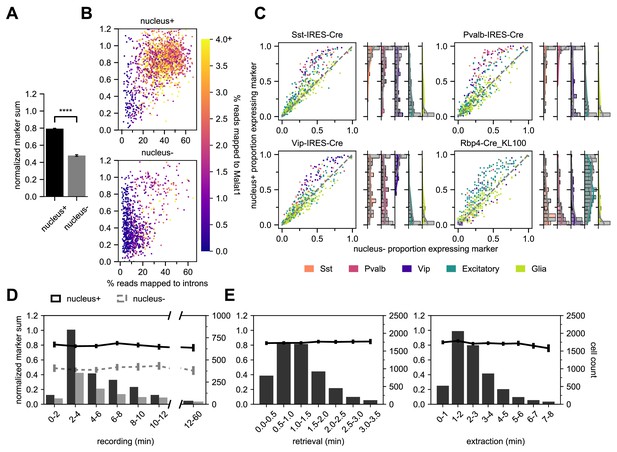
Nucleus extraction is key to transcriptomic success.
(A,B,D) examine an early dataset of Patch-seq samples with both nucleus+ and nucleus- outcomes (N = 1908 nucleus+ samples; N = 897 nucleus- samples). The bar plots in (A) represent the difference in NMS scores. (B) A scatter plot viewing the distribution of % reads mapped to introns and how they related to the NMS score for nucleus+ (top) and nucleus- (bottom). Color ramp indicates the % reads mapped to Malat1. (C) Scatter plots examining the relationship between nucleus+ or nucleus- and the presence of subclass-specific marker gene expression in a subset of data from (A,B,D) that includes four different Cre lines. The colors of dots represent the subclass to which the gene is specific (N = 50 Sst, N = 50 Pvalb, N = 50 Vip, N = 193 Excitatory, N = 146 Glia genes). Data pooled from N = 386/131 Sst-Cre, N = 97/55 Pvalb-Cre, N = 204/95 Vip-Cre and N = 84/38 Rbp4-Cre neurons (nucleus+/nucleus-). The histograms are the quantification of subclass-specific genes for each of the respective Cre lines for nucleus+ samples only. The gray bars are FACS data (N = 356 Sst-Cre, N = 532 Pvalb-Cre, N = 345 Vip-Cre and N = 771 Rbp4-Cre). In (D), the histograms represent the binned time spent and the neuron count for recording duration, the darker bars plot successful extraction of the nucleus (nucleus+) whereas the lighter bars plot failed extraction of nucleus (nucleus-). Solid line represents the NMS score for nucleus+ samples as a function of time, whereas gray dotted line represents nucleus- samples. (E) represents the binned time spent and neuron count for nucleus retrieval (left) and nucleus extraction (right) (N = 5930). The solid line represents the NMS score as a function of time.
-
Figure 5—source data 1
Subclass-specific gene expression for nucleus+ and nucleus- specimens from Sst-Cre, Pvalb-Cre, Vip-Cre and Rbp4_KL100_Cre mice.
- https://cdn.elifesciences.org/articles/65482/elife-65482-fig5-data1-v2.csv
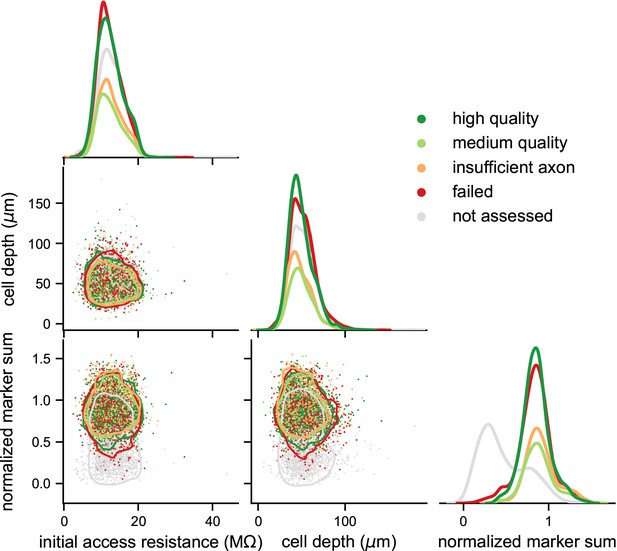
Initial access resistance and cell depth in slice are not associated with different transcriptomic or morphological outcomes.
Pairwise relationships between initial access resistance, cell depth and normalized marker sum, with colors mapping to morphological quality outcomes; N = 6230. Off-diagonal scatterplots are overlayed with a kernel density estimates (KDE) for each morphological outcome; 10% of the probability mass lies outside of each contour. Diagonal is a KDE showing the smoothed distribution of values for each measurement.
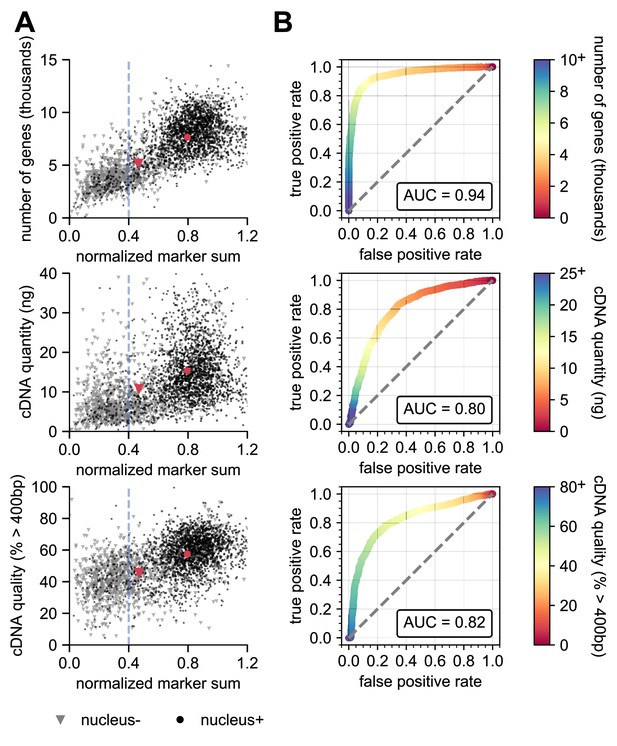
Nucleus+ is predictive for successful sequencing and cDNA data.
Scatter plots (A) showing the distribution of values of the NMS and ROC analyses (B) of the number of genes detected (top), cDNA quantity (ng) (middle), or cDNA quality (% > 400 b.p.) (bottom). In the scatter plots, gray triangles are nucleus- samples, whereas black circles are nucleus+ samples. The larger red symbols represent the means for each group. The dashed blue line is at the NMS pass/fail threshold of 0.4. N = 1950 nucleus+; N = 953 nucleus-. Gray dashed line in ROC analyses is the line of identity.
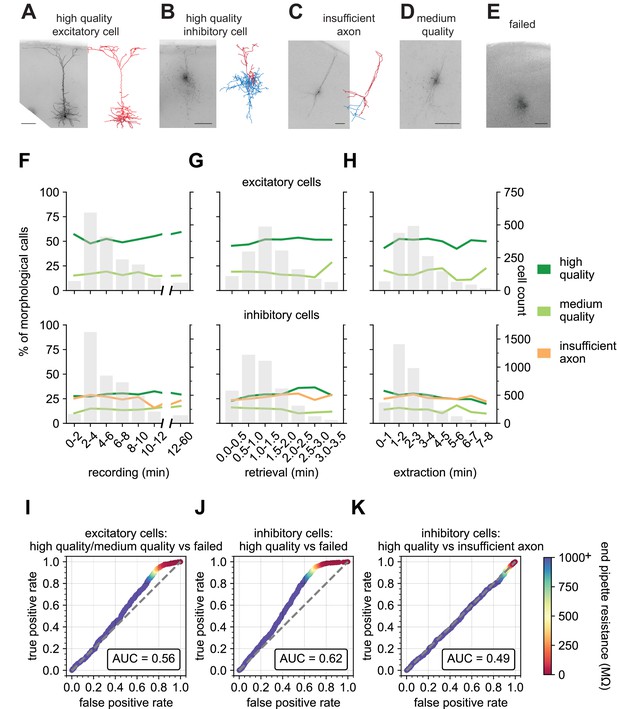
End pipette resistance, but not recording duration, is a significant predictor of morphology success.
Example biocytin recovery and subsequent morphological reconstructions of a high quality filled Rbp4-Cre+ excitatory (A) and a Vip-Cre+ inhibitory (B) neuron. (C) A Vip-Cre+ inhibitory neuron that failed due to an insufficiently filled axon. (D) An interneuron that was weakly filled and was classified as medium quality. (E) A failed fill where no processes are visible and biocytin leakage is apparent. (A–E) are high-resolution 63 x stack minimum image projection (MIP) images and (A–C) have the corresponding morphological reconstruction with dendrites in red and axon in blue. Line plots demonstrate the time spent for each of the Patch-seq phases: (F) recording (N = 1666 excitatory; N = 4214 inhibitory), (G) nucleus retrieval (N = 1514 excitatory; N = 4040 inhibitory), and (H) nucleus extraction (N = 1537 excitatory; N = 3752 inhibitory), and how they relate to the morphological recovery. ROC analyses comparing high/medium quality versus failed morphology outcomes for patched (I) excitatory (N = 1064 high/medium quality; N = 531 failed) and (J) inhibitory neurons (N = 1121 high quality; N = 1288 failed). (K) ROC analysis comparing N = 1121 high quality versus N = 1041 insufficient axon morphology outcomes for patched inhibitory neurons. Heat map of the ROC curve is the end pipette resistance (MΩ) measured at the conclusion of nucleus extraction. Gray dashed line in ROC analyses is the line of identity. Cell IDs are: high-quality excitatory neuron – 841854478, high-quality inhibitory neuron – 755592855, insufficient axon – 642494910.
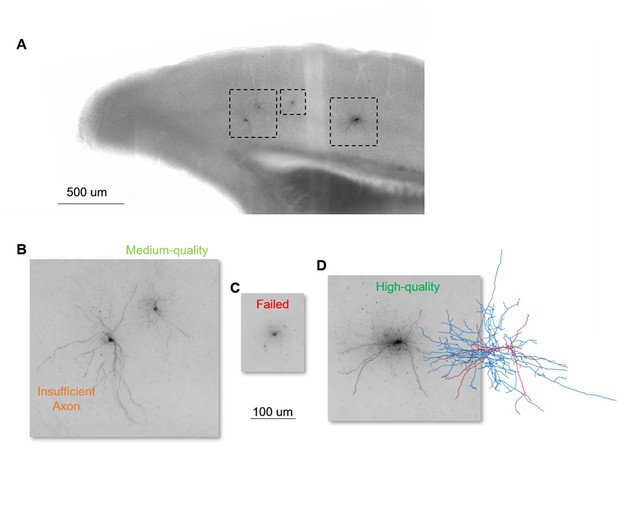
Example biocytin recovery and morphological calls.
(A) is a 20 x brightfield image of a coronal slice from a Sst-Cre line containing biocytin fills from four Patch-seq recordings. (B–D) are 63 x MIP images from the regions identified in (A) and their subsequent morphology outcome. (B) contains insufficient axon, left, and medium quality, right, fills; (C) is a failed fill, and (D) is a high-quality fill with corresponding morphological reconstruction with dendrites in red and axon in blue. Cell IDs from left to right are: 723612109, 723632259, 723947788, 723949579.
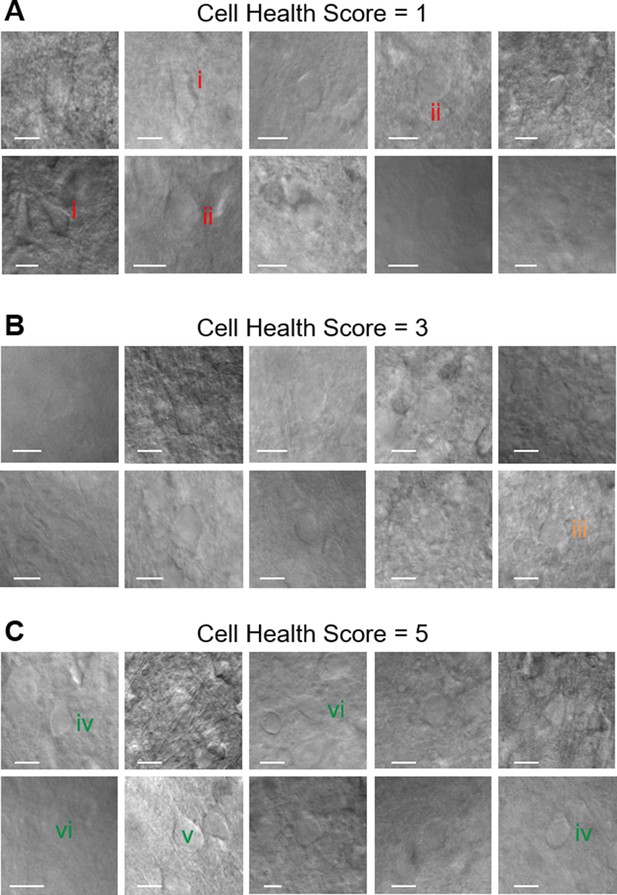
Examples of cell health ranking.
Examples of cell health calls, their corresponding images under brightfield ×40 magnification and representative features. (A) Cell health score = 1 features include: (i) shriveled/dark edge or (ii) flat and swollen appearance. (B) Cell health score = 3 features include: (iii) smoother, more defined soma. (C) Cell health score = 5 features include: (iv) well defined soma, (v) plump, 3D shape and/or (vi) soft and lighter/dim in appearance.
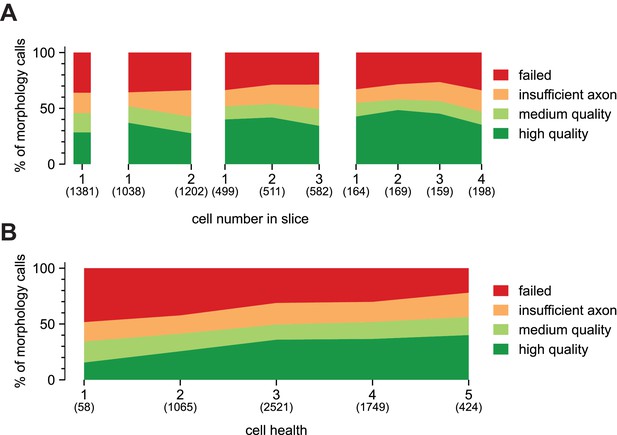
Patching sequence and cell health and how it relates to morphological recovery.
(A) A stacked area plot displaying the percentage of morphology outcomes and how they relate to cell health. Cell health is based on a qualitative assessment of visual appearance and recording quality, with a score of 1 designated as worst ranging to five as best. Numbers below, in parentheses, are sample sizes. (B) A stacked area plot displaying the percentage of morphology outcomes and how they relate to the cell number in a slice. Blocks shown are slices that had 1, 2, 3, or 4 recordings per slice before fixation in PFA. Numbers below, in parentheses, are sample sizes.
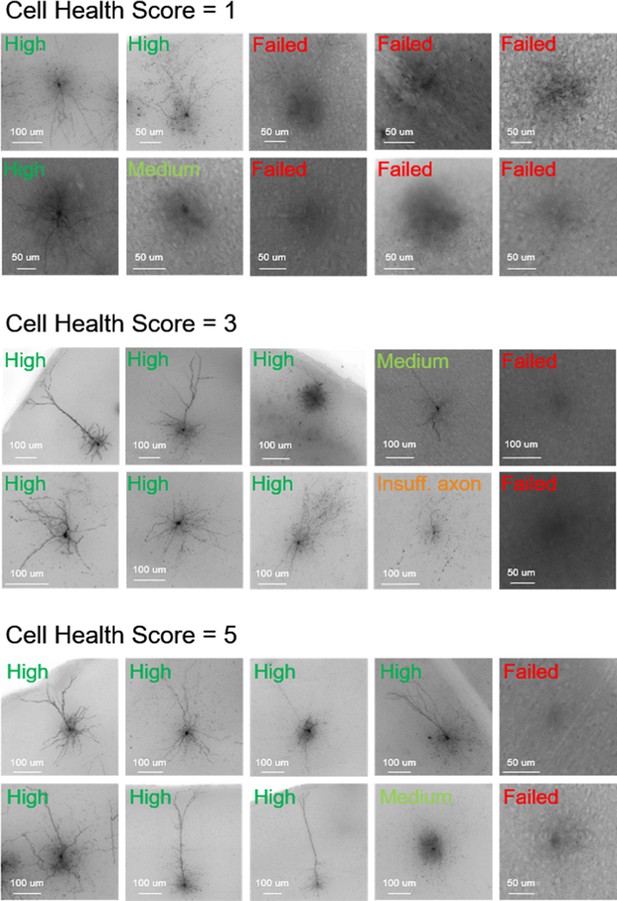
Examples of cell health and morphological outcome examples.
Cell health scores of 1, 3, or 5 and high-resolution 63 x MIPs of the biocytin-stained fills. Their subsequent morphological outcome is listed in each image. Cell IDs clockwise starting in upper left corner Cell health score 1: 661331024, 750807290, 869512878, 882454517, 652956871, 665602299, 855823375, 816110624, 730878829, 745040951 Cell health score 3: 841854478, 904059133, 914412754, 932264861, 880817724, 898967534, 825552023, 836592858, 875021432, 863585604 Cell health score 5: 761304075, 759948010, 720318238, 823868835, 920542161, 852202575, 809113130, 963287636, 759948010, 831175106.
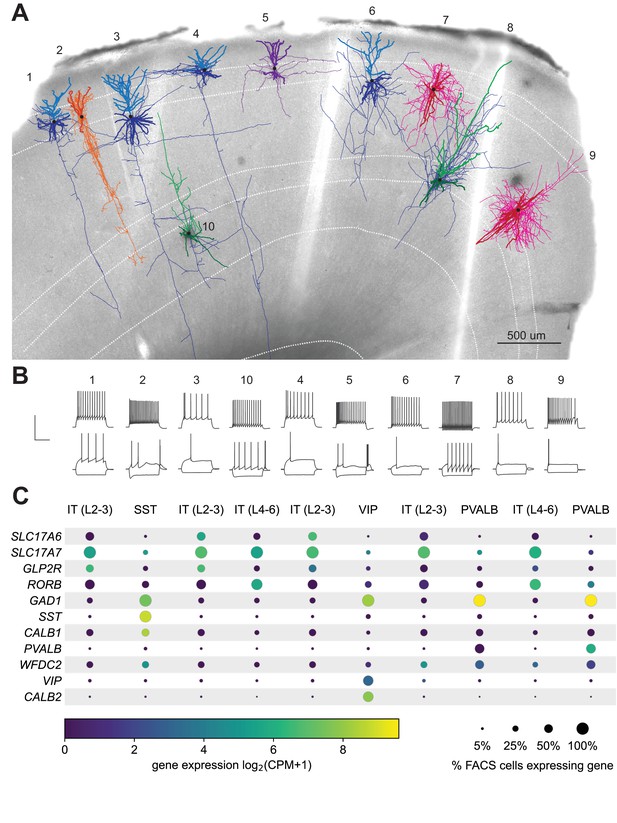
Exemplar human neocortical slice with 10 successful Patch-seq recordings.
(A) is a 20 x bright field image of a human acute neocortical slice containing biocytin reacted fills with overlaid digital reconstructions from ten Patch-seq recordings. Reconstructions are colored by their corresponding mapped subclass: IT (L2-3) - blue, IT (L4-6) - green, SST - orange, VIP - purple and PVALB - pink. Axons are light-weighted lines and the basal/apical dendrites for excitatory neurons are distinguishable by darker and lighter colors, respectively. Neurons are numbered in the order in which they were patched. Scale bar is 500 µm. (B) Voltage responses of corresponding neuron to 1 s-long current steps equal to –70 pA and rheobase (bottom traces) and rheobase +80 pA (top trace). Scale bar, vertical 80 mV, horizontal 500 ms. (C) Dot plot of marker genes that are differentially expressed across human cortical neurons. Expression values (color) for each neuron in slice pictured in (A) are log2-transformed. Percentages calculated from N = 3706 IT (L2-3); N = 5636 IT (L4-6); N = 809 SST; N = 1298 VIP; N = 809 PVALB dissociated neurons. Cell IDs, from left to right are: 689300528, 689306781, 689309060, 689371684, 689312221, 689319155, 689323433, 689331391, 689338841, 689360084.
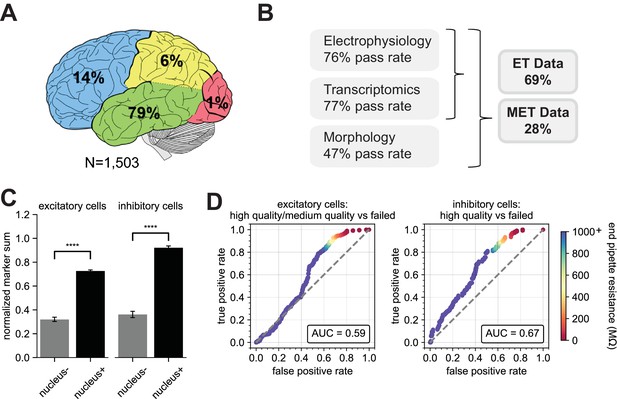
Application of the Patch-seq protocol to human neocortical cells.
(A) Depiction showing the distribution of human neocortical Patch-seq recordings and the lobes which the tissue was surgically excised from. In (B) are the pass rates for each of the independent modalities, ET characterization and MET characterization (N = 1,402). The bar plots in (C) represent the difference in NMS scores in nucleus- and nucleus+ samples from excitatory (N = 165 nucleus-; N = 720 nucleus+) and inhibitory (N = 129 nucleus-; N = 489 nucleus+) classes. Nucleus+ have significantly higher NMS scores than nucleus- samples, t(883) = 19.81 for excitatory; t(616) = 17.32 for inhibitory, p < 0.0001 ROC analyses comparing high/medium quality versus failed morphology outcomes for patched (D) excitatory (N = 269 high/medium quality; N = 321 failed) and inhibitory neurons (N = 106 high quality; N = 232 failed). Gray dashed line in ROC analyses is the line of identity.
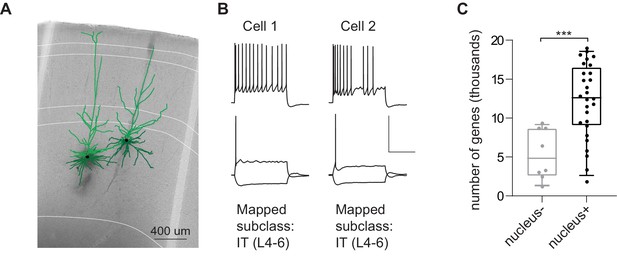
Application of the Patch-seq protocol to macaque neocortical neurons.
(A) is a 20 x bright field image of a macaque acute neocortical slice containing biocytin reacted fills with overlaid digital reconstructions from two Patch-seq recordings. Axons are light weight lines and the basal/apical dendrites for excitatory neurons are distinguishable by darker and lighter colors, respectively. Scale bar is 400 µm. (B) Voltage responses of corresponding neuron to 1 s-long current steps equal to –70 pA and rheobase (bottom traces) and rheobase +80 pA (top trace). Scale bar, vertical 80 mV, horizontal 500 ms. Both neuron 1 and 2 mapped to the excitatory subclass IT (L4-6). (C) Box plots showing the distribution of values for the number of genes detected for nucleus- and nucleus+ samples, t(34)=3.80, p < 0.001.
Videos
MIES and a successful experiment. A video demonstrating the efficiency and speed of MIES at acquiring intrinsic features from a whole-cell electrophysiology experiment.
MIES and the Rheobase analysis function. A video demonstrating the automated functionality of determining rheobase (PSQ_Rheobase) and spike detection. At time point = 0:00 – 0:35, the X3LP_Rheobase_DA_0 stimulus set is ran to search for the Rheobase with a 1 sec square depolarizing pulse. 30, 20, 22, 24, 26, 28, 30 pA steps are administered and ultimately fails to detect an action potential and ultimately determine rheobase - ‘Failure as we were note able to find the correct on/off spike pattern’. At time point = 0:52, X3LP_Rheobase_DA_0 ran again with MIES automatically adjusting the current steps to 30, 40, 38, and 40 pA, with successful action potential detection and determination of rheobase.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (M. musculus) | B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J, Ai14 | The Jackson Laboratory | RRID: IMSR_JAX: 007914 | |
| Strain, strain background (M. musculus) | B6; Batf3tm1(flpo)Hze/J, Batf3-IRES2-FlpO-neo | The Jackson Laboratory | RRID: IMSR_JAX: 034301 | |
| Strain, strain background (M. musculus) | B6.Cg-Calb1tm2.1(cre)Hze/J,Calb1-IRES2-Cre | The Jackson Laboratory | RRID: IMSR_JAX: 028532 | |
| Strain, strain background (M. musculus) | STOCK Ccktm1.1(cre)Zjh/J,Cck-IRES-Cre | The Jackson Laboratory | RRID: IMSR_JAX:012706 | |
| Strain, strain background (M. musculus) | B6;129S6-Chattm2(cre)Lowl/J,Chat-IRES-Cre | The Jackson Laboratory | RRID: IMSR_JAX:006410 | |
| Strain, strain background (M. musculus) | STOCK Tg(Chrna2-cre)OE25Gsat/Mmucd, Chrna2-Cre_OE25 | MMRRC | RRID: MMRRC_036502-UCD | |
| Strain, strain background (M. musculus) | B6; Chrna6tm1(flpo)Hze /J, Chrna6-IRES2-FlpO-neo | The Jackson Laboratory | RRID: IMSR_JAX:034302 | |
| Strain, strain background (M. musculus) | B6.Cg-Ccn2tm1.1(folA/cre)Hze/J,Ctgf-T2A-dgCre | The Jackson Laboratory | RRID: IMSR_JAX:028535 | |
| Strain, strain background (M. musculus) | B6;129S6-Ctxn3em1(flpo)Hze/J, Ctxn3-IRES2-FlpO-neo | The Jackson Laboratory | RRID: IMSR_JAX:034303 | |
| Strain, strain background (M. musculus) | B6(Cg)-Cux2tm3.1(cre/ERT2)Mull/Mmmh,Cux2-CreERT2 | MMRRC | RRID: MMRRC_032779-MU | |
| Strain, strain background (M. musculus) | B6;129S-Esr2tm1.1(cre)Hze/J, Esr2-IRES2-Cre | The Jackson Laboratory | RRID: IMSR_JAX: 030158 | |
| Strain, strain background (M. musculus) | B6(Cg)-Etv1tm1.1(cre/ERT2)Zjh/J,Etv1-CreERT2 | The Jackson Laboratory | RRID: IMSR_JAX:013048 | |
| Strain, strain background (M. musculus) | B6J.Cg-Gad2tm2(cre)Zjh/MwarJ, Gad2-IRES-Cre | The Jackson Laboratory | RRID: IMSR_JAX:028867 | |
| Strain, strain background (M. musculus) | STOCK Tg(Gad1-EGFP)94Agmo/J, Gad67-GFP_X94 | The Jackson Laboratory | RRID: IMSR_JAX:006334 | |
| Strain, strain background (M. musculus) | STOCK Tg(Colgalt2-cre)NF107Gsat/Mmucd, Glt25d2-Cre_NF107 | MMRRC | RRID: MMRRC_036504-UCD | |
| Strain, strain background (M. musculus) | B6;129S6-Gpr139tm1.1(flpo)Hze/J, Gpr139-IRES2-FlpO-WPRE-neo | The Jackson Laboratory | RRID: IMSR_JAX:034304 | |
| Strain, strain background (M. musculus) | STOCK Tg(Htr3a-cre)NO152Gsat/Mmucd, Htr3a-Cre_NO152 | MMRRC | RRID: MMRRC_036680-UCD | |
| Strain, strain background (M. musculus) | B6.Cg-Ndnftm1.1(folA/cre)Hze/J,Ndnf-IRES2-dgCre | The Jackson Laboratory | RRID: IMSR_JAX:028536 | |
| Strain, strain background (M. musculus) | STOCK Nkx2-1tm1.1(cre/ERT2)Zjh/J,Nkx2.1-CreERT2 | The Jackson Laboratory | RRID: IMSR_JAX:014552 | |
| Strain, strain background (M. musculus) | B6;129S-Nos1tm1.1(cre/ERT2)Zjh/J,Nos1-CreERT2 | The Jackson Laboratory | RRID: IMSR_JAX:014541 | |
| Strain, strain background (M. musculus) | B6;129S-Npr3tm1.1(cre)Hze/J, Npr3-IRES2-Cre | The Jackson Laboratory | RRID: IMSR_JAX: 031333 | |
| Strain, strain background (M. musculus) | FVB-Tg(Nr5a1-cre)2Lowl/J,Nr5a1-Cre | The Jackson Laboratory | RRID: IMSR_JAX:006364 | |
| Strain, strain background (M. musculus) | B6.FVB(Cg)-Tg(Ntsr1-cre)GN220Gsat/Mmucd, Ntsr1-Cre_GN220 | The Jackson Laboratory | RRID: MMRRC_030648-UCD | |
| Strain, strain background (M. musculus) | B6;129S-Oxtrtm1.1(cre)Hze/J, Oxtr-T2A-Cre | The Jackson Laboratory | RRID: IMSR_JAX: 031303 | |
| Strain, strain background (M. musculus) | B6;129S-Pdyntm1.1(cre/ERT2)Hze/J, Pdyn-T2A-CreERT2 | The Jackson Laboratory | RRID: IMSR_JAX: 030197 | |
| Strain, strain background (M. musculus) | B6;129P2-Pvalbtm1(cre)Arbr/J,Pvalb-IRES-Cre | The Jackson Laboratory | RRID: IMSR_JAX:008069 | |
| Strain, strain background (M. musculus) | B6.Cg-Pvalbtm1.1(cre/ERT2)Hze/J, Pvalb-T2A-CreERT2 | The Jackson Laboratory | RRID: IMSR_JAX: 021189 | |
| Strain, strain background (M. musculus) | STOCK Tg(Rbp4-cre)KL100Gsat/Mmucd, Rbp4-Cre_KL100 | MMRRC | RRID: MMRRC_031125-UCD | |
| Strain, strain background (M. musculus) | B6.Cg-Rorbtm3.1(flpo)Hze/J, Rorb-IRES2-Cre | The Jackson Laboratory | RRID: IMSR_JAX: 029590 | |
| Strain, strain background (M. musculus) | B6;C3-Tg(Scnn1a-cre)1Aibs/J, Scnn1a-Tg1-Cre | The Jackson Laboratory | RRID: IMSR_JAX: 009111 | |
| Strain, strain background (M. musculus) | B6;C3-Tg(Scnn1a-cre)2Aibs/J,Scnn1a-Tg2-Cre | The Jackson Laboratory | RRID: IMSR_JAX:009112 | |
| Strain, strain background (M. musculus) | B6;C3-Tg(Scnn1a-cre)3Aibs/J,Scnn1a-Tg3-Cre | The Jackson Laboratory | RRID: IMSR_JAX:009613 | |
| Strain, strain background (M. musculus) | STOCK Tg(Sim1-cre)KJ18Gsat/Mmucd, Sim1-Cre_KJ18 | MMRRC | RRID: MMRRC_031742-UCD | |
| Strain, strain background (M. musculus) | B6J.129S6(FVB)-Slc17a6tm2(cre)Lowl/MwarJ,Slc17a6-IRES-Cre | The Jackson Laboratory | RRID: IMSR_JAX:028863 | |
| Strain, strain background (M. musculus) | B6;129S-Slc17a7tm1.1(cre)Hze/J,Slc17a7-IRES2-Cre | The Jackson Laboratory | RRID: IMSR_JAX:023527 | |
| Strain, strain background (M. musculus) | STOCK Tg(Slc17a8-icre)1Edw/SealJ, Slc17a8-iCre | The Jackson Laboratory | RRID: IMSR_JAX:018147 | |
| Strain, strain background (M. musculus) | B6;129S-Slc17a8tm1.1(cre)Hze/J, Slc17a8-IRES2-Cre | The Jackson Laboratory | RRID: IMSR_JAX: 028534 | |
| Strain, strain background (M. musculus) | B6.Cg-Slc32a1tm1.1(flpo)Hze/J, Slc32a1-IRES2-FlpO | The Jackson Laboratory | RRID: IMSR_JAX: 031331 | |
| Strain, strain background (M. musculus) | B6J.129S6(FVB)-Slc32a1tm2(cre)Lowl/MwarJ,Slc32a1-IRES-Cre | The Jackson Laboratory | RRID: IMSR_JAX:028862 | |
| Strain, strain background (M. musculus) | B6;129S6-Sncgem1(flpo)Hze/J, Sncg-IRES2-FlpO-neo | The Jackson Laboratory | RRID: IMSR_JAX:034424 | |
| Strain, strain background (M. musculus) | B6J.Cg-Ssttm2.1(cre)Zjh/MwarJ,Sst-IRES-Cre | The Jackson Laboratory | RRID: IMSR_JAX:028864 | |
| Strain, strain background (M. musculus) | B6;129S-Tac1tm1.1(cre)Hze/J,Tac1-IRES2-Cre | The Jackson Laboratory | RRID: IMSR_JAX:021877 | |
| Strain, strain background (M. musculus) | B6.FVB(Cg)-Tg(Th-cre)FI172Gsat/Mmucd, Th-Cre_FI172 | MMRRC | RRID: MMRRC_031029-UCD | |
| Strain, strain background (M. musculus) | C57BL/6N-Thtm1Awar/Mmmh, Th-P2A-FlpO or TH-2A-Flpo | MMRRC | RRID: MMRRC_050618-MU | |
| Strain, strain background (M. musculus) | B6.FVB(Cg)-Tg(Tlx3-cre)PL56Gsat/Mmucd, Tlx3-Cre_PL56 | MMRRC | RRID: MMRRC_041158-UCD | |
| Strain, strain background (M. musculus) | B6.FVB(Cg)-Tg(Tlx3-cre)PL56Gsat/Mmucd, Tlx3-Cre_PL56 | MMRRCC | RRID: MMRRC_041158-UCD | |
| Strain, strain background (M. musculus) | B6J.Cg-Viptm1(cre)Zjh/AreckJ,Vip-IRES-Cre | The Jackson Laboratory | RRID: IMSR_JAX:031628 | |
| Strain, strain background (M. musculus) | B6.Cg-Npytm1.1(flpo)Hze/J,Npy-IRES2-FlpO | The Jackson Laboratory | RRID: IMSR_JAX:030211 | |
| Strain, strain background (M. musculus) | B6;129S-Vipr2tm1.1(cre)Hze/J, Vipr2-IRES2-Cre | The Jackson Laboratory | RRID: IMSR_JAX: 031332 | |
| Commercial assay or kit | SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing | Takara | 634,894 | |
| Commercial assay or kit | Nextera XT Index Kit V2 Set A-d | Nextera | FC-131–2001, 2002, 2003, 2004 | |
| Software, algorithm | IGOR pro | Wavemetrics | RRID:SCR_000325 | |
| Software, algorithm | MIES: Software for Electrophysiology functions | Allen Institute | RRID:SCR_016443 | |
| Software, algorithm | Vaa3d | Peng et al., 2010 | https://alleninstitute.org/what-we-do/brain-science/research/products-tools/vaa3d/ | |
| Software, algorithm | IPFX | Allen Institute | https://github.com/alleninstitute/ipfx | Matt, 2021 |
| Software, algorithm | DRCME | Allen Institute, Gouwens et al., 2019 | https://github.com/alleninstitute/drcme | Nathan W, 2021 |
Additional files
-
Supplementary file 1
Patch-seq workflow states and QC definitions.
The purpose of this table is to list the different parameters and purpose for each stage within the Patch-seq protocols, the metrics measured, the subsequent data and range of data obtained. The criteria that the Allen Institute uses for Patch-seq can be adopted, relaxed, or disregarded. 1 Electrophysiology, morphology and transcriptomic parameters and metrics can also be found in the technical white papers at http://celltypes.brain-map.org.
- https://cdn.elifesciences.org/articles/65482/elife-65482-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65482/elife-65482-transrepform1-v2.docx
-
Source data 1
Human MET metadata.
- https://cdn.elifesciences.org/articles/65482/elife-65482-supp2-v2.csv





