Human Erbb2-induced Erk activity robustly stimulates cycling and functional remodeling of rat and human cardiomyocytes
Figures
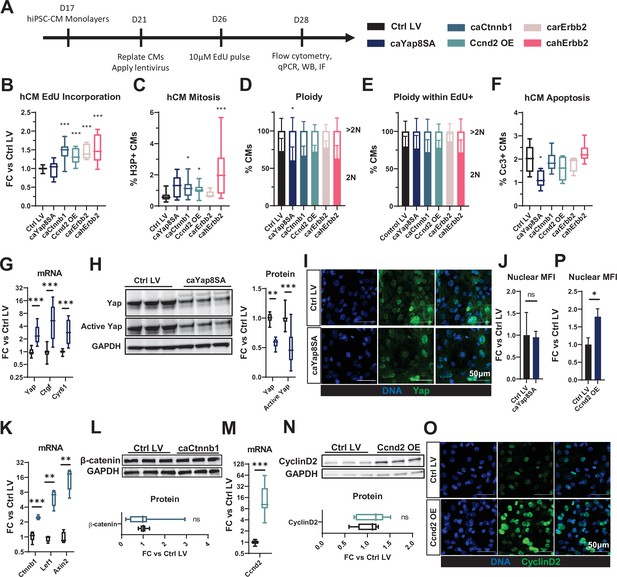
LV-delivered mitogens drive hiPSC-CM proliferation in monolayers.
(A) Schematic of experimental design in hiPSC-CM monolayers. (B–F) Flow cytometry analysis of mCherry+ hiPSC-CMs showing (B) fold-change (FC) in EdU incorporation relative to control LV-transduced hiPSC-CMs, (C) percentage of H3P+ CMs, (D, E) percentage of 2N and >2N cells in all CMs (D) and EdU+ CMs (E), and (F) percentage of apoptotic Cc3+ CMs. (G, H) Analysis of relative (G) gene expression of Yap and its targets Ctgf and Cyr61, and (H) total and active Yap protein abundance in caYap8SA-transduced vs. control hiPSC-CMs. (I,J) Representative immunostaining images (I) and quantified nuclear mean fluorescence intensity (MFI; J) of YAP in caYap8SA-transduced versus control hiPSC-CMs. (K, L) Analysis of relative (K) expression of Ctnnb1 and Wnt-signaling genes Lef1 and Axin2 and (L) Ctnnb1 protein abundance in caCtnnb1-transduced versus control hiPSC-CMs. (M, N) Analysis of relative Ccnd2 (M) gene and (N) protein expression and (O, P) representative immunostaining images (O) and quantified nuclear MFI (P) of Ccnd2 in Ccnd2-transduced versus control hiPSC-CMs. Data: box and whiskers showing distribution and min to max. Column graphs showing mean+ SD (*p<0.05, **p<0.01, ***p<0.001 vs. Ctrl LV). See Supplementary file 1 for sample numbers and complete statistical information for all figures. Cc3, cleaved caspase-3; hiPSC-CM, human-induced pluripotent stem cell-derived cardiomyocyte; LV, lentiviral vector.
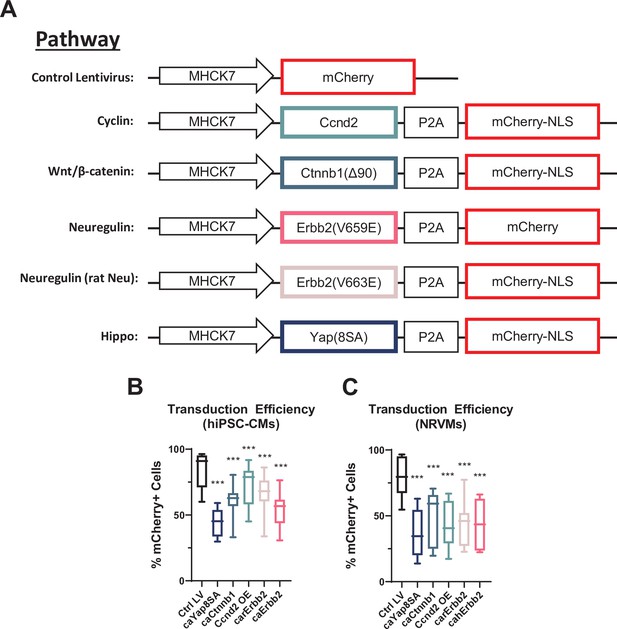
Lentiviral design and transduction efficiency in CM monolayers.
(A) Schematic depicting compositions of lentiviral constructs used to perturb specified pathways. (B–E) Flow cytometry analysis of mCherry signal was used to quantify lentiviral transduction efficiency in (B) hiPSC-CM and (C) NRVM monolayers. Data: box and whiskers showing distribution and min to max (*p<0.05, **p< 0.01, ***p<0.001 vs. Ctrl LV). hiPSC-CM, human-induced pluripotent stem cell-derived cardiomyocyte; LV, lentiviral vector; NRVM, neonatal rat ventricular myocyte.
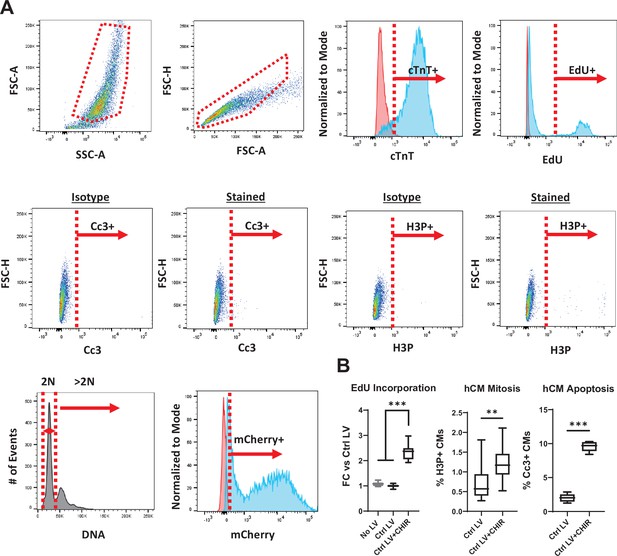
Flow cytometry method for analysis of CM proliferation and apoptosis.
(A) Representative flow cytometry analysis plots showing gating strategies for various stains. All samples used the same indicated FSC-A by SSC-A and FSC-H by FSC-A gating to gate all single cells while excluding doublets. For cTnT staining to verify differentiation purity, the isotype is in red and the stained sample is in blue. For EdU staining, the unstained sample is in red and the stained sample is in blue. For mCherry detection, the non-transduced sample is in red and the transduced sample is in blue. Plots shown for hiPSC-CMs; NRVM analysis was conducted in the same manner. (B) Validation of the flow cytometry analysis method in hiPSC-CMs exposed for 48 hr to a known mitogen, the GSK-3 inhibitor CHIR99021 (2 µM), and simultaneously incubated with EdU. Data: box and whiskers showing distribution and min to max (*p<0.05, **p<0.01, ***p<0.001 vs. Ctrl LV). hiPSC-CM, human-induced pluripotent stem cell-derived cardiomyocyte; LV, lentiviral vector; NRVM, neonatal rat ventricular myocyte.
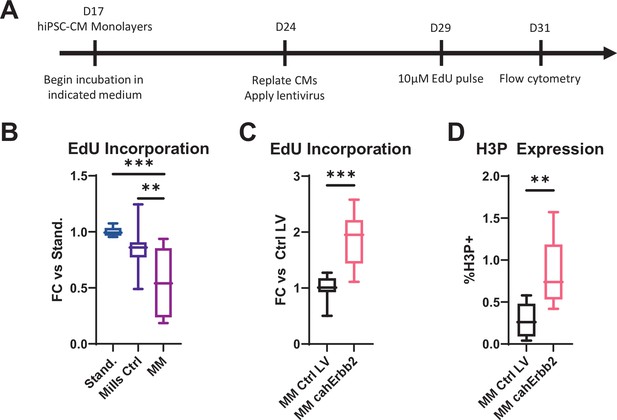
cahErbb2 induces cycling in low-glucose, palmitate-containing maturation culture media.
(A) Schematic of experimental design in hiPSC-CM monolayers. (B) Flow cytometry analysis of control mCherry+ hiPSC-CMs, showing the fold-change (FC) in CM EdU incorporation in a maturation media (MM) and control media (Mills Ctrl) from Mills et al. relative to our standard media used in Figure 1 (Stand). (C, D) Flow cytometry analysis of hiPSC-CMs transduced with cahErbb2 or control lentivirus (LV) and cultured in MM, showing (C) relative FC in CM EdU incorporation or (D) H3P expression. Data: box and whiskers showing distribution and min to max (*p<0.05, **p<0.01, ***p<0.001 vs. Ctrl LV). hiPSC-CM, human-induced pluripotent stem cell-derived cardiomyocyte.
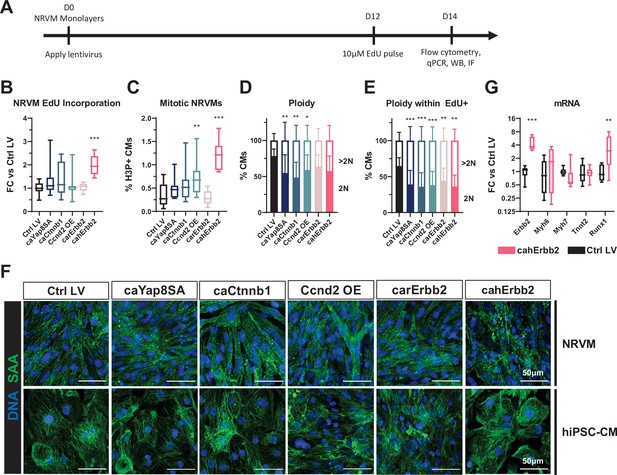
cahErbb2 induces NRVM cycle entry in monolayers and promotes sarcomere disassembly in NRVMs and hiPSC-CMs.
(A) Schematic of experimental design in NRVM monolayers. (B–E) Flow cytometry analysis of mCherry+ NRVMs showing (B) fold-change (FC) in EdU incorporation relative to control LV-treated NRVMs, (C) percentage of H3P+ CMs, and (D, E) percentage of 2N and >2N cells in all CMs (D) and EdU+ CMs (E). (F) Representative immunostaining images of sarcomeric α-actinin showing sarcomeric structure in LV transduced NRVMs and hiPSC-CMs. (G) Relative expression of Erbb2, sarcomeric genes (Myh6, Myh7, and Tnnt2), and dedifferentiation marker Runx1 in cahErbb2-transduced versus control hiPSC-CMs. Data: box and whiskers showing distribution and min to max. Column graphs showing mean+ SD (*p<0.05, **p<0.01, ***p<0.001 vs. Ctrl LV). hiPSC-CM, human-induced pluripotent stem cell-derived cardiomyocyte; LV, lentiviral vector; NRVM, neonatal rat ventricular myocyte.
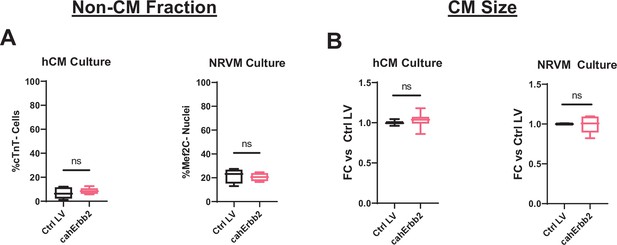
cahErbb2 expression does not affect non-CM abundance or CM size in hiPSC-CM and NRVM monolayers.
(A) Quantification of non-CM abundance based on cTnT staining and flow cytometry analysis of hiPSC-CMs (left) and Mef2c expression in immunostained NRVMs (right). (B) Relative CM size in hiPSC-CMs (left) and NRVMs (right) quantified from flow cytometry forward scatters. Data: box and whiskers showing distribution and min to max (*p<0.05, **p<0.01, ***p<0.001 vs. Ctrl LV). hiPSC-CM, human-induced pluripotent stem cell-derived cardiomyocyte; LV, lentiviral vector; NRVM, neonatal rat ventricular myocyte.
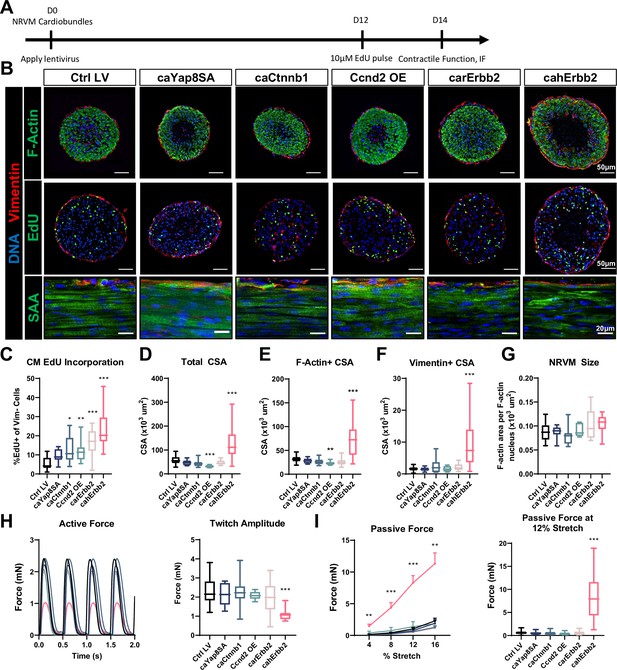
cahErbb2 induces NRVM cycle entry in cardiobundles and promotes sarcomere disassembly and contractile dysfunction.
(A) Schematic of experimental design in NRVM cardiobundles. (B) Representative immunostaining images of cardiobundle cross-sections showing morphology (top), EdU incorporation (middle), and whole-mount tissue showing sarcomere structure (bottom). (C–G) Quantification of immunostained cardiobundle cross-sections for (C) NRVM EdU incorporation and (D) total, (E) F- actin+, (F) vimentin+ cross-sectional area (CSA), and (G) Quantified F-actin+ CSA per nuclei number within this area, shown as a measure of CM size. (H, I) Force analysis in LV-transduced cardiobundles showing (H) representative twitch traces and quantified maximum twitch amplitude and (I) passive force-length and force amplitude at 12% stretch (0% stretch is culture length of 7 mm). Data: box and whiskers showing distribution and min to max. Line plot showing mean+ SEM (*p<0.05, **p<0.01, ***p<0.001 vs. Ctrl LV). LV, lentiviral vector; NRVM, neonatal rat ventricular myocyte.
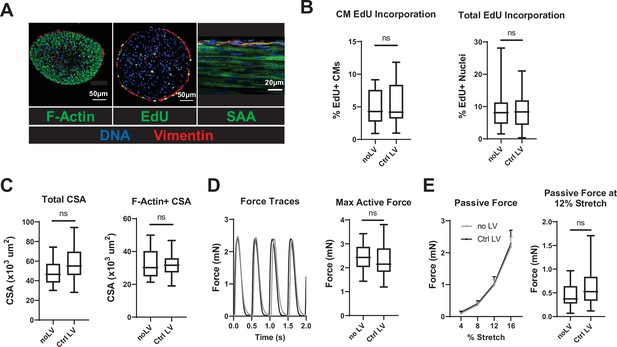
LV transduction does not affect NRVM cardiobundles.
(A) Representative immunostaining images of cardiobundle cross-sections showing morphology (left), EdU incorporation (middle), and whole-mount tissue showing sarcomere structure (right). (B, C) Quantification of immunostained cardiobundle cross-sections for (B) CM and total EdU incorporation, and (C) total and F- actin+ CSA. (D, E) Force analysis in cardiobundles showing (D) representative twitch traces and quantified maximum twitch amplitude and (E) passive force-length and force amplitude at 12% stretch (0% stretch is culture length of 7 mm). Data: box and whiskers showing distribution and min to max. Line plot showing mean+ SEM (*p<0.05, **p<0.01, ***p<0.001 vs. Ctrl LV). CM, cardiomyocyte; CSA, cross-sectional area; LV, lentiviral vector.

Twitch kinetics and unloaded contractile force in cardiobundles.
(A–C) Twitch kinetics in cardiobundles characterized by (A) total twitch duration, (B) time to peak, and (C) relaxation time. (D, E) Passive force-length relationship shown from the slack length where tissues were fully unloaded (passive force=0)—16% stretch (D, 0% stretch is culture length of 7 mm) and corresponding twitch forces measured at a slack length (E). Data: box and whiskers showing distribution and min to max. Line plot showing mean+ SEM (*p<0.05, **p<0.01, ***p<0.001 vs. Ctrl LV). LV, lentiviral vector.
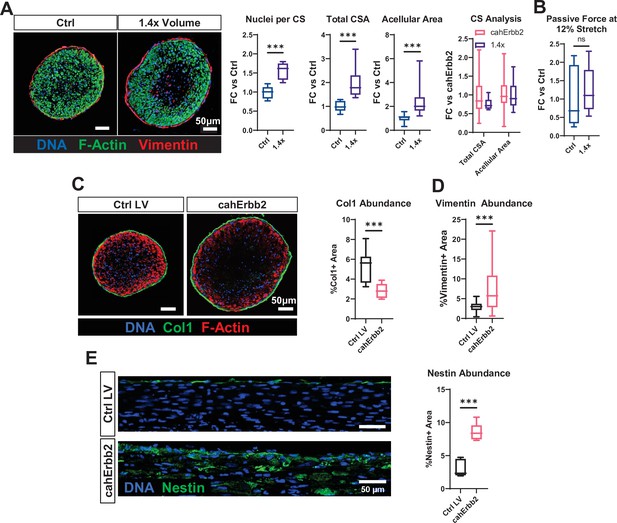
Underlying mechanisms of increased passive tension in cahErbb2-expressing cardiobundles.
(A) Representative cross-sectional immunostaining images of cardiobundles made using standard (Ctrl) or 1.4× larger (1.4×) cell/hydrogel volume and corresponding quantifications of nuclei per cross-section (CS), and total and acellular CS area (CSA) shown relative to Ctrl group, as well as total and acellular CSA shown relative to CahErbb2 group. (B) Passive force in 1.4× cardiobundles quantified at 12% stretch shown relative to Ctrl group. (C) Representative cross-sectional immunostaining images and corresponding quantification of collagen I (Coll1) abundance. (D) Vimentin abundance quantified from cardiobundle cross-section images in Figure 3B. (E) Longitudinal tissue sections and corresponding quantification of nestin abundance. Data: box and whiskers showing distribution and min to max (*p<0.05, **p<0.01, ***p<0.001 vs. Ctrl LV or Ctrl). LV, lentiviral vector.
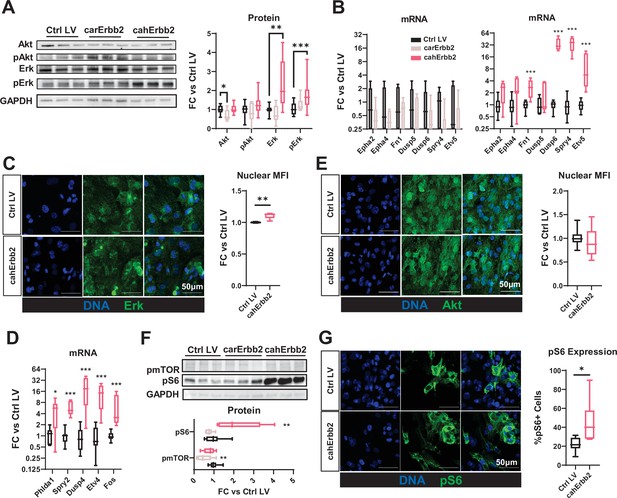
cahErbb2 but not carErbb2 activates Erk signaling to drive proliferation in CMs.
(A–B) Representative Western blots and quantified relative protein (A) and Erk target gene expression (B) in carErbb2- or cahErbb2-transduced versus control hiPSC-CM monolayers. (C) Representative immunostaining images and quantified nuclear MFI of Erk in cahErbb2-transduced versus control hiPSC-CMs. (D) Quantified relative Erk target gene expression in cahErbb2-transduced versus control hiPSC-CM monolayers. (E) Representative immunostaining images and quantified nuclear MFI of Akt in cahErbb2-transduced versus control hiPSC-CMs. (F) Representative Western blots and quantified relative phosphorylated mTOR (pmTOR) and ribosomal protein S6 (pS6) expression in carErbb2- or cahErbb2-transduced versus control hiPSC-CMs. (G) Representative immunostaining images and quantified nuclear MFI of pS6 in cahErbb2-transduced versus control hiPSC-CMs. Data: box and whiskers showing distribution and min to max. Column graphs showing mean+ SD (*p<0.05, **p<0.01, ***p<0.001 vs. Ctrl LV). hiPSC-CM, human-induced pluripotent stem cell-derived cardiomyocyte; LV, lentiviral vector MFI, mean fluorescence intensity.
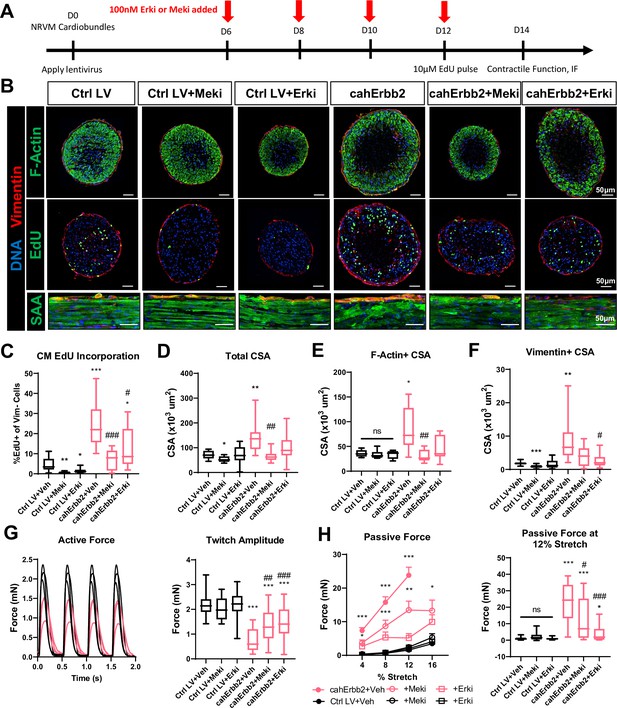
Erk or Mek inhibition attenuates cahErbb2-induced effects in NRVM cardiobundles.
(A) Schematic of experimental design in cahErbb2 and Ctrl LV NRVM cardiobundles. (B) Representative immunostaining images of cardiobundle cross-sections showing morphology (top), EdU incorporation (middle), and whole-mount tissue showing sarcomere structure (bottom). (C–F) Quantification of immunostained cardiobundle cross-sections for (C) NRVM EdU incorporation and (D) total, (E) F- actin+, and (F) vimentin+ cross-sectional area (CSA). (G, H) Force analysis in cardiobundles showing (G) representative twitch traces and quantified maximum twitch amplitude and (H) passive force-length and force amplitude at 12% stretch (0% stretch is culture length of 7 mm). Data: box and whiskers showing distribution and min to max. Line plot showing mean+ SEM (*p<0.05, **p<0.01, ***p<0.001 vs. Ctrl LV; #p<0.05, ##p<0.01, ###p<0.001 vs. cahErbb2). LV, lentiviral vector; NRVM, neonatal rat ventricular myocyte.
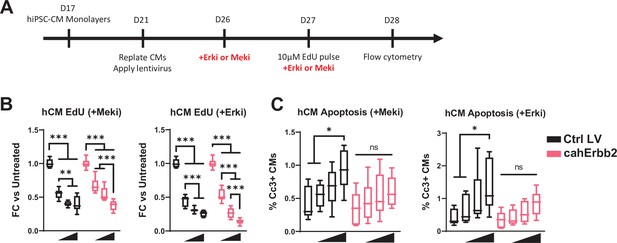
Erk or Mek inhibition attenuates cahErbb2-induced proliferation in hiPSC-CMs.
(A) Schematic of experimental design in hiPSC-CM monolayers. (B, C) Flow cytometry analysis of cahErbb2-transduced and control mCherry+ hiPSC-CMs treated for 48 hr with vehicle control or increasing doses (10 nM, 100 nM, and 1 µM) of Erk (SCH772984) or Mek (PD0325901) inhibitors showing (B) the fold-change in CM EdU incorporation relative to corresponding vehicle-treated CMs and (C) Cc3 expression. Data: box and whiskers showing distribution and min to max (*p<0.05, **p<0.01, ***p<0.001 vs. Ctrl LV). Cc3, cleaved caspase-3; hiPSC-CM, human-induced pluripotent stem cell-derived cardiomyocyte; LV, lentiviral vector.

Twitch kinetics of NRVM cardiobundles treated with Erki or Meki.
(A–C) Twitch kinetics in cardiobundles characterized by (A) total twitch duration, (B) time to peak, and (C) relaxation time. Data: box and whiskers showing distribution and min to max (*p<0.05, **p<0.01, ***p<0.001 vs. Ctrl LV; #p<0.05, ##p<0.01, ###p<0.001 vs. cahErbb2). NRVM, neonatal rat ventricular myocyte.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-Cardiac Troponin T (Rabbit polyclonal) | Abcam | ab45932 | FC (1:200) |
| Antibody | Anti-Sarcomeric Alpha Actinin (Mouse monoclonal) | Sigma-Aldrich | A7811 | IF (1:200) |
| Antibody | Cleaved Caspase-3 (Asp175) (Rabbit polyclonal) | Cell Signaling Technology | 9661 | FC (1:800) |
| Antibody | GAPDH (Mouse monoclonal) | Santa Cruz Biotechnology | sc-47724 | WB (1:1000) |
| Antibody | Mouse IgG1, kappa monoclonal [15-6E10A7] - Isotype Control | Abcam | ab170190 | FC (1:2000) |
| Antibody | Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (Rabbit polyclonal) | Cell Signaling Technology | 9101 | WB (1:1000) |
| Antibody | p44/42 MAPK (Erk1/2) (Total Erk) (Rabbit polyclonal) | Cell Signaling Technology | 9102 | WB (1:1000) |
| Antibody | Phospho-Histone H3 (Ser10) (6G3) (Mouse monoclonal) | Cell Signaling Technology | 9706 | FC (1:1000) |
| Antibody | Phospho-mTOR (Ser2448) (Rabbit polyclonal) | Cell Signaling Technology | 2971 | WB (1:1000) |
| Antibody | Phospho-S6 Ribosomal Protein (Ser235/236) (Rabbit polyclonal) | Cell Signaling Technology | 2211 | WB (1:1000)IF (1:100) |
| Antibody | Rabbit IgG, polyclonal - Isotype Control | Abcam | ab37415 | FC (1:2000) |
| Antibody | Recombinant Anti-active YAP1 [EPR19812] (Rabbit polyclonal) | Abcam | ab205270 | WB (1:1000)IF (1:500) |
| Antibody | Recombinant Anti-AKT1 (phospho S473) [EP2109Y] (Rabbit monoclonal) | Abcam | ab81283 | WB (1:5000) |
| Antibody | Recombinant Anti-AKT1+ AKT2+ AKT3 [EPR16798] (Rabbit monoclonal) | Abcam | ab179463 | WB (1:10,000)IF (1:100) |
| Antibody | Recombinant Anti-Cyclin D2 (Rabbit monoclonal) | Abcam | ab207604 | WB (1:1000)IF (1:100) |
| Antibody | Recombinant Anti-Vimentin [EPR3776] (Rabbit monoclonal) | Abcam | ab92547 | IF (1:500) |
| Antibody | YAP (D8H1X) (Total Yap) (Rabbit monoclonal) | Cell Signaling Technology | 14074 | WB (1:1000)IF (1:200) |
| Antibody | β-Catenin (Carboxy-terminal Antigen) (Rabbit polyclonal) | Cell Signaling Technology | 9587 | WB (1:1000) |
| Chemical compound, drug | SCH772984 | Cayman Chemical | 19166 | |
| Chemical compound, drug | PD0325901 | Sigma-Aldrich | PZ0162-5MG | |
| Commercial assay, kit | Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit | Thermo Fisher Scientific | C10419 | |
| Commercial assay, kit | Click-iT EdU Alexa Fluor 488 Imaging Kit | Thermo Fisher Scientific | C10337 | |
| Cell line (Homo sapiens, male) | DU11 iPSC Line | Duke University Stem Cell Core | ||
| Cell line (H. sapiens) | Hek293T | ATCC | CRL-3216 | |
| Strain, strain background (Rattus Norvegicus) | P2 Sprague-Dawley Rat Pups | Charles River | ||
| Recombinant DNA reagent | pLV-beta-catenin deltaN90 | pLV-beta-catenin deltaN90 was a gift from Bob Weinberg | Addgene plasmid # 36985 | Used to generate new plasmids |
| Recombinant DNA reagent | HER2 CA (V659E) | HER2 CA (V659E) was a gift from Mien-Chie Hung | Addgene plasmid # 16259 | Used to generate new plasmids |
| Recombinant DNA reagent | R777-E020 Hs.CCND2-nostop | R777-E020 Hs.CCND2-nostop was a gift from Dominic Esposito | Addgene plasmid # 70304 | Used to generate new plasmids |
| Recombinant DNA reagent | pSV2 neuNT | pSV2 neuNT was a gift from Bob Weinberg | Addgene plasmid # 10,918 | Used to generate new plasmids |
| Recombinant DNA reagent | pCMV-flag YAP2 5SA | pCMV-flag YAP2 5SA was a gift from Kunliang Guan | Addgene plasmid # 27371 | Used to generate new plasmids |
| Recombinant DNA reagent | Pax2 | psPAX2 was a gift from Didier Trono | Addgene plasmid # 12260 | Used to generate lentivirus in HEK293Ts |
| Recombinant DNA reagent | VSVG | pMD2.G was a gift from Didier Trono | Addgene plasmid # 12259 | Used to generate lentivirus in HEK293Ts |
| Recombinant DNA reagent | Control Lentivirus (MHCK7-mCherry) | This manuscript | See Materials and methods section | |
| Recombinant DNA reagent | Cyclin Lentivirus (MHCK7-Ccnd2-P2A-mCherry-NLS) | This manuscript | See Materials and methods section | |
| Recombinant DNA reagent | B-catenin Lentivirus (MHCK7- Ctnnb1(∆90)-P2A-mCherry-NLS) | This manuscript | See Materials and methods section | |
| Recombinant DNA reagent | Human Erbb2 Lentivirus (MHCK7-Erbb2(V659E-P2A-mCherry)) | This manuscript | See Materials and methods section | |
| Recombinant DNA reagent | Rat Erbb2 Lentivirus (MHCK7-Erbb2(V663E-P2A-mCherry-NLS)) | This manuscript | See Materials and methods section | |
| Recombinant DNA reagent | Yap Lentivirus (MHCK7-Yap8SA-P2A-mCherry-NLS) | This manuscript | See Materials and methods section | |
| Software, algorithm | CellProfiler | McQuin et al., 2018 | https://cellprofiler.org/ | |
| Software, algorithm | ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ | |
| Software, algorithm | FlowJo Software | The FlowJo team, 2021 | ||
| Software, algorithm | GraphPad Prism | https://www.graphpad.com/ | ||
| Software, algorithm | Matlab | |||
| Sequence-based reagent | Yap1 | PrimerBank ID: 303523510c1 | F:TAGCCCTGCGTAGCCAGTTAR:TCATGCTTAGTCCACTGTCTGT | |
| Sequence-based reagent | Myh6 | NCBI PrimerBlast | F:GCCCTTTGACATTCGCACTGR:GGTTTCAGCAATGACCTTGCC | |
| Sequence-based reagent | Myh7 | PrimerBank ID: 115496168c1 | F:ACTGCCGAGACCGAGTATGR:GCGATCCTTGAGGTTGTAGAGC | |
| Sequence-based reagent | Ctgf | PrimerBank ID: 98986335c1 | F:CAGCATGGACGTTCGTCTGR:AACCACGGTTTGGTCCTTGG | |
| Sequence-based reagent | Cyr61 | PrimerBank ID: 197313774c3 | F:CAGCATGGACGTTCGTCTGR:AACCACGGTTTGGTCCTTGG | |
| Sequence-based reagent | Ctnnb1 | PrimerBank ID: 148233337c2 | F:CATCTACACAGTTTGATGCTGCTR:GCAGTTTTGTCAGTTCAGGGA | |
| Sequence-based reagent | Lef1 | PrimerBank ID: 260656055c1 | F:AGAACACCCCGATGACGGAR:GGCATCATTATGTACCCGGAAT | |
| Sequence-based reagent | Axin2 | PrimerBank ID: 195927058c1 | F:CAACACCAGGCGGAACGAAR:GCCCAATAAGGAGTGTAAGGACT | |
| Sequence-based reagent | Ccnd2 | PrimerBank ID: 209969683c3 | F:TTTGCCATGTACCCACCGTCR:AGGGCATCACAAGTGAGCG | |
| Sequence-based reagent | Erbb2 | PrimerBank ID: 54792097c2 | F:TGTGACTGCCTGTCCCTACAAR:CCAGACCATAGCACACTCGG | |
| Sequence-based reagent | Epha2 | PrimerBank ID: 296010835c2 | F:AGAGGCTGAGCGTATCTTCATR:GGTCCGACTCGGCATAGTAGA | |
| Sequence-based reagent | Epha4 | PrimerBank ID: 45439363c3 | F:GCAAGGAGACGTTTAACCTGTR:CTTGGGTGAAGCTCTCATCAG | |
| Sequence-based reagent | Fn1 | PrimerBank ID: 47132556c2 | F:AGGAAGCCGAGGTTTTAACTGR:AGGACGCTCATAAGTGTCACC | |
| Sequence-based reagent | Dusp5 | PrimerBank ID: 62865889c2 | F:GCCAGCTTATGACCAGGGTGR:GTCCGTCGGGAGACATTCAG | |
| Sequence-based reagent | Dusp6 | PrimerBank ID: 42764682c1 | F:GAAATGGCGATCAGCAAGACGR:CGACGACTCGTATAGCTCCTG | |
| Sequence-based reagent | Spry4 | PrimerBank ID: 188595696c1 | F:TCTGACCAACGGCTCTTAGACR:GTGCCATAGTTGACCAGAGTC | |
| Sequence-based reagent | Etv5 | PrimerBank ID: 194018465c1 | F:TCAGCAAGTCCCTTTTATGGTCR:GCTCTTCAGAATCGTGAGCCA | |
| Sequence-based reagent | Phlda1 | PrimerBank ID: 83977458c1 | F:GAAGATGGCCCATTCAAAAGCGR:GAGGAGGCTAACACGCAGG | |
| Sequence-based reagent | Spry2 | PrimerBank ID: 22209007c1 | F:CCTACTGTCGTCCCAAGACCTR:GGGGCTCGTGCAGAAGAAT | |
| Sequence-based reagent | Dusp4 | PrimerBank ID: 325651887c1 | F:GGCGGCTATGAGAGGTTTTCCR:TGGTCGTGTAGTGGGGTCC | |
| Sequence-based reagent | Etv4 | PrimerBank ID: 118918427c2 | F:CAGTGCCTTTACTCCAGTGCCR:CTCAGGAAATTCCGTTGCTCT | |
| Sequence-based reagent | Fos | PrimerBank ID: 254750707c2 | F:GGGGCAAGGTGGAACAGTTATR:CCGCTTGGAGTGTATCAGTCA | |
| Sequence-based reagent | Dab2 | PrimerBank ID: 349585059c1 | F:GTAGAAACAAGTGCAACCAATGGR:GCCTTTGAACCTTGCTAAGAGA | |
| Sequence-based reagent | Pdgfa | PrimerBank ID: 197333758c1 | F:GCAAGACCAGGACGGTCATTTR:GGCACTTGACACTGCTCGT | |
| Sequence-based reagent | Runx1 | PrimerBank ID: 169790826c1 | F:CTGCCCATCGCTTTCAAGGTR:GCCGAGTAGTTTTCATCATTGCC | |
| Sequence-based reagent | Hprt1 | PrimerBank ID: 164518913c1 | F:CCTGGCGTCGTGATTAGTGR:AGACGTTCAGTCCTGTCCATAA |
Additional files
-
Supplementary file 1
Statistical information for all figures.
- https://cdn.elifesciences.org/articles/65512/elife-65512-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65512/elife-65512-transrepform1-v2.pdf
-
Source data 1
Source data used to generate figures.
- https://cdn.elifesciences.org/articles/65512/elife-65512-supp2-v2.xlsx
-
Source data 2
Unedited, labeled western blots for all figures.
- https://cdn.elifesciences.org/articles/65512/elife-65512-supp3-v2.docx
-
Source data 3
Unedited, uncropped western blots for all figures.
- https://cdn.elifesciences.org/articles/65512/elife-65512-supp4-v2.zip
-
Source data 4
Plasmid maps for constructs generated in this study.
- https://cdn.elifesciences.org/articles/65512/elife-65512-supp5-v2.zip





