NICEdrug.ch, a workflow for rational drug design and systems-level analysis of drug metabolism
Figures
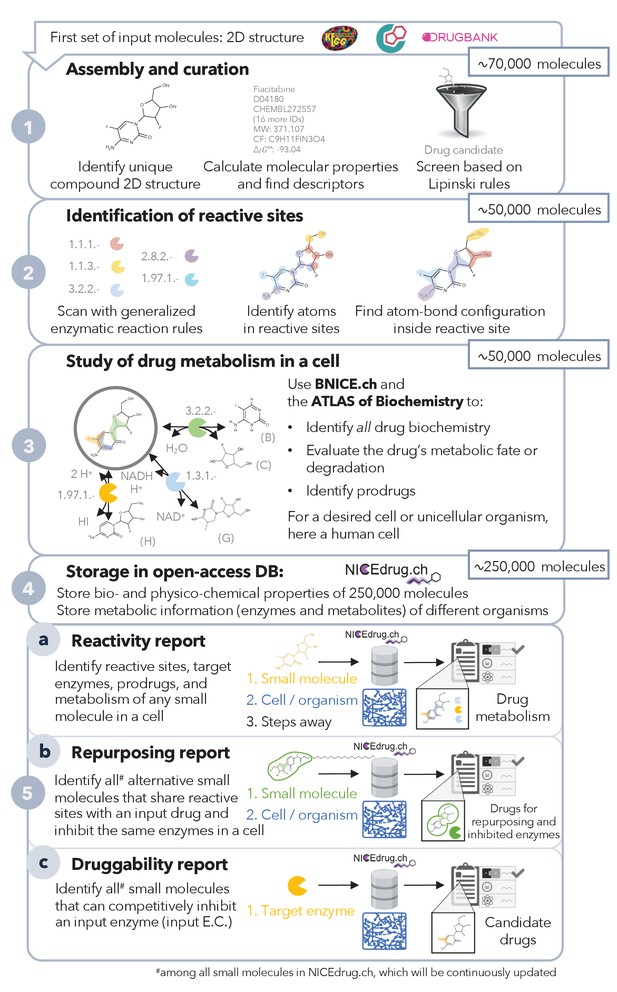
Pipeline to construct and use the NICEdrug.ch database.
NICEdrug.ch (1) curates available information and calculates the properties of an input compound; (2) identifies the reactive sites of that compound; (3) explores the hypothetical metabolism of the compound in a cell; (4) stores all functional, reactive, bio-, and physico-chemical properties in open-source database; and (5) allows generation of reports to evaluate (5a) reactivity of a small molecule, (5b) drug repurposing, and (5c) druggability of an enzymatic target. See also Figure 1—figure supplement 1; Figure 1—figure supplement 2; Figure 1—figure supplement 3, and Supplementary file 1 .
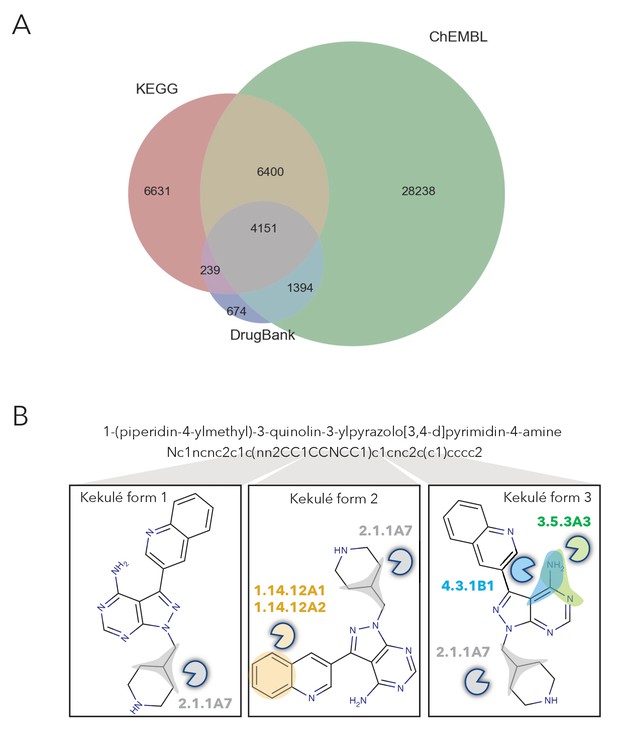
Overview of number of molecules in NICEdrug.ch and their structural curation.
(A) Venn diagram showing the number of compounds in NICEdrug.ch and their source database: KEGG, DrugBank, ChEMBLE NTD, and ChEMBLE. (B) Representation on how different kekulé forms affect the identification of reactive sites and prediction of biological activity for an example molecule.
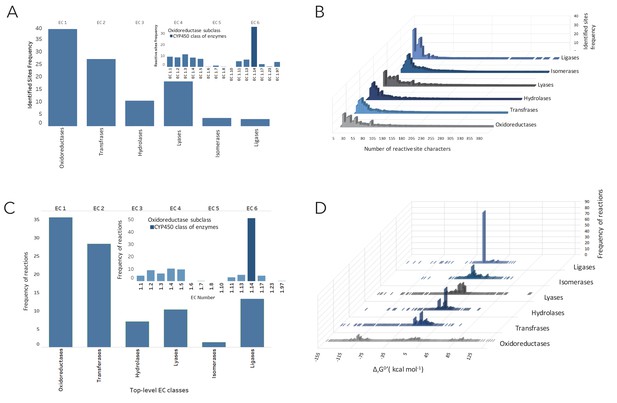
Distribution of reactive sites and metabolic reactions as of EC numbers linked to all molecules in NICEdrug.ch.
(A) Distribution of reactive sites identified in all molecules of NICEdrug.ch among classes of EC numbers. (B) Specificity of reactive sites identified in drugs based on length and types of participating atoms. (C) Distribution of drug metabolic reactions based on class of EC number. (D) Distribution of Gibbs free energy for the drug metabolic reactions, which are the reactions linked to all molecules of NICEdrug.ch.
Description of NICEdrug score.
Example of NICEdrug score calculation. The NICEdrug score takes into account the structure of a molecule’s reactive site and its seven-atom-away neighborhood for similarity evaluation, analogous to BridgIT.
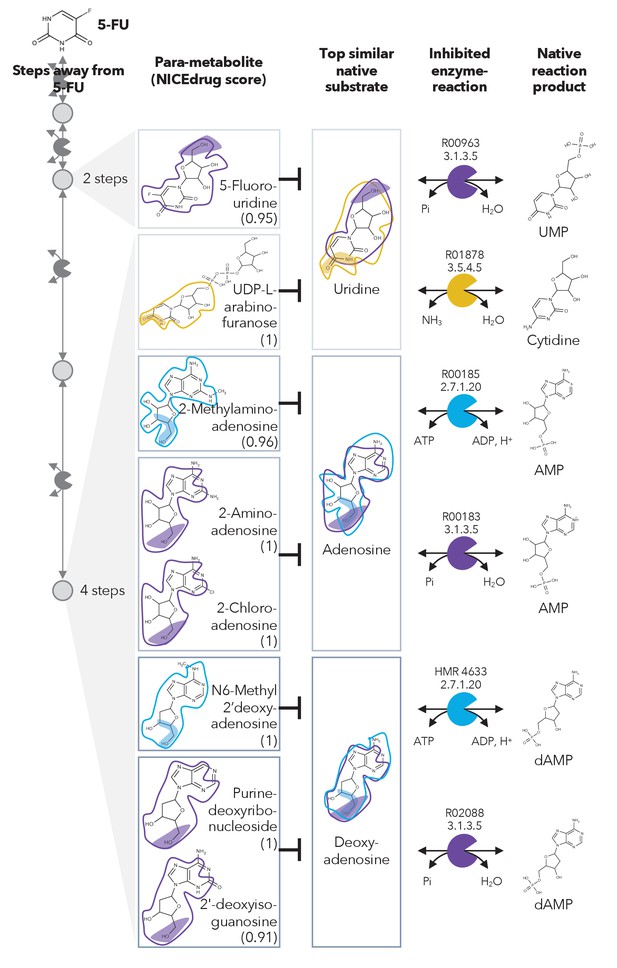
Similarity in reactive site and neighborhood defines para-metabolites in 5-FU metabolism and inhibited human metabolic enzymes.
Eight para-metabolites in the 5-FU metabolic neighborhood (represented as defined in ‘Materials and methods’). We show the most similar native human metabolites, inhibited enzymes, and native products of the reactions. See also Supplementary file 3 .
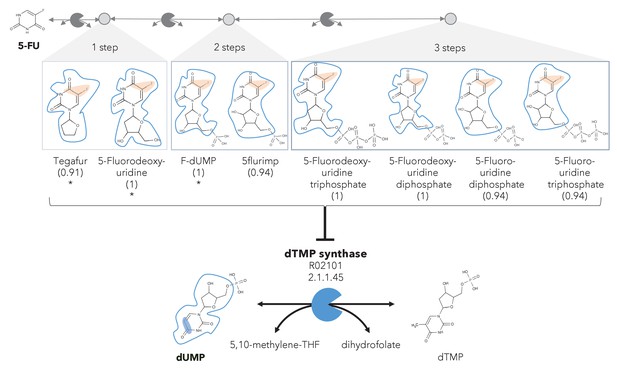
A different reactive site but similar neighborhood defines top anti-metabolites in 5-FU metabolism and inhibited human metabolic enzyme.
Eight anti-metabolites of dUMP in the 5-FU metabolic neighborhood (represented as defined in ‘Materials and methods’). Note that the reactive site of the anti-metabolites is different than the one of the native human metabolite, but the neighborhood is highly similar, which determines the high NICEdrug score (value in parenthesis). We show the inhibited human enzyme (dTMP synthase) and reaction, and its native product. See also Supplementary file 3
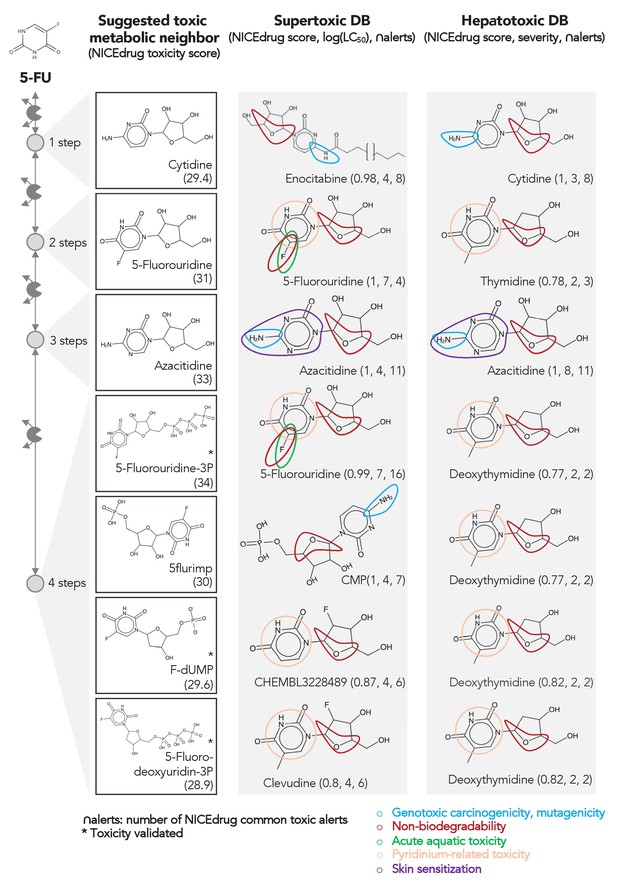
Comparing downstream products to known toxic molecules and analyzing their common structural toxic alerts explains metabolic toxicity of 5-FU.
Example of six suggested toxic molecules in the 5-FU metabolic neighborhood (represented as defined in ‘Materials and methods’). We show toxic compounds from the supertoxic and hepatotoxic databases that lead to the highest NICEdrug toxicity score (number under toxic intermediate name, ‘Materials and methods’). We highlight functional groups linked to five NICEdrug toxic alerts (legend bottom right). See also Supplementary file 3.
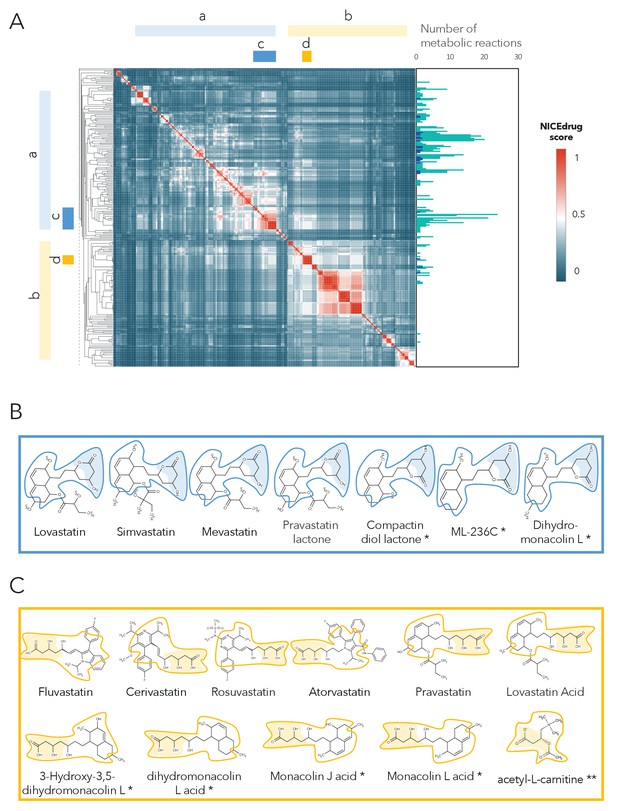
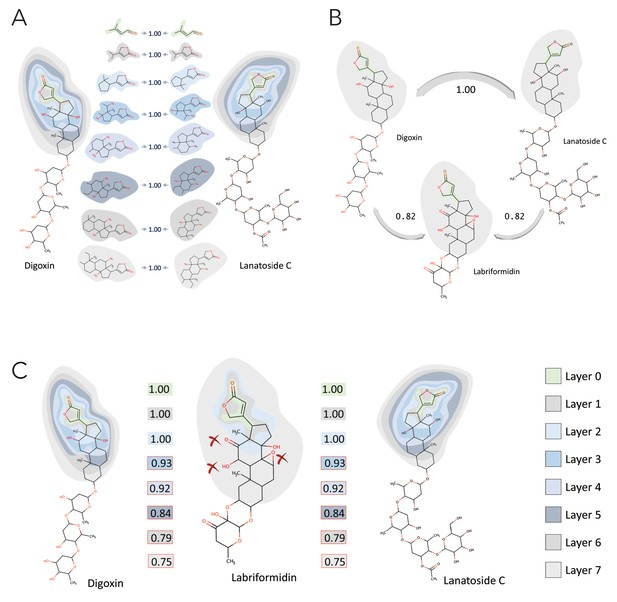
Clustering of molecules with statin reactive sites based on NICEdrug score suggests drugs for repurposing.
(A) Pairwise NICEdrug score between all molecules with statin reactive sites (heat map) and number of metabolic reactions in which they participate (right). We highlight clusters of statins of type 1 (cluster a) and type 2 (cluster b), and clusters of most similar molecules to type one statins (cluster c) and type two statins (cluster d). Within the metabolic reactions, we indicate the total number of reactions (dark color) and the number of reactions that involve the statin reactive site (light color). (B) Examples of statins and Mevastatin analogues of type one from cluster c (blue) and of type two from cluster d (gold). We left the known statins unmarked, which are appropriately clustered together based on the NICEdrug score, and we mark with * new molecules that cluster with statins and that NICEdrug.ch suggests could be repurposed to act as statins. Reactive sites in type one statins and type two statins are colored in blue and orange, respectively. The reactive site neighborhood as considered in the NICEdrug score is also marked. See also; Figure 5—figure supplement 1 , and Supplementary file 4.
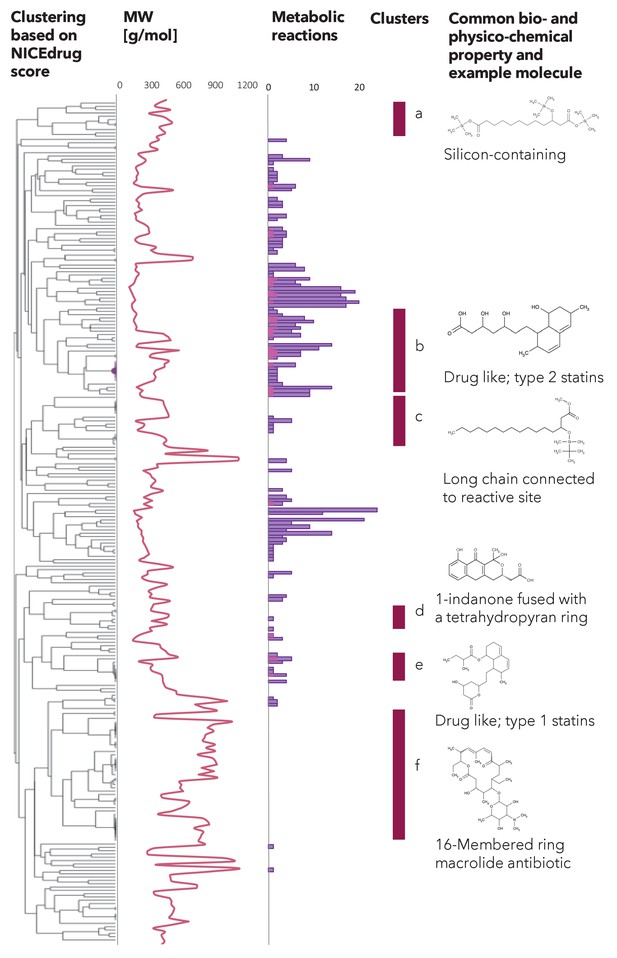
Clustering based on NICEdrug score, molecular weight, and reactivity of statin like molecules.
Hierarchical clustering based on the NICEdrug score of all molecules in NICEdrug.ch that contain statin reactive site (left). We report the molecules’ molecular weight (middle left) and number of drug metabolic reactions or reactions in which these drugs participate (middle). The molecular weight seems to be inversely correlated with the number of drug metabolic reactions. We highlight six clusters of drugs (a–f, middle right) and an example representative molecule (left). Interestingly, these clusters also group molecules based on bio- or physico-chemical properties: ‘cluster a’ involves a range of silicon-containing chemical molecules, ‘cluster b’ are drug-like molecules of type two statins, ‘cluster c’ includes chemical molecules with a long chain connected to the reactive site, ‘cluster d’ involves molecules with 1-indanone fused with a tetrahydropyran ring, ‘cluster e’ comprises drug-like molecules of type one statins, and ‘cluster f’ are 16-membered ring macrolide antibiotics.
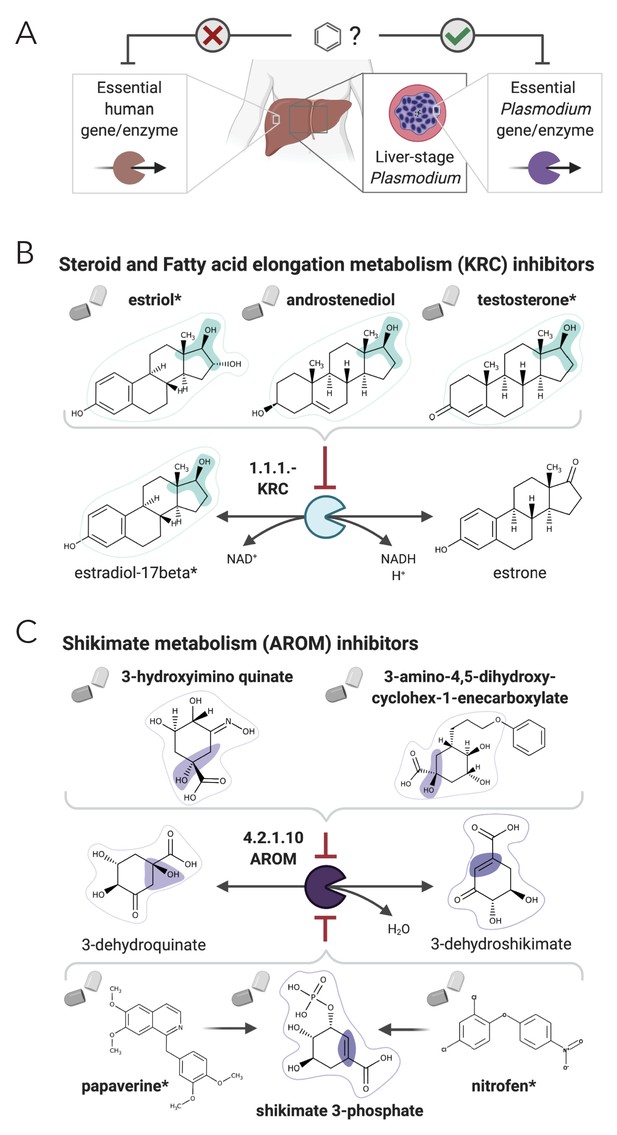
NICEdrug.ch suggests shikimate 3-phosphate as a top candidate to target liver-stage malaria and minimize side effects in host human cells.
(A) Schema of ideal scenario to target malaria, wherein a drug efficiently inhibits an essential enzyme for malaria parasite survival and does not inhibit essential enzymes in the host human cell to prevent side effects. (B) Shikimate 3-phosphate inhibits enzymes in the Plasmodium shikimate metabolism, which is essential for liver-stage development of the parasite. Shikimate 3-phosphate does not inhibit any enzyme in the human host cell since it is not a native human metabolite, and it does not show similarity to any native human metabolite. (C) Mechanistic details of inhibition of aroC by shikimate 3-phosphate and other NICEdrug candidates. See also Supplementary file 5; Supplementary file 6.
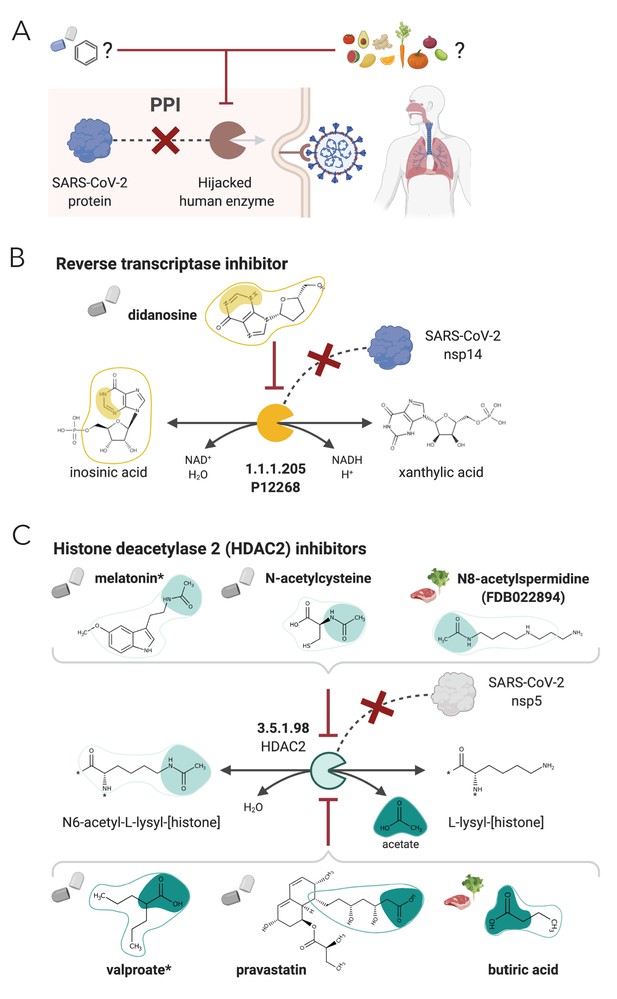
NICEdrug.ch strategy to fight COVID-19, and NICEdrug.ch candidate inhibitors of SARS-CoV-2 host factors: reverse transcriptase and HDAC2.
(A) Schema of NICEdrug strategy to target COVID-19, wherein a drug (top-left) or molecules in food (top-right) efficiently inhibit a human enzyme hijacked by SARS-CoV-2. Inhibition of this host factor reduces or abolishes protein–protein interactions (PPI) with a viral protein and prevents SARS-CoV-2 proliferation. (B) Inhibition of the reverse transcriptase (EC: 1.1.1.205 or P12268) and the PPI with SARS-CoV-nsp14 by didanosine based on NICEdrug.ch. (C) Inhibition of the HDAC2 (EC: 3.5.1.98) and the PPI with SARS-CoV-nsp5 by molecules containing acetyl moiety (like melatonin, N-acetylcysteine, and N8-acetylspermidine), and molecules containing carboxylate moiety (like valproate, stains, and butyrate) based on NICEdrug.ch. See also Figure 7—figure supplement 1; Figure 7—figure supplement 2, Supplementary file 7; Supplementary file 8.
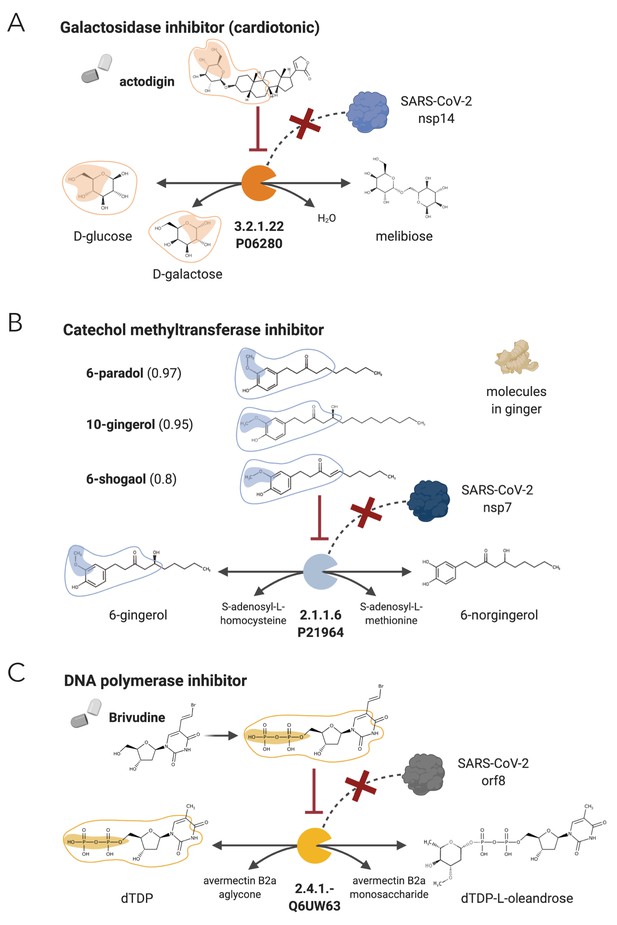
NICEdrug candidate inhibitors of SARS-CoV-2 host factors: galactosidase, catechol methyltransferase, and DNA polymerase.
(A) Inhibition of the galactosidase (EC: 3.2.1.22 or P06280) and the PPI with SARS-CoV-2 nsp14 by actodigin based on NICEdrug.ch. (B) Inhibition of the catechol methyltransferase (EC: 2.1.1.6 or P21964) and the PPI with SARS-CoV-2 nsp7 by 6-paradol, 10-gingerol, and 6-shogaol, which are molecules in ginger, based on NICEdrug.ch. (C) Inhibition of the DNA polymerase (EC: 2.4.1.-) and the PPI with SARS-CoV-2 nsp8 by brivudine based on NICEdrug.ch.
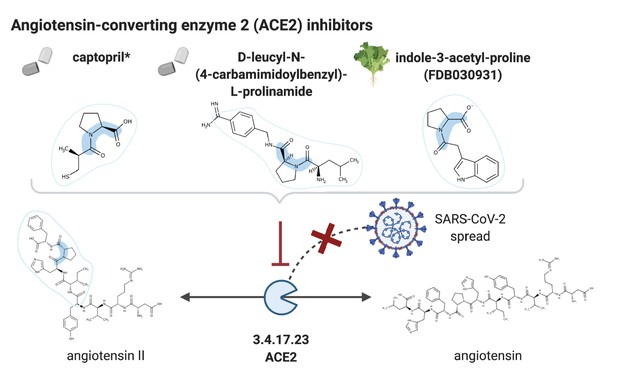
NICEdrug candidate inhibitors of ACE2.
Inhibition of the ACE2 (EC: 3.4.17.23), a putative host factor of SARS-CoV-2, by the known inhibitor captopril, and NICEdrug candidates D-leucyl-N-(4-carbamimidoylbenzyl)-l-prolinamide and indole-3-acetyl-proline.
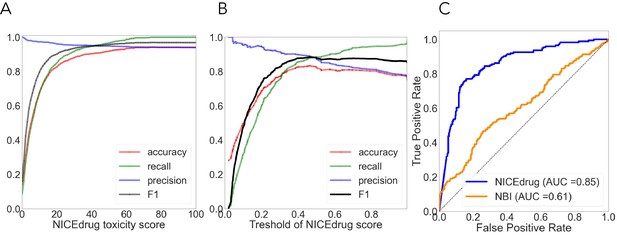
Quantitative comparison of drug toxicity (A) and drug–enzyme pairs (B, C).
(A) Evaluation of NICEdrug.ch toxicity predictions on the test dataset of 1777 drug candidates from PubChem. Y-axis represents the value of a metric, namely accuracy (red), precision (green), recall (blue), and F1 (yellow) based on putative threshold for NICEdrug toxicity score. X-axis represents the predicted NICEdrug.ch toxicity score. (B) Evaluation of overall performance of NICEdrug.ch and NBI tool in predicting interaction of enzymes and drug pairs in terms of ROC curves and value of AUC. (C) Quality of enzymes and drug pairs predictions using NICEdrug.ch in terms of statistical measures including: precision, recall, F1, and accuracy.
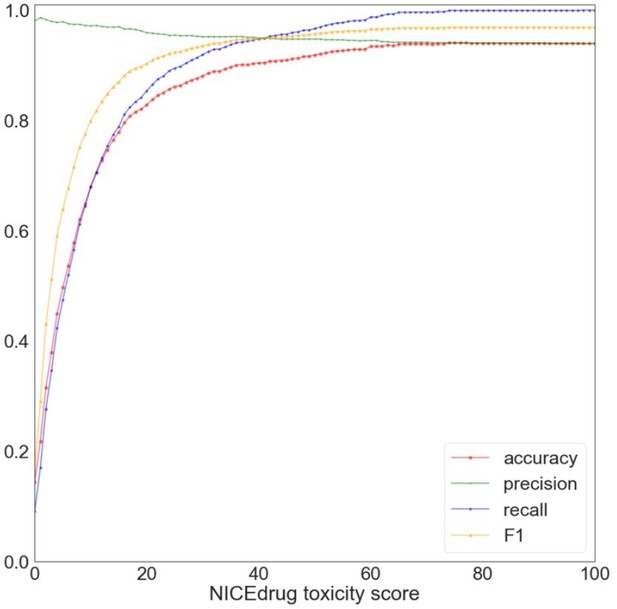
Evaluation of NICEdrug.
ch toxicity predictions on the test dataset of 1,777 drug candidates from PubChem. Y-axis represents the value of a metric, namely accuracy (red), precision (green), recall (blue), and F1 (yellow) based on putative threshold for NICEdrug toxicity score. X-axis represents the predicted NICEdrug.ch toxicity score.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Software, algorithm | OpenBabel 2.4.1 | doi:10.1186/1758-2946-3-33 | ||
| Software, algorithm | BridgIT | doi:10.1073/pnas.1818877116 | ||
| Software, algorithm | ATLAS of biochemistry | doi:10.1021/acssynbio.6b00054; doi:10.1021/acssynbio.0c00052 | ||
| Software, algorithm | MORPHEUS | https://clue.io/morpheus | ||
| Software, algorithm | NICEdrug.ch (curated bioactive molecules and analysis of drug metabolism) | This paper; http://nicedrug.ch/ | See Materials and methods |
Additional files
-
Supplementary file 1
Information of human metabolism considered in this study, related to Figures 2–6.
(A) List of cofactors, (B) list of metabolites, and (C) list of EC numbers considered in BNICE.ch for the generation of reactions in the analysis of drug metabolism in a human cell (‘Materials and methods’).
- https://cdn.elifesciences.org/articles/65543/elife-65543-supp1-v1.xlsx
-
Supplementary file 2
Comparing NICEdrug.ch against other prediction tools for drug metabolism and available biochemical assays, related to Figure 8.
(A) Comparison of available computational tools to predict drug metabolism. (B) Evaluation of NICEdrug.ch potential to predict the druggability (through competitive inhibition) of an enzyme by a small molecule based on available biochemical assays and high-throughput compound screenings results. (C) Detailed comparison of NICEdrug.ch druggability results for tolcapone (a neuropsychiatric agent), pravastatin (a cardiovascular agent), oxfenicine (a vasodilator), dopamine (a cardiovascular agent) and acarbose (an antidiabetic agent) against available biochemical assays. (D) Result of large-scale quantitative comparison of drug-metabolite, toxicity and drug-enzyme predictions using NICEdrug and other drug discovery tools. (E) Literature derived examples demonstrating NICEdrug performance in predicting pathways of drug metabolism.
- https://cdn.elifesciences.org/articles/65543/elife-65543-supp2-v1.xlsx
-
Supplementary file 3
Metabolic neighborhood of 5-FU, related to Figures 2–4.
(1) List of compounds in the 5-FU metabolic neighborhood including up to four reactions or steps away. (2) Description of reactions in the 5-FU metabolic neighborhood including up to four reactions or steps away.
- https://cdn.elifesciences.org/articles/65543/elife-65543-supp3-v1.xlsx
-
Supplementary file 4
NICEdrug analysis for all molecules with reactive site of statins in NICEdrug.ch, related to Figure 5.
(A) Matrix of NICEdrug score between each pair of the whole set of 254 molecules in NICEdrug.ch with reactive site of statins. (B) Description of nine drugs candidates for repurposing to replace statins based on NICEdrug.ch. These drugs can act as competitive inhibitors of HMG-CoA reductase like statins.
- https://cdn.elifesciences.org/articles/65543/elife-65543-supp4-v1.xlsx
-
Supplementary file 5
Essential genes or enzymes and linked metabolites in liver-stage Plasmodium and a human cell, related to Figure 6.
(A) List of essential genes and associated reactions in liver-stage Plasmodium, as obtained from the study (Stanway et al., 2019). (B) List of essential genes and associated reactions in a human cell, as obtained from the study (Wang et al., 2015). (C) List of metabolites linked to essential genes in liver-stage Plasmodium. (D) List of metabolites linked to essential genes in a human cell.
- https://cdn.elifesciences.org/articles/65543/elife-65543-supp5-v1.xlsx
-
Supplementary file 6
Description of drugs, prodrugs, metabolites and enzymes analyzed in the study of malaria, related to Figure 6.
(A) NICEdrug druggability analysis of essential genes or enzymes in liver-stage Plasmodium: all drugs sharing reactive-site centric similarity with the Plasmodium metabolites and comparison with human metabolites. (B) NICEdrug druggability analysis of essential genes or enzymes in liver-stage Plasmodium: all prodrugs (up to three steps away of 346 drugs) sharing reactive-site centric similarity with the Plasmodium metabolites and comparison with human metabolites. (C) Description of drugs and prodrugs identified in the malaria analysis with NICEdrug.ch and validated in the study by Antonova-Koch et al., 2018 along with their similar Plasmodium metabolite and human metabolite.
- https://cdn.elifesciences.org/articles/65543/elife-65543-supp6-v1.xlsx
-
Supplementary file 7
Hijacked human enzymes by SARS-CoV-2, and drugs and food-based compounds that can inhibit them based on the NICEdrug score, related to Figure 7.
(A) Hijacked human proteins by SARS-CoV-2 as identified by Gordon et al., 2020 with an annotated enzymatic function (EC number), also called here ‘SARS-CoV-2 hijacked enzymes’. (B) NICEdrug druggability report for SARS-CoV-2 hijacked enzymes including all NICEdrug small molecules. (C) Best candidate drugs against COVID-19: NICEdrug druggability report for SARS-CoV-2 hijacked enzymes including drugs with NICEdrug score above 0.5 compared to the native human substrate. (D) Summary of NICEdrug best candidate drugs against COVID-19 and their classification according to the drug category in the KEGG database. (E) NICEdrug druggability report of SARS-CoV-2 hijacked enzymes including prodrugs (up to three steps away of any NICEdrug small molecule) with NICEdrug score above 0.5 compared to the native human substrate. (F) Best candidate food-based molecules against COVID-19: NICEdrug druggability report of SARS-CoV-2 hijacked enzymes including food-based molecules with NICEdrug score above 0.5 compared to the native human substrate. (G) Summary of the NICEdrug best candidate food-based molecules against COVID-19 and their classification according to the fooDB source.
- https://cdn.elifesciences.org/articles/65543/elife-65543-supp7-v1.xlsx
-
Supplementary file 8
NICEdrug analysis of inhibitory mechanisms of currently used anti SARS-CoV-2 drugs, related to Figure 7.
(A) All drug molecules and (B) prodrugs in NICEdrug.ch sharing reactive site with the native substrates of the human enzyme HDAC2 and their NICEdrug score with this substrate. (C) All molecules cataloged in fooDB sharing reactive site with the native substrates of the human enzyme HDAC2 and their NICEdrug score with this substrate. (D) All drug molecules and (E) prodrug molecules in NICEdrug.ch sharing reactive site with the native substrates of the human enzyme ACE2 and their NICEdrug score with this substrate. (F) All molecules cataloged in fooDB sharing reactive site with the native substrates of the human enzyme ACE2 and their NICEdrug score with this substrate. (G) All molecules in NICEdrug.ch or cataloged in fooDB sharing reactive site with the native substrates of the human enzyme DNA-directed RNA polymerase and their NICEdrug score with this substrate.
- https://cdn.elifesciences.org/articles/65543/elife-65543-supp8-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65543/elife-65543-transrepform-v1.docx





